Abstract
Polygalacturonase inhibiting proteins (PGIPs) are cell wall proteins that inhibit the pectin-depolymerizing activity of polygalacturonases secreted by microbial pathogens and insects. These ubiquitous inhibitors have a leucine-rich repeat structure that is strongly conserved in monocot and dicot plants. Previous reviews have summarized the importance of PGIP in plant defense and the structural basis of PG-PGIP interaction; here we update the current knowledge about PGIPs with the recent findings on the composition and evolution of pgip gene families, with a special emphasis on legume and cereal crops. We also update the information about the inhibition properties of single pgip gene products against microbial PGs and the results, including field tests, showing the capacity of PGIP to protect crop plants against fungal, oomycetes and bacterial pathogens.
Keywords: polygalacturonase inhibiting proteins (PGIPs), gene family, transgenic plants, plant protection, fungal pathogens, bacterial pathogens
Introduction
Successful colonization of plant tissues by microbial pathogens requires the overcoming of the cell wall. To this end, pathogens produce a wide array of plant cell wall degrading enzymes (CWDEs), among which endo-polygalacturonases (PGs; EC 3.2.1.15) are secreted at very early stages of the infection process (ten Have et al., 1998). PGs cleave the α-(1–4) linkages between the D-galacturonic acid residues of homogalacturonan, the main component of pectin, causing cell separation and maceration of the host tissue. To counteract the activity of PGs, plants deploy the cell wall polygalacturonase inhibiting proteins (PGIPs) that inhibit the pectin-depolymerizing activity of PGs. No plant species or mutants totally lacking PGIP activity have been characterized so far. The structure of PGIPs is typically formed by 10 imperfect leucine-rich repeats (LRRs) of 24 residues each, which are organized to form two β-sheets, one of which (sheet B1) occupies the concave inner side of the molecule and contains residues crucial for the interaction with PGs (Di Matteo et al., 2003). In addition to PG inhibition, the interaction between PGs and PGIPs promotes the formation of oligogalacturonides (OGs), which are elicitors of a variety of defense responses (Cervone et al., 1989; Ridley et al., 2001; Ferrari et al., 2013). Since many aspects of the PGIP biology have been already summarized in previous reviews (De Lorenzo et al., 2001; De Lorenzo and Ferrari, 2002; D'Ovidio et al., 2004a; Gomathi and Gnanamanickam, 2004; Shanmugam, 2005; Di Matteo et al., 2006; Federici et al., 2006; Cantu et al., 2008; Misas-Villamil and van der Hoorn, 2008; Protsenko et al., 2008; Reignault et al., 2008; Lagaert et al., 2009), here we present an overview of the recent findings on genome composition and evolution of pgip gene families and on the efficacy of PGIP to limit the development of diseases caused by microbial pathogens in crop plants.
PGIP genes and their genomic organization
Early characterization of a polygalacturonase-inhibiting activity was reported in 1970s (Albersheim and Anderson, 1971) and the first pgip gene was isolated 20 years later in French bean (Toubart et al., 1992). Since then, several PGIPs and a large number of pgip genes have been characterized. Up to now more than 170 complete or partial pgip genes from dicot and monocot plants have been deposited in nucleotide databases (e.g., http://www.ncbi.nlm.nih.gov/). Most of these genes have been identified as pgip genes on the basis of sequence identity but only a few of them have been shown to encode proteins with PG-inhibitory activity.
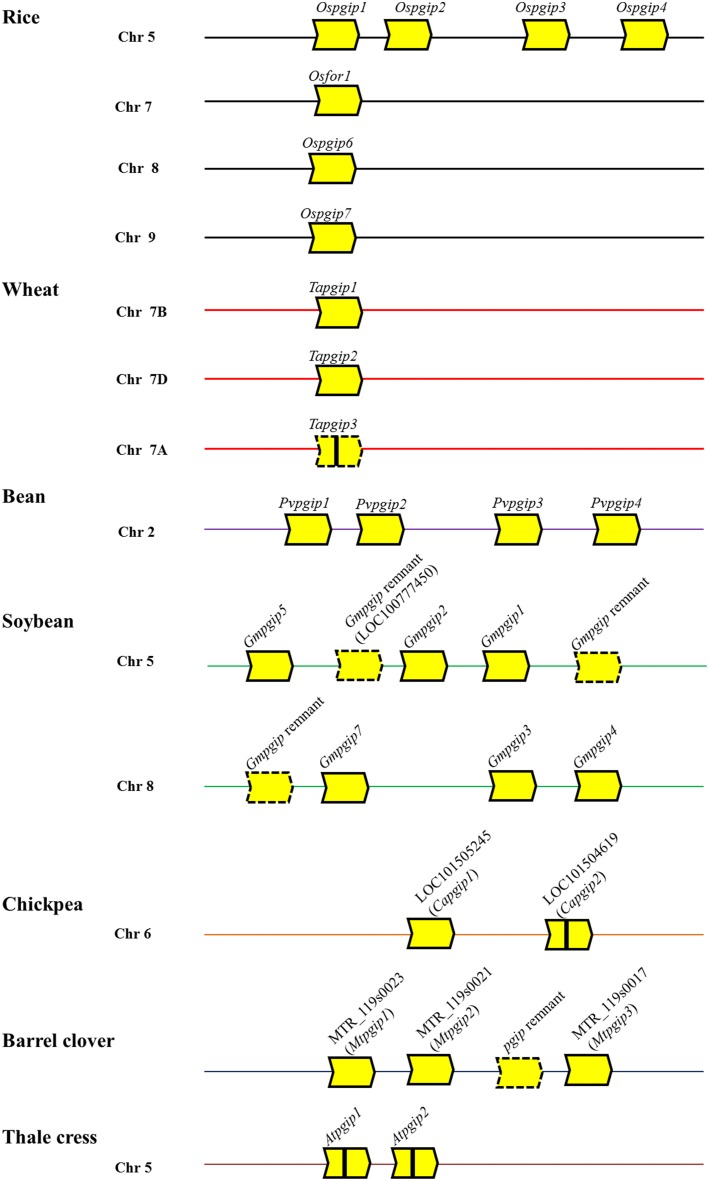
Genome analysis has shown that pgip genes did not undergo a large expansion and may exist as single genes, as in diploid wheat species (Di Giovanni et al., 2008), or organized into gene families, the members of which are organized in tandem and can vary from two, as in Arabidopsis thaliana (Ferrari et al., 2003), to sixteen, as in Brassica napus (Hegedus et al., 2008). The majority of pgip genes are intronless, however, some of them can contain a short intron as in Atpgip1 and Atpgip2 (Ferrari et al., 2003). Moreover, pgip genes can be inactivated by transposon elements as in cultivated and wild wheat where the occurrence of Copia-retrotransposon and Vacuna transposons has been reported (Di Giovanni et al., 2008). Characterized pgip loci are shown in Figure 1. Like other families of defense-related genes, pgip families show variation in the expression pattern of the different members, some of which are constitutive, others are tissue-specific and, in most cases, up-regulated following stress stimuli (see reviews indicated above; Table 1). At the protein level, members of a pgip family show both functional redundancy and sub-functionalization (De Lorenzo et al., 2001; Federici et al., 2006). As suggested previously, these features likely have an adaptive significance for combating more efficiently a broad array of pathogens (Ferrari et al., 2003) or responding more rapidly to diverse environmental stimuli (D'Ovidio et al., 2004b). In support of this view, a recent analysis of the genomic organization and composition of the legume pgip families suggested that the forces driving the evolution of the pgip genes follow the birth-and-death model (Kalunke et al., 2014), similarly to what proposed for the evolution of NBS-LRR-type R genes (Michelmore and Meyers, 1998). This possibility is based on genomic features that include inferred recent duplications, diversification as well as pseudogenization of pgip copies, as found in soybean, bean, barrel clover and chickpea (Kalunke et al., 2014). The organization of the pgip families therefore supports the view that tandem duplications are frequent in stress-related genes and are beneficial for survival in challenging environments (Oh et al., 2012).
Figure 1.
Schematic representation of the genomic organization pgip families in rice, wheat, bean, soybean, chickpea, barrel clover and thale cress. Each block-arrow with compound-type lines represents a predicted pgip gene and a block-arrow with dash type lines represents a predicted pseudo-gene or remnant gene. Vertical line within block-arrow indicates introns (Capgip2, Atpgip1, and Atpgip2) or a Copia retrotransposon (Tapgip3). The direction of the arrow indicates ATG to stop codon. The location of pgip genes of legume species are based on Kalunke et al. (2014), those of rice and wheat on Janni et al. (2006) and Di Giovanni et al. (2008), and those of thale crest on Ferrari et al. (2003). Chr, chromosome.
Table 1.
Treatments or stress stimuli affecting pgip expression in some plant species with a well characterized pgip family.
| Pgip family | Treatments or stress stimuli | References |
|---|---|---|
| Rice | Abscisic acid (ABA), brassinosteroid, gibberellic acid (GA), 3-indole acetic acid (IAA), jasmonic acid (JA), kinetin, naphthalene acetic acid (NAA), salicylic acid (SA); Rhizoctonia solani (necrotrophic fungus) | Janni et al., 2006; Lu et al., 2012 |
| Wheat | Bipolaris sorokiniana (necrotrophic fungus) and mechanical wounding | Janni et al., 2013 |
| Bean | Oligogalacturonides (OGs); mechanical wounding; Botrytis cinerea, Sclerotinia sclerotiorum (necrotrophic fungi); Colletotrichum lindemuthianum (hemibiotrophic fungus) | Bergmann et al., 1994; Nuss et al., 1996; Devoto et al., 1997; D'Ovidio et al., 2004b; Oliveira et al., 2010; Kalunke et al., 2011 |
| Soybean | Mechanical wounding; S. sclerotiorum (necrotrophic fungus) | D'Ovidio et al., 2006; Kalunke et al., 2014 |
| M. truncatula | JA, SA, ABA; Colletotrichum trifolii (hemibiotrophic fungus) | Song and Nam, 2005 |
| Rapeseed | JA, SA, mechanical wounding; S. sclerotiorum | Hegedus et al., 2008 |
| Pepper | SA, Methyl jasmonate (Me-JA), ABA, wounding, cold treatment | Wang et al., 2013 |
| Arabidopsis | OGs; JA; B. cinerea; Stemphylium solani (necrotrophic fungus); aluminum, low-pH, cold; geminivirus | Ferrari et al., 2003; Ascencio-Ibanez et al., 2008; Sawaki et al., 2009; Di et al., 2012; Kobayashi et al., 2014 |
Inhibition activity of PGIPs
A number of papers deals with the inhibition activity of PGIPs purified from several plant tissues. This aspect has been reviewed several years ago (De Lorenzo et al., 2001); here, we present an update of this information (Table 2). Because purified PGIPs may contain a mix of highly similar PGIP isoforms, the activity detected in a tissue may result from the contribution of the activities of different PGIPs expressed in that tissue. An appropriate approach to study the inhibition activity of individual PGIP isoforms is their expression in a heterologous system. However, only a few of the more than 170 pgip genes isolated so far from different plant species have been investigated. As reported in Table 3, individual heterologous expression and analysis of all members of a pgip family has been performed only for Arabidopsis (Ferrari et al., 2003), common bean (D'Ovidio et al., 2004b), soybean (D'Ovidio et al., 2006; Kalunke et al., 2014) and wheat (Janni et al., 2013). PGIPs have been expressed in prokaryotic systems, as a fusion with the maltose-binding protein (MBP) (Jang et al., 2003; Table 3) or using lower temperature for bacterial growth (Chen et al., 2011), in Pichia pastoris and in plants by stable transformation or, transiently, by virus-mediated expression (Table 3). In some cases, the proteins were successfully expressed, but did not show any inhibitory activity in vitro, as, for example, in the case of some GmPGIPs (D'Ovidio et al., 2006). GmPGIP3, but not GmPGIP1, GmPGIP2, and GmPGIP7 showed inhibitory activity, whereas no expression of GmPGIP5 was obtained (D'Ovidio et al., 2006; Kalunke et al., 2014). Similarly, TaPGIP1 and TaPGIP2, encoded by the two members of the wheat pgip family, were successfully expressed but showed no inhibition activity (Janni et al., 2013).
Table 2.
Bulk PGIP purified from plants and tested against microbial PGs. These data update those reported in De Lorenzo et al. (2001).
| Plant | Tissue | PGIP preparation | Polygalacturonases | References | |
|---|---|---|---|---|---|
| Inhibited | Not inhibited | ||||
| Tomato (Solanum lycopersici L.) | Stem | Crude extract | Ralstonia solanacearum | Schacht et al., 2011 | |
| Tobacco (Nicotiana tabacum L.) | Nectar | Botrytis cinerea | Thornburg et al., 2003 | ||
| Potato (Solanum tuberosum L.) | Gel chromatography | Aspergillus niger | Fusarium solani (isolate 3122) | Machinandiarena et al., 2001 | |
| Fusarium moniliforme§ | |||||
| Fusarilm solani isolate 1402 | |||||
| Common Bean (Phaseolus vulgaris L.) |
Leaves | PG-Sepharose chromatography | Fusarium anthophilum | Fusarium verticillioides | Raiola et al., 2008 |
| Fusarium circinatum | Fusarium proliferatum ISPAVEmc 1189 | ||||
| Fusarium subglutinans. | Fusarium nygamai | ||||
|
Fusarium proliferatum isolate 1152 |
|||||
|
Fusarium proliferatum PVS-Fu 64 |
|||||
| Fusarium sacchari | |||||
| Fusarium fujikuroi | |||||
| F. thapsinum | |||||
|
Fusarium moniliforme§ FC-10 |
Fusarium moniliforme§ PD |
Sella et al., 2004 | |||
| Leek (Allium ampeloprasum L.) | Basal leaves | Mono-S chromatography | Fusarium anthophilum | Raiola et al., 2008 | |
| Fusarium circinatum | |||||
| Fusarium subglutinans | |||||
| Fusarium proliferatum | |||||
| Fusarium sacchari | |||||
| Fusarium fujikuroi | |||||
| Fusarium verticillioides | |||||
|
Fusarium proliferatum ISPAVEmc 1189 |
|||||
| Fusarium nygamai | |||||
| Asparagus (Asparagus officinalis L.) | White spear | Mono-S chromatography | Fusarium anthophilum | Raiola et al., 2008 | |
| Fusarium circinatum | |||||
| Fusarium subglutinans. | |||||
| Fusarium proliferatum | |||||
| Fusarium sacchari | |||||
| Fusarium fujikuroi | |||||
| Fusarium verticillioides | |||||
|
Fusarium proliferatum ISPAVEmc 1189 |
|||||
| Fusarium nygamai | |||||
| Pepper (Capsicum annuum L.) | Fruit | Ion-exchange chromatography | Colletotrichum gleosporoides, | Shivashankar et al., 2010 | |
| Colletotrichum capsici, | |||||
| Colletotrichum lindemuthianum | |||||
| Sclerotium rolfsi | |||||
| Fusarium moniliforme§ | |||||
| Guava (Psidium guajava L.) | Fruit | Purified using a Sephadex G-100 | Aspergillus niger | Deo and Shastri, 2003 | |
| “Oroblanco” grapefruit hybrid (Citrus grandis × C. paradisi Macf.) | Fruit | Anion exchange chromatography | Penicillium italicum | D'hallewin et al., 2004 | |
| Botrytis cinerea | |||||
| Apple (Malus domestica L.) | Fruit | Colletotrichum acutatum | Gregori et al., 2008 | ||
| Fruit skin | Partial purified | Botryosphaeria dothidea | Glomerella cingulata | Lee et al., 2006 | |
| Parenchymal tissues | Partial purified | Monilia fructigena | Buza et al., 2004 | ||
| Cantaloupe (Cucumis melo L.) | Fruit | Cation exchange chromatography | Phomopsis cucurbitae | Didymella bryoniae | Fish and Davis, 2004 |
| Aspergillus niger | Rhizopus PG | ||||
| Fusarium solani | Fusarium verticillioides | ||||
| Cotton (Gossypium hirsutum L.) | Stem | PG-affinity chromatography | Aspergilus niger | James and Dubery, 2001 | |
| Pear (Pyrus communis L.) | Fruit | Partial purified | Verticillium dahliae | Ladu et al., 2012; Faize et al., 2003 | |
| Botrytis cinerea | |||||
| Venturia nashicola | |||||
| Pearl millets (Pennisetum glaucum (L) R. Br.) | Seedlings | Crude extract | Aspergilus niger | Prabhu et al., 2012 | |
| Grass pea (Lathyrus sativus L.) | Seeds | Gel-filtration chromatography | Aspergilus niger | Tamburino et al., 2012 | |
| Rhizopus spp | |||||
| Orange (Citrus reticulate L.) | Fruit | Partial purified | Diaprepes abbreviatus | Doostdar et al., 1997 | |
| Blue mustard (Chorispora bungeana) | Leaves, stem, root | Partial purified | Aspergillus niger | Di et al., 2009 | |
| Stemphylium solani | |||||
| Ginseng (Panax ginseng L.) | Crude extract | Colletotrichum gloeosporioides | Sathiyaraj et al., 2010 | ||
| Phythium ultimum | |||||
| Fusarium oxysporum | |||||
| Rhizoctonia solani | |||||
| Bread wheat (Triticum aestivum L.) | Leaves | Cation exchange chromatography | Cochliobolus sativus | Aspergillus niger (EPG I and II) | Kemp et al., 2003 |
| Cryphonectria parasitica | |||||
| Postia placenta | |||||
| Fusarium moniliforme§ | |||||
| Colletotrichum lindemuthianum | |||||
| Aspergillus niger exopolygalacturonase | |||||
| Ralstonia solanacearum | |||||
| Durum wheat (Triticum turgidum ssp. dicoccoides | Leaves | Crude extract | Fusarium graminearum | Fusarium phyllophylum | Janni et al., 2013 |
| Bipolaris sorokiniana | |||||
| Stenocarpella maydis | |||||
Reclassified as Fusarium phyllophilum (Mariotti et al., 2008).
Table 3.
Pgip genes individually expressed in plants or in heterologous systems and tested for inhibition activity against microbial PGs.
| Species | Gene | Heterologous systems | Origin of purified PG | References | |
|---|---|---|---|---|---|
| Inhibited | Not inhibited | ||||
| Common bean (Phaseolus vulgaris L.) | PvPGIP1 | Transgenic tomato | Fusarium oxysporum | Desiderio et al., 1997 | |
| Botrytis cinerea | |||||
| Alternaria solani | |||||
| Stenocarpella maydis | Berger et al., 2000 | ||||
| Aspergillus niger | |||||
| PvPGIP1 PvPGIP2 PvPGIP3 PvPGIP4 |
PVX/Nicotiana benthamiana | Aspergillus niger | Lygus rugulipennis | D'Ovidio et al., 2006; Frati et al., 2006 | |
| Fusarium moniliforme§ | Adelphocoris lineolatus | ||||
| Stenocarpella maydis | Orthops kalmi | ||||
| Colletotrichum acutatum | Closterotomus norwegicus | ||||
| Botrytis cinerea | |||||
| PvPGIP2 | Transgenic wheat | Bipolaris sorokiniana | Claviceps purpurea | Janni et al., 2008; Volpi et al., 2013 | |
| F. graminearum | |||||
| Transgenic Brassica napus | Rhizoctonia solani | Akhgari et al., 2012 | |||
| Transgenic sugarbeet | Fusarium phyllophilum FC10 | Mohammadzadeh et al., 2012 | |||
| PVX/Nicotiana benthamiana | Fusarium phyllophilum FC-10 | Fusarium phyllophilum 25305 | Mariotti et al., 2008 | ||
| Fusarium phyllophilum 10241 | Fusarium verticillioides 62264 | ||||
| Fusarium phyllophilum 25219 | Fusarium verticillioides PD | ||||
| Fusarium phyllophilum 25218 | |||||
| Runner bean (Phaseolus coccineus L.) | PcPGIP2 | PVX/Nicotiana benthamiana | Fusarium moniliforme§ | Farina et al., 2009 | |
| Aspergillus niger | |||||
| Colletotrichum lupini | |||||
| Botrytis cinerea | |||||
| Tepary bean (Phaseolus acutifolius L.) | PaPGIP2 | PVX/Nicotiana benthamiana | Fusarium moniliforme§ | Farina et al., 2009 | |
| Aspergillus niger | |||||
| Colletotrichum lupini | |||||
| Botrytis cinerea | |||||
| Lima bean (Phaseolus lunatus L.) | PlPGIP2 | PVX/Nicotiana benthamiana | Fusarium moniliforme§ | Farina et al., 2009 | |
| Aspergillus niger | |||||
| Colletotrichum lupini | |||||
| Botrytis cinerea | |||||
| Soybean (Glycine max L.) | GmPGIP1 GmPGIP2 |
PVX/Nicotiana benthamiana | Sclerotinia sclerotiorum PGb | D'Ovidio et al., 2006; Frati et al., 2006 | |
| Sclerotinia sclerotiorum PGa | |||||
| Fusarium moniliforme§ | |||||
| Botrytis aclada | |||||
| Aspergillus niger | |||||
| Botrytis cinerea | |||||
| Colletotrichum acutatum | |||||
| Fusarium graminearum | |||||
| Lygus rugulipennis | |||||
| Adelphocoris lineolatus | |||||
| Orthops kalmi | |||||
| Closterotomus norwegicus | |||||
| GmPGIP3 | PVX/Nicotiana benthamiana | Sclerotinia sclerotiorum PGb | D'Ovidio et al., 2006; Frati et al., 2006 | ||
| Sclerotinia sclerotiorum PGa | |||||
| Fusarium moniliforme§ | |||||
| Botrytis aclada | |||||
| Aspergillus niger | |||||
| Botrytis cinerea | |||||
| Colletotrichum acutatum | |||||
| Fusarium graminearum | |||||
| GmPGIP4 | PVX/Nicotiana benthamiana | Sclerotinia sclerotiorum PGb | D'Ovidio et al., 2006; Frati et al., 2006 | ||
| Sclerotinia sclerotiorum PGa | |||||
| Fusarium moniliforme§ | |||||
| Botrytis aclada | |||||
| Aspergillus niger | |||||
| Botrytis cinerea | |||||
| Colletotrichum acutatum | |||||
| Fusarium graminearum | |||||
| GmPGIP7 | PVX/Nicotiana benthamiana | Sclerotinia sclerotiorum | |||
| Fusarium graminearum | Kalunke et al., 2014 | ||||
| Colletotrichum acutatum | |||||
| Aspergillus niger | |||||
| Pepper (Capsicum annum L.) | CaPGIP1 CaPGIP2 | Escherichia coli | Alternaria alternata | Wang et al., 2013 | |
| Colletotrichum nicotianae | |||||
| Rapeseed (Brassica napus L.) | BnPGIP1 | Pichia pastoris | Sclerotinia sclerotiorum PG6 | Bashi et al., 2013 | |
| Chinese cabbage (Brassica rapa L.) | BrPGIP2 | Transgenic Brassica rapa | Pectobacterium carotovorum | Hwang et al., 2010 | |
| Botryosphaeria dothidea | |||||
| BrPGIP2 | Escherichia coli | Sclerotinia sclerotiorum | HuangFu et al., 2014 | ||
| Grapevine (Vitis vinifera L.) | VvPGIP1 | Transgenic tobacco | Botrytis cinerea PGI | Botrytis cinerea PG3 | Joubert et al., 2006 |
| Botrytis cinerea PG4 | Aspergillus niger PGII | ||||
| Botrytis cinerea PG6 | |||||
| Aspergillus. niger PGA | |||||
| Aspergillus niger PGB | |||||
| Aspergillus niger PGI | Botrytis cinerea PG2 | Joubert et al., 2007 | |||
| Apple (Malus domestica Borkh.) | MdPGIP1 | Transgenic tobacco | Colletotrichum lupini | Aspergillus niger | Oelofse et al., 2006 |
| Botryosphaeria obtusa | |||||
| Diaporthe ambigua | |||||
| Transgenic potato | Verticillium dahliae | Gazendam et al., 2004 | |||
| Pear (Pyrus communis L.) | PpPGIP | Transgenic grape | Botrytis cinerea | Agüero et al., 2005 | |
| Transgenic tomato | Botrytis cinerea | Powell et al., 2000 | |||
| Transgenic persimmon | Botrytis cinerea | Tamura et al., 2004 | |||
| Raspberry (Rubus idaeus L.) | RiPGIP | Transgenic pea | Stenocarpella maydis | Richter et al., 2006 | |
| Colletotrichum lupini | |||||
| Wheat (Triticum aestivum L.) | TaPGIP1 TaPGIP2 |
PVX/Nicotiana benthamiana | Fusarium phyllophylu | Janni et al., 2013 | |
| Stenocarpella maydis | |||||
| Bipolaris sorokiniana | |||||
| Fusarium graminearum | |||||
| Rice (Oryza sativa L.) | OsPGIP1 | PVX/Nicotiana benthamiana | Sclerotinia sclerotiorum | Janni et al., 2006 | |
| Fusarium moniliforme§ | |||||
| Fusarium graminearum | |||||
| Aspergillus niger | |||||
| Botrytis cinerea | |||||
| OsFOR1 | Escherichia coli BL21 | Aspergillus niger PG | Jang et al., 2003 | ||
| Pearl millet [Pennisetum glaucum (L.) R. Br.] | PglPGIP1 | Escherichia coli SHuffle® T7 Express | Aspergillus niger, AnPGII | Fusarium moniliforme, FmPGIII | Prabhu et al., 2014 |
| Arabidopsis thaliana | AtPGIP1 AtPGIP2 |
Transgenic Arabidopsis | Colletotrichum gloeosporioides | Aspergillus niger | Frati et al., 2006; Ferrari et al., 2012, 2003 |
| Stenocarpella maydis | Fusarium moniliforme§ | ||||
| Botrytis cinerea | Lygus rugulipennis | ||||
| Fusarium graminearum | Adelphocoris lineolatus | ||||
| Orthops kalmi | |||||
| Closterotomus norwegicus | |||||
Reclassified as Fusarium phyllophilum FC10 (Mariotti et al., 2008).
The absence of inhibition activity in vitro may also reflect the possibility that some PGIPs are active only in the in planta environment, as suggested by Joubert et al. (2006) in the case of the Botrytis cinerea BcPG2 and VvPGIP1 from grapevine (Vitis vinifera L.). These proteins do not interact in vitro, although VvPGIP1 reduces symptoms caused by BcPG2 upon co-infiltration in leaves. The number and sources of PGs tested is also limited; only a few studies have been carried out against PGs of bacteria and insects (Doostdar et al., 1997; D'Ovidio et al., 2004b; Frati et al., 2006; Hwang et al., 2010; Schacht et al., 2011; Kirsch et al., 2012). The limitations of data prevents to draw conclusions about correlations between PGIPs of specific plant families and specific pathogens. Notably, PG produced by a highly detrimental pathogen, Fusarium verticillioides, is not inhibited by any known PGIP (see Table 2). This PG has been a target of an unsuccessful attempt to render PvPGIP2 an efficient inhibitor against this PG (see below, Benedetti et al., 2011a).
The utilization of pgip genes for crop protection relies on the identification of inhibitors with broad specificities against the many PGs produced by phytopathogens and/or the construction of novel PGIPs with stronger and broader inhibitor activity. Many more PGIPs than those reported in Tables 2, 3 exist in nature and are likely to have different specificities against microbial PGs, considering that single amino acid changes are able to change specificity of the inhibitors (Leckie et al., 1999). Searching for PGIPs with novel specificities may allow to count on a much larger reservoir of possible genes for crop protection. A direct and simple strategy to isolate PGIPs with recognition capability against a given PG may be based on affinity chromathography methods, similar to that originally used to purify PGIP from P. vulgaris (Cervone et al., 1987), and mass spectrometry. Attempts to drive in vitro evolution of PGIPs to generate proteins with improved inhibition properties have not been successful yet (Benedetti et al., 2011a).
The occurrence of PG-inhibiting activity in crude leaf protein extracts of tetraploid wild wheat (T. dicoccoides) possessing non functional pgip genes (Di Giovanni et al., 2008) suggested the existance of pgip genes with a sequence divergent from the classical one. This possibility, which deserves further investigation, is also supported by the finding that the wheat tissue contains PG-inhibiting proteins with N-terminal sequences (Lin and Li, 2002; Kemp et al., 2003) different from TaPGIP1 and TaPGIP2 (Janni et al., 2013) and from the pgip sequences reported so far (http://www.ncbi.nlm.nih.gov/nucleotide/). Recently, a wheat gene with some sequence similarity to pgip genes has been reported and was shown to be involved in the defense response against Fusarium graminearum (Hou et al., 2014).
Structural studies on the PG-PGIP interaction
Thus, the possibility of engineering new forms of PGIPs depends on the detailed structural knowledge of the PG-PGIP interaction. Several structural studies have been performed (Mattei et al., 2001; King et al., 2002; Benedetti et al., 2011b, 2013; Gutierrez-Sanchez et al., 2012), but a high resolution 3D-structure of the PG-PGIP complex is still missing. The enzyme-inhibitor combinations that have been more extensively investigated, are those that PGIP2 from Phaseolus vulgaris (PvPGIP2) forms with PG from A. niger (AnPGII), F. phyllophilum (FpPG) and C. lupini (ClPG). Site-directed mutagenesis has shown that the residues involved in the interaction are located in the concave surface of the inhibitor (Leckie et al., 1999; Federici et al., 2001; Spinelli et al., 2009; Benedetti et al., 2011b, 2013). Computational methods such as the Codon Substitution Model in combination with the Desolvation Energy Calculation and the Repeat Conservation Mapping (RCM; Helft et al., 2011) have pinpointed several residues of PvPGIP2 responsible for the PG-inhibiting activity (Casasoli et al., 2009).
On the other hand, residues of PG that are critical for the interaction with PGIP have been also studied. FvPG is 92.5% identical to FpPG, but is inhibited by neither PvPGIP2 nor other known PGIPs. By both loss- and gain-of-function site-directed mutations, a single amino acid at position 274 of both FvPG and FpPG was demonstrated to act as a switch for recognition by PvPGIP2 (Raiola et al., 2008; Benedetti et al., 2013). Unfortunately, the lack of high-resolution structural information on the PG-PGIP complex does not allow to precisely identify the contacting residue in PGIP. Moreover, both PGs and PGIPs are glycosylated proteins (Caprari et al., 1993; Lim et al., 2009); however, whether glycosylation plays a role in the PGIP-PG interaction requires further investigation. For example, glycosylation in pearl millet PGIP was found to affect pH and temperature stability of the protein but not its capability of inhibiting AnPGII (Prabhu et al., 2015).
A single PGIP may display different mechanisms of PG inhibition (competitive, non competitive and mixed) suggesting that the protein is highly versatile in recognizing different epitopes of various PGs (Federici et al., 2001; King et al., 2002; Sicilia et al., 2005; Bonivento et al., 2008). Consequently, many 3D-models based on docking predictions have been proposed so far (Sicilia et al., 2005; Maulik et al., 2009; Prabhu et al., 2014). Techniques such as the mass amide exchange mass spectrometry in the case of AnPGII and FpPG and the Small Angle X-ray Scattering (SAXS) in the case of FpPG and ClPG have produced models that, in some cases, are discordant. For example, while the mass amide exchange mass spectrometry predicts that the area of FpPG in contact with PvPGIP2 is located at the N-terminus and predominantly on the underside of the enzyme beta-barrel structures (Gutierrez-Sanchez et al., 2012), the SAXS analysis indicates that the protein region in contact with PvPGIP2 is located at the C-terminus of the enzyme and includes the loops surrounding the active site cleft. A site-directed mutagenesis analysis has been used to validate this second view (Benedetti et al., 2013). In general, low resolution techniques such as SAXS analysis or mass amide exchange mass spectrometry require validation by site-directed mutagenesis to locate the contacting residues in a protein complex.
The X-ray crystallography, successfully used to solve several high-resolution structures of PGs (van Santen et al., 1999; Federici et al., 2001; Bonivento et al., 2008) and that of PvPGIP2 (Di Matteo et al., 2003), was so far unsuccessful in the case of the PG-PGIP complex. This is probably due to the intrinsic instability of the PG-PGIP interaction, which only occurs, under apoplastic conditions of pH and ionic strength, through the contact of only a few, sometimes only one, residues (Leckie et al., 1999). The use of a cross-linker for stabilizing the PG-PGIP complex coupled to techniques that allow the protein analysis directly in solution, such as SAXS and NMR spectroscopy (Wand and Englander, 1996; Nietlispach et al., 2004), may be a valid alternative in order to obtain a detailed map of the contacting residues but this requires a subsequent validation by site-directed mutagenesis.
PGIPs engineered in dicot crops
The important role of PGIP in plant defense has been demonstrated by overexpressing pgip genes in several plant species. In these experiments, the source of the used genes was either the same plant species utilized for transformation or a different one (Table 4). The transformation of the model plant A. thaliana has been particularly useful to highlight the potentiality of several pgip genes, namely the endogenous Atpgip1 and Atpgip2, the bean Pvpgip2 and the rapeseed (Brassica napus) Bnpgip1 or Bnpgip2. Arabidopsis plants overexpressing Atpgip1 or Atpgip2 showed a significant reduction of disease symptoms caused by B. cinerea (Ferrari et al., 2003) and were less susceptible against the hemibiotrophic fungal pathogen F. graminearum (Ferrari et al., 2012), the major causal agent of Fusarium head blight (FHB). Conversely, silencing of their expression using an antisense Atpgip, led to enhanced susceptibility (Ferrari et al., 2006). Arabidopsis plants expressing Pvpgip2, encoding an efficient inhibitor of the B. cinerea PG (ten Have et al., 1998), showed reduction of disease symptoms caused by B. cinerea and those expressing the rapeseed genes Bnpgip1 and Bnpgip2 delayed the symptoms caused by S. sclerotiorum (Bashi et al., 2013).
Table 4.
List of transgenic crops produced using the gene coding for PGIP and their response to fungal, oomycetes or bacterial phytopathogens.
| Transgenic crops | PGIP genec | Tested against fungal, oomycetes or bacterial phytopathogens | References |
|---|---|---|---|
| Tomatoa (Solanum lycopersicum L.) | PcPGIP | Botrytis cinerea* | Powell et al., 2000, 1994 |
| PvPGIP1 | Fusarium oxysporum f.sp. lycopersici† | Desiderio et al., 1997 | |
| Botrytis cinerea† | |||
| Alternaria solani† | |||
| Tobaccoa (Nicotiana tabacum L.) | PvPGIP2 | Botrytis cinerea* | Manfredini et al., 2005 |
| Rhizoctonia solani* | Borras-Hidalgo et al., 2012 | ||
| Phytophthora parasitica* | |||
| Peronospora hyoscyami* | |||
| CaPGIP1 | Alternaria alternata* | Wang et al., 2013 | |
| Colletotrichum nicotianae* | |||
| VvPGIP1 | Botrytis cinerea* | Joubert et al., 2006 | |
| BrPGIP2 | Pectobacterium carotovorum* | Hwang et al., 2010 | |
| Potatoa (Solanum tuberosum L.) | MdPGIP1 StPGIP | Verticillium dahliae† Verticillium dahliae* | Gazendam et al., 2004; Guo et al., 2014 |
| Brassica rapaa | BrPGIP2 | Pectobacterium carotovorum* | Hwang et al., 2010 |
| Rapeseeda (Brassica napus L.) | BnPGIP2 | Sclerotinia sclerotiorum* | HuangFu et al., 2014 |
| Peaa (Pisum sativum L.) | RiPGIP | Glomus intraradicesΨ | Hassan et al., 2012 |
| Grapevinea (Vitis vinifera L.) Ricea (Oriza sativa L.) | PcPGIP OsPGIP1 | Botrytis cinerea* | Agüero et al., 2005; Wang et al., 2014b |
| Xylella fastidiosa* Rhizoctonia solani | |||
| Wheatb (Triticum aestivum L., Triticum durum Desf.) | PvPGIP2 GmPGIP3 | Bipolaris sorokiniana* | Janni et al., 2008 |
| Fusarium graminearum* | Ferrari et al., 2012 | ||
| Claviceps purpurea† Bipolaris sorokiniana* Gaeumannomyces graminis var. tritici* | Volpi et al., 2013; Wang et al., 2014a | ||
| Arabidopsis thaliana L.a | PvPGIP2 | Botrytis cinerea* | Manfredini et al., 2005 |
| AtPGIP1 AtPGIP2 | Fusarium graminearum* | Ferrari et al., 2012 | |
| BnPGIP1 BnPGIP2 | Sclerotinia sclerotiorum* | Bashi et al., 2013 |
The transgenic gene was under control of CaMV 35S promoter.
The transgenic gene was under control of Ubiquitin promoter.
Pc, Pyrus communis; Pv, Phaseolus vulgaris; Ca, Capsicum annum; Vv, Vitis vinifera; Br, Brassica rapa; Md, Malus domestica; St, Solanum torvum; Ri, Rubus idaeus; Ac, Actinidia deliciosa; At, Arabidopsis thaliana; Bn, Brassica napa.
Showed enhanced resistance.
No evidence of enhanced resistance.
No effect on mycorrhization.
The protective potential of pgip genes has also been demonstrated in transgenic crops. The first transgenic crop plant obtained by using a pgip gene and tested against pathogenic microorganisms were tomatos expressing PvPGIP1 from P. vulgaris. These plants, however, did not show any increased resistance against Fusarium oxysporum f. sp. lycopersici, B. cinerea, and Alternaria solani. The negative result was due to the inability of PvPGIP1 to inhibit the PGs secreted by these fungi, as shown by in vitro inhibition assays and led to discovery of other forms of PGIPs and eventually to the existence of a complex PGIP family in French bean (Desiderio et al., 1997). A few years later, transgenic tomato plants expressing a pear (Pyrus communis L.) PGIP (PcPGIP) capable of inhibiting the PGs secreted by B. cinerea, showed a reduction of disease lesions caused by this fungus both on ripening fruit (15% reduction) and leaves (about 25% reduction). The initial establishment of infection was not affected in the transgenic plants but the later colonization of the host tissue was significantly reduced (Powell et al., 2000).
Tobacco has been the most used crop plant for testing the effect of PGIP expression on resistance to pathogens. Constitutive and high-level expression of Pvpgip2 (from P. vulgaris), Vvpgip1 (from V. vinifera), Capgip1 [from pepper (Capsicum annum)] and Brpgip2 (from B. rapa) have been obtained in transgenic tobacco. Plants expressing PvPGIP2 showed about 35% reduction of symptoms caused by B. cinerea (Manfredini et al., 2005) and, more recently, were shown to display reduced disease symptoms against Rhizoctonia solani and two oomycete pathogens, Phytophthora parasitica var. nicotianae and the blue mold-causing agent Peronospora hyoscyami f. sp. tabacina (Borras-Hidalgo et al., 2012). Notably, the experiments against P. hyoscyami f.sp. tabacina were performed in the field during seasonal conditions that favor the pathogen spreading. In agreement with what observed under controlled conditions, resistance of transgenic plants was comparable to that exhibited by Nicotiana species (N. rustica, N. debneyi and N. megalosiphon) that are highly resistant to blue mold disease. These transgenic plants expressing PvPGIP2 represented the first example of PGIP-expressing plants subjected to field trails. Recently, transgenic rice expressing OsPGIP1 showed also improved resistance against Rhizoctonia solani in field experiments (Wang et al., 2014b).
Transgenic tobacco plants expressing the grapevine pgip gene Vvpgip1 (Joubert et al., 2006) also showed a reduced (from 47 to 69%) disease susceptibility to B. cinerea infection. As for plants expressing PvPGIP2, the resistance phenotype correlated with the accumulation of VvPGIP1 as well as with its capability of inhibiting the activity of PG secreted by B. cinerea, namely BcPG1, BcPG3, and BcPG6. Several observations, however, suggest that PGIP may improve resistance by mechanisms other than classical PGIP-PG inhibition. For example, non-infected transgenic tobacco plants expressing Vvpgip1 show modified expression patterns of genes involved in various metabolic pathways (Alexandersson et al., 2011) and an altered cell wall structure (Nguema-Ona et al., 2013). In these plants, lignin accumulation and arabinoxyloglucan-cellulose re-organization leads to a general strengthening/reinforcing of the cell wall that may contribute to an improved resistance against B. cinerea.
A reduction of disease symptoms (about 50%) caused by Alternaria alternata and Colletotrichum nicotianae was also observed in transgenic tobacco lines expressing the pepper CaPGIP1 and, once again, resistance correlated with the inhibition capacity of purified CaPGIP1 against PG activity of both fungal pathogens (Wang et al., 2013).
Within the Solanaceae family, transgenic potato (Solanum tuberosum) plants expressing the gene StPGIP1 from S. torvum showed a 50% reduction of wilt disease symptoms caused by Verticillium dahliae and a normal plant growth (Guo et al., 2014). Transgenic potato plants overexpressing the apple pgip1 gene showed protection against the same fungal pathogen but displayed an extended juvenile phase (Gazendam et al., 2004).
Transgenic grapevine (V. vinifera) plants constitutively expressing the pear PcPGIP gene represent an interesting example of the potential of PGIP for protection against pathogens other than fungi and oomycetes. These plants show a delayed development of the Pierce's disease (PD) caused by bacterial pathogen Xylella fastidiosa (Agüero et al., 2005). Not only leaf scorching and Xylella titre were reduced but also plants showed a better re-growth after pruning compared to infected untransformed controls. Moreover, an inverse dose-effect relationship was shown between development of PD and levels of PcPGIP activity in the tissues. The improved resistance of the grapevine plants expressing PcPGIP against a bacterial pathogen was unexpected, because until then the PGIP inhibition activity was thought to be limited to fungal and insect PGs (Cervone et al., 1990; Johnston et al., 1993; D'Ovidio et al., 2004b). It was later shown that pear PcPGIP inhibits the PG encoded by X. fastidiosa and that PG activity is a virulence factor of this pathogen (Roper et al., 2007; Pérez-Donoso et al., 2010). The observation that PcPGIP is present in xylem exudates of non-transgenic scions grafted on transgenic rootstocks expressing PcPGIP suggests that grafting of non transgenic varieties on transgenic rootstocks represents, in this case, a useful agronomical practice for plant protection (Agüero et al., 2005).
The results obtained with X. fastidiosa prompted further investigations on the capability of PGIP of controlling bacterial diseases (summarized in Table 4). Transgenic tobacco plants expressing B. rapa BrPGIP2 were resistant against Pectobacterium carotovorum, the causal agent of the soft rot disease, with a strong reduction (66–88%) of the symptoms as compared to wild-type plants (Hwang et al., 2010). The resistance correlated with the inhibitory activity against P. carotovorum PG activity found in the total protein extracts of the transgenic plants (Hwang et al., 2010). Also chinese cabbage (B. rapa ssp. pekinensis) plants overexpressing BrPGIP2 showed higher resistance against P. carotovorum and produced normal looking pods-like structures with no viable seeds. Combination of crossing with non-transgenic plants did not restore fertility of the transgenic plants, suggesting that mechanisms such as ploidy changes occurring during the tissue culture stage or changes in cell-wall architecture of sexual organs are responsible for the abnormality (Hwang et al., 2010).
No phenotypic abnormalities were, instead, found in transgenic tobacco plants expressing BrPGIP2 (Hwang et al., 2010), nor in rapeseed plants overexpressing the B. napus Bnpgip2. The latter plants displayed a significant reduction of rot caused by the necrotrophic fungal pathogen S. sclerotiorum (HuangFu et al., 2014).
Additional PGIP-transgenic crops include pea (Pisum sativum L.), transformed with Ripgip from raspberry (Rubus idaeus L.) (Richter et al., 2006), persimmon (Diospyros kaki L.) and apple (Malus domestica Borkh.) transformed with pear PcPGIP (Szankowski et al., 2003; Tamura et al., 2004), sugarbeet (Beta vulgaris L.) transformed with bean Pvpgip2 (Mohammadzadeh et al., 2012), chickpea transformed with either Ripgip or a pgip gene from kiwi fruit (Senthil et al., 2004), tobacco transformed with PpPGIP gene from Pyrus pyrifolia Nakai (Liu et al., 2013) and maize (Zea mays L.) transformed with bean Pvpgip1 (O'Kennedy et al., 2001). The response of these plants to pathogens has not been reported yet. Transgenic pea plants expressing RiPGIP were instead evaluated for their response to beneficial microorganisms. Glomus intraradices, an arbuscular mycorrhizal fungus, colonized roots of transgenic plants at an extend comparable to that observed in control non transgenic plants, indicating that the expression of RiPGIP does not affect mycorrhization (Hassan et al., 2012).
PGIPs engineered in monocot crops
Although the low pectin content of cereal species like wheat and rice indicates that this cell wall component may have a marginal role during infection, results show that the expression of PGIP in transgenic plants limits some diseases caused by fungal pathogens (Janni et al., 2008; Ferrari et al., 2012; Wang et al., 2014a,b). In our labs, the bean Pvpgip2 gene was used under the constitutive promoter of the maize unbiquitin gene (Ubi-1) to transform both durum and bread wheat by particle bombardment. PvPGIP2 was correctly targeted to the apoplast and the transgenic plants did not show any major morphological and growth defects. Transgenic wheat showed a significant reduction (46–50%) of foliar spot blotch symptoms caused by the hemibiotrophic fungal pathogen Bipolaris sorokiniana and improved resistance (25–30%) against the hemibiotrophic fungal pathogen F. graminearum (Ferrari et al., 2012), the major causal agent of FHB in wheat. A reduced degradability of the transgenic tissue by PG treatments correlated with the capacity of PvPGIP2 to inhibit PG activity of B. sorokiniana and less strongly PG of F. graminearum (Janni et al., 2008; Ferrari et al., 2012). An interesting aspect of the wheat plants expressing PvPGIP2 is that, under moderate infection with F. graminearum, the reduced FHB symptoms are concomitant with a greater amount of total starch in the grains as compared to control plants (D'Ovidio et al., 2012). On the other hand, wheat plants expressing PvPGIP2 were susceptible to the biotrophic fungal pathogen Claviceps purpurea, the causal agent of ergot disease probably because PvPGIP2 is not able to inhibit the activity of C. purpurea CpPG1 and CpPG2 (Volpi et al., 2013). Recently, transgenic wheat expressing the soybean GmPGIP3 was shown to be resistant to both take-all and common root rot diseases caused by the fungal pathogen Gaeumannomyces graminis var. tritici and B. sorokiniana, respectively; symptoms were reduced of about 47–83% and 42–60%, respectively (Wang et al., 2014a). Similarly, the expression of OsPGIP1 in transgenic rice enhanced resistence against Rhizoctonia solani in field tests and resistance was related with the expression levels of OsPGIP1 (Wang et al., 2014b).
Concluding remarks and future challenges
The results reported in this review clearly indicate that PGIP is useful to improve resistance in different crop species. High-level expression of PGIP does not prevent infection but limits significantly the colonization of the host tissue with a consequent positive impact on crop yield and product quality. The efficacy of PGIP to control diseases has been demonstrated against fungi, oomycetes and bacteria and is equally efficient against necrorophic and hemibiotrophic pathogens. The experiments performed with biotrophs do not allow to draw any clear conclusion since the only fungal biotrophic pathogen analyzed, C. purpures, produced PG activity that was not inhibited by the PGIP expressed in the transgenic plants (Volpi et al., 2013). The identification and development of PGIPs with stronger and broader inhibitory capacities may be useful to utilize these proteins in crop protection. Germplasm analysis to identify novel PGIPs is still limited (Farina et al., 2009) and the initial attempts to drive in vitro evolution of PGIP to generate proteins with improved inhibition properties have not been particularly successful (Benedetti et al., 2011a). Structural studies should be implemented in order to obtain a detailed map of the contacts between various PGs and PGIPs. This is necessary not only for constructing novel inhibitors with stronger activities but also for future programs of genome editing in which the existing genes of a plant species may be ameliorated to better adapt to new virulent strains of microorganisms evolving in nature.
The available results support the notion that inhibition of the microbial PG by PGIP is a prerequisite of the inhibitors to confer resistance to transgenic plants against microbes. The delay of symptoms is often related to the capacity of PGIP to inhibit the PG activity secreted by the pathogens and, consequently, to reduce both tissue maceration and favor the release of OGs, as summarized in Figure 2. However, this aspect of the PGIP's biology needs further investigation. In some cases PGIP has been reported to confer resistance without any evidence of PG-inhibition in vitro (Joubert et al., 2006). Moreover, some evidence suggests that the capability of reducing tissue maceration is associated with the property of PGIP to bind pectin, likely shielding this component of the cell wall from PG activity (Spadoni et al., 2006). In this regard the observation that transgenic plants expressing PGIPs exhibit an altered gene expression and cell wall composition is also intriguing. It is not yet clear the mechanism that links the ectopic expression of PGIP to alteration of gene expression and whether this contributes to disease resistance (Alexandersson et al., 2011; Nguema-Ona et al., 2013).
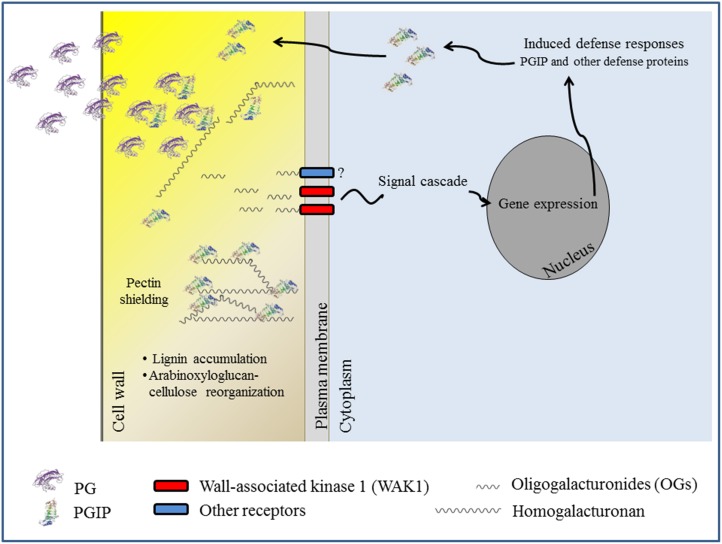
Figure 2.
A model for the role of PGIP in the defense response against pathogens. Delay of symptoms is related to the inhibitory activity of PGIP toward PGs secreted by the pathogens and likely to the accumulation of oligogalacturonide (OG) elicitors, which are recognized by WAK1 and likely other receptors not yet characterized. Cell wall modification and pectin shielding could also play a role. Signaling cascades activated by OGs are described in Ferrari et al. (2013).
An important but very little explored aspect of the PGIP biology is its possible role in processes of growth and development. Although plants overexpressing PGIPs do not show obvious morphological alterations, indeed several reports point to PGIP as a player in development. PGIP are induced, not only by phosphate deficiency, but also by auxin treatment and in mutants defective in SIZ1, a SUMO (small ubiquitin-related modifier) E3 ligase that is involved in several stress responses, including Pi starvation, and flowering (Sato and Miura, 2011). Suppression of PGIPs under the control ABA insensitive 5 (ABI5) transcription factor accompanies promotion of seed germination by the peroxisomal ABC transporter PED3 (Kanai et al., 2010). Upregulation of PGIP2 correlates with the acquisition of competence to form green callus in an auxin-rich callus induction medium (Che et al., 2007) and occurs in Arabidopsis tissue culture lines in which the expression of the peroxidases PRX33 and PRX34 is knocked down by antisense expression (O'Brien et al., 2012), whereas PGIP1 was identified in a proteomic study performed on Arabidopsis etiolated hypocotyls used as a model of cells undergoing elongation followed by growth arrest within a short time (Irshad et al., 2008). Finally, both PGIP1 and PGIP2 are associated with cell wall stabilization at low pH under the control of the zinc-finger protein STOP1 (Sensitive to Proton Rhizotoxicity 1) and STOP2 (Kobayashi et al., 2014). A role of PGIP not only in defense but also in growth and development implies that the inhibitor may affect one or more of the many endogenous PGs expressed by plants. This is also an unexplored aspect of the PGIP biology and, at the moment, only one very old evidence is available showing that PGIP may have a plant-derived PG partner (Cervone et al., 1990).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Italian Ministry of University and Research (PRIN 2010–2011) to RD.
References
- Agüero C. B., Uratsu S. L., Greve C., Powell A. L. T., Labavitch J. M., Meredith C. P., et al. (2005). Evaluation of tolerance to Pierce's disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 6, 43–51. 10.1111/j.1364-3703.2004.00262.x [DOI] [PubMed] [Google Scholar]
- Akhgari A., Motallebi B., Zamani M. (2012). Bean polygalacturonase-inhibiting protein expressed in transgenic Brassica napus inhibits polygalacturonase from its fungal pathogen Rhizoctonia solani. Plant Prot. Sci. 48, 1–9. [Google Scholar]
- Albersheim P., Anderson A. J. (1971). Proteins from plant cell walls inhibit polygalacturonases secreted by plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 68, 1815–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersson E., Becker J. V. W., Jacobson D., Nguema-Ona E., Steyn C., Denby K. J., et al. (2011). Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res. Notes 4:493. 10.1186/1756-0500-4-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibanez J. T., Sozzani R., Lee T.-J., Chu T.-M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. 10.1104/pp.108.121038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashi Z. D., Rimmer S. R., Khachatourians G. G., Hegedus D. D. (2013). Brassica napus polygalacturonase inhibitor proteins inhibit Sclerotinia sclerotiorum polygalacturonase enzymatic and necrotizing activities and delay symptoms in transgenic plants. Can. J. Microbiol. 59, 79–86. 10.1139/cjm-2012-0352 [DOI] [PubMed] [Google Scholar]
- Benedetti M., Andreani F., Leggio C., Galantini L., Di Matteo A., Pavel N. V., et al. (2013). A single amino-acid substitution allows endo-polygalacturonase of Fusarium verticillioides to acquire recognition by PGIP2 from Phaseolus vulgaris. PLoS ONE 8: e80610. 10.1371/journal.pone.0080610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti M., Bastianelli E., Salvi G., De Lorenzo G., Caprari C. (2011a). Artificial evolution corrects a repulsive amino acid in polygalacturonase inhibiting proteins (PGIPs). J. Plant Pathol. 93, 89–95 10.4454/jpp.v93i1.277 [DOI] [Google Scholar]
- Benedetti M., Leggio C., Federici L., De Lorenzo G., Pavel N. V., Cervone F. (2011b). Structural resolution of the complex between a fungal polygalacturonase and a plant polygalacturonase-inhibiting protein by small-angle X-ray scattering. Plant Physiol. 157, 599–607. 10.1104/pp.111.181057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger D. K., Oelofse D., Arendse M. S., Du Plessis E., Dubery I. A. (2000). Bean polygalacturonase inhibitor protein-1 (PGIP-1) inhibits polygalacturonases from Stenocarpella maydis. Physiol. Mol. Plant Pathol. 57, 5–14 10.1006/pmpp.2000.0274 [DOI] [Google Scholar]
- Bergmann C. W., Ito Y., Singer D., Albersheim P., Darvill A. G., Benhamou N., et al. (1994). Polygalacturonase-inhibiting protein accumulates in Phaseolus vulgaris L. in response to wounding, elicitors and fungal infection. Plant J. 5, 625–634. 10.1111/j.1365-313X.1994.00625.x [DOI] [PubMed] [Google Scholar]
- Bonivento D., Pontiggia D., Di Matteo A., Fernandez-Recio J., Salvi G., Tsernoglou D., et al. (2008). Crystal structure of the endopolygalacturonase from the phytopathogenic fungus Colletotrichum lupini and its interaction with polygalacturonase inhibiting proteins. Proteins 70, 294–299. 10.1002/prot.21610 [DOI] [PubMed] [Google Scholar]
- Borras-Hidalgo O., Caprari C., Hernandez-Estevez I., De Lorenzo G., Cervone F. (2012). A gene for plant protection: expression of a bean polygalacturonase inhibitor in tobacco confers a strong resistance against Rhizoctonia solani and two oomycetes. Front. Plant Sci. 3:268. 10.3389/fpls.2012.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buza N. L., Krinitsyna A. A., Protsenko M. A., Vartapetyan V. V. (2004). Role of the polygalacturonidase inhibitor Protein in the ripening of apples and their resistance to Monilia fructigena, a causative agent of fruit rot. Appl. Biochem. Microbiol. 40, 89–92. 10.1023/B:ABIM.0000010361.48129.6e [DOI] [PubMed] [Google Scholar]
- Cantu D., Vicente A. R., Labavitch J. M., Bennett A. B., Powell A. L. (2008). Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends Plant Sci. 13, 610–617. 10.1016/j.tplants.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Caprari C., Bergmann C., Migheli Q., Salvi G., Albersheim P., Darvill A., et al. (1993). Fusarium moniliforme secretes four endopolygalacturonases derived from a single gene product. Physiol. Mol. Plant Pathol. 43, 453–462 10.1006/pmpp.1993.1073 [DOI] [Google Scholar]
- Casasoli M., Federici L., Spinelli F., Di Matteo A., Vella N., Scaloni F., et al. (2009). Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proc. Natl. Acad. Sci. U.S.A. 106, 7666–7671. 10.1073/pnas.0812625106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervone F., De Lorenzo G., Degrà L., Salvi G., Bergami M. (1987). Purification and characterization of a polygalacturonase-inhibiting protein from Phaseolus vulgaris L. Plant Physiol. 85, 631–637. 10.1104/pp.85.3.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervone F., De Lorenzo G., Pressey R., Darvill A. G., Albersheim P. (1990). Can Phaseolus PGIP inhibit pectic enzymes from microbes and plants? Phytochemistry 29, 447–449 10.1016/0031-9422(90)85094-V [DOI] [Google Scholar]
- Cervone F., Hahn M. G., De Lorenzo G., Darvill A., Albersheim P. (1989). Host-pathogen interactions. XXXIII. A plant protein converts a fungal pathogenesis factor into an elicitor of plant defense responses. Plant Physiol. 90, 542–548. 10.1104/pp.90.2.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Lall S., Howell S. H. (2007). Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226, 1183–1194. 10.1007/s00425-007-0565-4 [DOI] [PubMed] [Google Scholar]
- Chen X., Liu X., Zuo S., Ma Y., Tong Y., Pan X., et al. (2011). Prokaryotic expression of rice Ospgip1 gene and bioinformatic analysis of encoded product. Rice Sci. 18, 250–256 10.1016/S1672-6308(12)60002-X [DOI] [Google Scholar]
- D'hallewin G., Schirra M., Powell A. L., Greve L. C., Labavitch J. M. (2004). Properties of a polygalacturonase-inhibiting protein isolated from 'Oroblanco' grapefruit. Physiol. Plant. 120, 395–404. 10.1111/j.0031-9317.2004.00264.x [DOI] [PubMed] [Google Scholar]
- D'Ovidio R., Laino P., Janni M., Botticella E., Di Carli M. S., Benvenuto E., et al. (2012). Proteomic analysis of mature kernels of Fusarium graminearum-infected transgenic bread wheat expressing PGIP, in Proceedings of 11th International Gluten Workshop, eds He Z., Wang D. (Beijing: ), 18. [Google Scholar]
- D'Ovidio R., Mattei B., Roberti S., Bellincampi D. (2004a). Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim. Biophys. Acta 1696, 237–244. 10.1016/j.bbapap.2003.08.012 [DOI] [PubMed] [Google Scholar]
- D'Ovidio R., Raiola A., Capodicasa C., Devoto A., Pontiggia D., Roberti S., et al. (2004b). Characterization of the complex locus of bean encoding polygalacturonase-inhibiting proteins reveals subfunctionalization for defense against fungi and insects. Plant Physiol. 135, 2424–2435. 10.1104/pp.104.044644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ovidio R., Roberti S., Giovanni M. D., Capodicasa C., Melaragni M., Sella L., et al. (2006). The characterization of the soybean polygalacturonase-inhibiting proteins (Pgip) gene family reveals that a single member is responsible for the activity detected in soybean tissues. Planta 224, 633–645. 10.1007/s00425-006-0235-y [DOI] [PubMed] [Google Scholar]
- De Lorenzo G., D'Ovidio R., Cervone F. (2001). The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 39, 313–335. 10.1146/annurev.phyto.39.1.313 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G., Ferrari S. (2002). Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 5, 295–299. 10.1016/S1369-5266(02)00271-6 [DOI] [PubMed] [Google Scholar]
- Deo A., Shastri N. V. (2003). Purification and characterization of polygalacturonase-inhibitory proteins from Psidium guajava Linn. (guava) fruit. Plant Sci. 164, 147–156 10.1016/S0168-9452(02)00337-0 [DOI] [Google Scholar]
- Desiderio A., Aracri B., Leckie F., Mattei B., Salvi G., Tigelaar H., et al. (1997). Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol. Plant Microbe Interact. 10, 852–860. 10.1094/MPMI.1997.10.7.852 [DOI] [PubMed] [Google Scholar]
- Devoto A., Clark A. J., Nuss L., Cervone F., De Lorenzo G. (1997). Developmental and pathogen-induced accumulation of transcripts of polygalacturonase-inhibiting protein in Phaseolus vulgaris L. Planta 202, 284–292 10.1007/s004250050130 [DOI] [Google Scholar]
- Di C., Li M., Long F., Bai M., Zheng X., Xu S., et al. (2009). Molecular cloning, functional analysis and localization of a novel gene encoding polygalacturonase-inhibiting protein in Chorispora bungeana. Planta 231, 169–178. 10.1007/s00425-009-1039-7 [DOI] [PubMed] [Google Scholar]
- Di C. X., Zhang H., Sun Z. L., Jia H. L., Yang L. N., Si J., et al. (2012). Spatial distribution of polygalacturonase-inhibiting proteins in Arabidopsis and their expression induced by Stemphylium solani infection. Gene 506, 150–155. 10.1016/j.gene.2012.06.085 [DOI] [PubMed] [Google Scholar]
- Di Giovanni M., Cenci A., Janni M., D'Ovidio R. (2008). A LTR copia retrotransposon and Mutator transposons interrupt pgip genes in cultivated and wild wheats. Theor. Appl. Genet. 116, 859–867. 10.1007/s00122-008-0719-1 [DOI] [PubMed] [Google Scholar]
- Di Matteo A., Bonivento D., Tsernoglou D., Federici L., Cervone F. (2006). Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry 67, 528–533. 10.1016/j.phytochem.2005.12.025 [DOI] [PubMed] [Google Scholar]
- Di Matteo A., Federici L., Mattei B., Salvi G., Johnson K. A., Savino C., et al. (2003). The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc. Natl. Acad. Sci. U.S.A. 100, 10124–10128. 10.1073/pnas.1733690100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doostdar H., McCollum T. G., Mayer R. T. (1997). Purification and characterization of an endo-polygalacturonase from the gut of West Indies sugarcane rootstalk borer weevil (Diaprepes abbreviatus L.) larvae. Comp. Biochem. Physiol. Part B Biochem. Mol. Bio. 118, 861–867. [Google Scholar]
- Faize M., Sugiyama T., Faize L., Ishii H. (2003). Polygalacturonase-inhibiting protein (PGIP) from Japanese pear: possible involvement in resistance against scab. Physiol. Mol. Plant Pathol. 63, 319–327 10.1016/j.pmpp.2004.03.006 [DOI] [Google Scholar]
- Farina A., Rocchi V., Janni M., Benedettelli S., De Lorenzo G., D'Ovidio R. (2009). The bean polygalacturonase-inhibiting protein 2 (PvPGIP2) is highly conserved in common bean (Phaseolus vulgaris L.) germplasm and related species. Theor. Appl. Genet. 118, 1371–1379. 10.1007/s00122-009-0987-4 [DOI] [PubMed] [Google Scholar]
- Federici L., Caprari C., Mattei B., Savino C., Di Matteo A., De Lorenzo G., et al. (2001). Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc. Natl. Acad. Sci. U.S.A. 98, 13425–13430. 10.1073/pnas.231473698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici L., Di Matteo A., Fernandez-Recio J., Tsernoglou D., Cervone F. (2006). Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 11, 65–70. 10.1016/j.tplants.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Galletti R., Vairo D., Cervone F., De Lorenzo G. (2006). Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol. Plant Microbe Interact. 19, 931–936. 10.1094/MPMI-19-0931 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Savatin D. V., Sicilia F., Gramegna G., Cervone F., De Lorenzo G. (2013). Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4:49. 10.3389/fpls.2013.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Sella L., Janni M., De Lorenzo G., Favaron F., D'Ovidio R. (2012). Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. 14, 31–38. 10.1111/j.1438-8677.2011.00449.x [DOI] [PubMed] [Google Scholar]
- Ferrari S., Vairo D., Ausubel F. M., Cervone F., De Lorenzo G. (2003). Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell Online 15, 93–106 10.1105/tpc.005165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish W. W., Davis A. R. (2004). The purification, physical/chemical characterization, and cDNA sequence of cantaloupe fruit polygalacturonase-inhibiting protein. Phytopathology 94, 337–344. 10.1094/PHYTO.2004.94.4.337 [DOI] [PubMed] [Google Scholar]
- Frati F., Galletti R., De Lorenzo G., Salarino G., Conti E. (2006). Activity of endo-polygalacturonases in mirid bugs (Heteroptera: Miridae) and their inhibition by plant cell wall proteins (PGIPs). Eur. J. Entomol. 103, 515–522 10.14411/eje.2006.067 [DOI] [Google Scholar]
- Gazendam I., Oelofse D., Berger D. K. (2004). High-level expression of apple PGIP1 is not sufficient to protect transgenic potato against Verticillium dahliae. Physiol. Mol. Plant Pathol. 65, 145–155 10.1016/j.pmpp.2005.01.002 [DOI] [Google Scholar]
- Gomathi V., Gnanamanickam S. S. (2004). Polygalacturonase-inhibiting proteins in plant defence. Curr. Sci. 87, 1211–1217. [Google Scholar]
- Gregori R., Mari M., Bertolini P., Barajas J. A., Tian J. B., Labavitch J. M. (2008). Reduction of Colletotrichum acutatum infection by a polygalacturonase inhibitor protein extracted from apple. Postharvest. Biol. Technol. 48, 309–313 10.1016/j.postharvbio.2007.10.006 [DOI] [Google Scholar]
- Guo J.-L., Zhu Y.-P., Shi K., Jue D.-W., Liu S.-P., Hong Y.-B., et al. (2014). Over-expression of Solanum torvum PGIP enhances resistance to Verticillium dahliae in transgenic potato plants. Bothalia Pretoria. 44, 392–404. [Google Scholar]
- Gutierrez-Sanchez G., King D., Kemp G., Bergmann C. (2012). SPR and differential proteolysis/MS provide further insight into the interaction between PGIP2 and EPGs. Fungal Biol. 116, 737–746. 10.1016/j.funbio.2012.04.010 [DOI] [PubMed] [Google Scholar]
- Hassan F., Noorian M. S., Jacobsen H. J. (2012). Effect of antifungal genes expressed in transgenic pea (Pisum sativum L.) on root colonization with Glomus intraradices. GM Crops Food 3, 301–309. 10.4161/gmcr.21897 [DOI] [PubMed] [Google Scholar]
- Hegedus D. D., Li R., Buchwaldt L., Parkin I., Whitwill S., Coutu C., et al. (2008). Brassica napus possesses an expanded set of polygalacturonase inhibitor protein genes that are differentially regulated in response to Sclerotinia sclerotiorum infection, wounding and defense hormone treatment. Planta 228, 241–253. 10.1007/s00425-008-0733-1 [DOI] [PubMed] [Google Scholar]
- Helft L., Reddy V., Chen X., Koller T., Federici L., Recio J. F., et al. (2011). LRR Conservation mapping to predict functional sites within protein leucine-rich repeat domains. PLoS ONE 6:e21614. 10.1371/journal.pone.0021614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W., Mu J., Li A., Wang H., Kong L. (2014). Identification of a wheat polygalacturonase-inhibiting protein involved in Fusarium head blight resistance. Eur. J. Plant Pathol. 141, 731–745 10.1007/s10658-014-0574-7 [DOI] [Google Scholar]
- HuangFu H., Guan C., Jin F., Yin C. (2014). Prokaryotic expression and protein function of Brassica napus PGIP2 and its genetic transformation. Plant Biotechnol. Rep. 8, 171–181 10.1007/s11816-013-0307-y [DOI] [Google Scholar]
- Hwang B. H., Bae H., Lim H. S., Kim K. B., Kim S. J., Im, et al. (2010). Overexpression of polygalacturonase-inhibiting protein 2 (PGIP2) of Chinese cabbage (Brassica rapa ssp. pekinensis) increased resistance to the bacterial pathogen Pectobacterium carotovorum ssp. carotovorum. Plant Cell Tiss. Organ Cult. 103, 293–305 10.1007/s11240-010-9779-4 [DOI] [Google Scholar]
- Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. (2008). A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 8:94. 10.1186/1471-2229-8-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J. T., Dubery I. A. (2001). Inhibition of polygalacturonase from Verticillium dahliae by a polygalacturonase inhibiting protein from cotton. Phytochemistry 57, 149–156. 10.1016/S0031-9422(01)00024-3 [DOI] [PubMed] [Google Scholar]
- Jang S., Lee B., Kim C., Kim S. J., Yim J., Han J. J., et al. (2003). The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Mol. Biol. 53, 357–372. 10.1023/B:PLAN.0000006940.89955.f1 [DOI] [PubMed] [Google Scholar]
- Janni M., Bozzini T., Moscetti I., Volpi C., D'Ovidio R. (2013). Functional characterisation of wheat Pgip genes reveals their involvement in the local response to wounding. Plant Biol. Stuttg. Ger. 15, 1019–1024. 10.1111/plb.12002 [DOI] [PubMed] [Google Scholar]
- Janni M., Giovanni M., Roberti S., Capodicasa C., D'Ovidio R. (2006). Characterization of expressed Pgip genes in rice and wheat reveals similar extent of sequence variation to dicot PGIPs and identifies an active PGIP lacking an entire LRR repeat. Theor. Appl. Genet. 113, 1233–1245. 10.1007/s00122-006-0378-z [DOI] [PubMed] [Google Scholar]
- Janni M., Sella L., Favaron F., Blechl A. E., De Lorenzo G., D'Ovidio R. (2008). The expression of a bean PGIP in transgenic wheat confers increased resistance to the fungal pathogen Bipolaris sorokiniana. Mol. Plant Microbe Interact. 21, 171–177. 10.1094/MPMI-21-2-0171 [DOI] [PubMed] [Google Scholar]
- Johnston D. J., Ramanathan V., Williamson B. (1993). A protein from immature raspberry fruits which inhibits endopolygalacturonases from Botrytis cinerea and other micro-organisms. J. Exp. Bot. 44, 971–976 10.1093/jxb/44.5.971 [DOI] [Google Scholar]
- Joubert D. A., Kars I., Wagemakers L., Bergmann C., Kemp G., Vivier M. A., et al. (2007). A polygalacturonase-inhibiting protein from grapevine reduces the symptoms of the endopolygalacturonase BcPG2 from Botrytis cinerea in Nicotiana benthamiana leaves without any evidence for in vitro interaction. Mol. Plant Microbe Interact. 20, 392–402. 10.1094/MPMI-20-4-0392 [DOI] [PubMed] [Google Scholar]
- Joubert D. A., Slaughter A. R., Kemp G., Becker J. V. W., Krooshof G. H., Bergmann C., et al. (2006). The grapevine polygalacturonase-inhibiting protein (VvPGIP1) reduces Botrytis cinerea susceptibility in transgenic tobacco and differentially inhibits fungal polygalacturonases. Transgenic Res. 15, 687–702. 10.1007/s11248-006-9019-1 [DOI] [PubMed] [Google Scholar]
- Kalunke R. M., Cenci A., Volpi C., O'Sullivan D. M., Sella L., Favaron F., et al. (2014). The pgip family in soybean and three other legume species: evidence for a birth-and-death model of evolution. BMC Plant Biol. 14:189. 10.1186/s12870-014-0189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalunke R. M., Janni M., Sella L., David P., Geffroy V., Favaron F., et al. (2011). Transcript analysis of the bean polygalacturonase inhibiting protein gene family reveals that Pvpgip2 is expressed in the whole plant and is strongly induced by pathogen infection. J. Plant Pathol. 93, 141–148 10.4454/jpp.v93i1.284 [DOI] [Google Scholar]
- Kanai M., Nishimura M., Hayashi M. (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 62, 936–947. 10.1111/j.1365-313X.2010.04205.x [DOI] [PubMed] [Google Scholar]
- Kemp G., Bergmann C. W., Clay R., Van der Westhuizen A. J., Pretorius Z. A. (2003). Isolation of a polygalacturonase-inhibiting protein (PGIP) from wheat. Mol. Plant Microbe Interact. 16, 955–961. 10.1094/MPMI.2003.16.11.955 [DOI] [PubMed] [Google Scholar]
- King D., Bergmann C., Orlando R., Benen J. A., Kester H. C., Visser J. (2002). Use of amide exchange mass spectrometry to study conformational changes within the endopolygalacturonase II-homogalacturonan-polygalacturonase inhibiting protein system. Biochemistry 41, 10225–10233. 10.1021/bi020119f [DOI] [PubMed] [Google Scholar]
- Kirsch R., Wielsch N., Vogel H., Svatoš A., Heckel D. G., Pauchet Y. (2012). Combining proteomics and transcriptome sequencing to identify active plant-cell-wall-degrading enzymes in a leaf beetle. BMC Genomics 13:587. 10.1186/1471-2164-13-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Ohyama Y., Kobayashi Y., Ito H., Iuchi S., Fujita M., et al. (2014). STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 7, 311–322. 10.1093/mp/sst116 [DOI] [PubMed] [Google Scholar]
- Ladu G., Pani G., Venditti T., Dore A., Molinu M. G., D'Hallewin G. (2012). Natural resistance against pre- and post-harvest pathogens in Sardinian pears germoplasm. Commun. Agric. Appl. Biol. Sci. 77, 163–171. [PubMed] [Google Scholar]
- Lagaert S., Beliën T., Volckaert G. (2009). Plant cell walls: protecting the barrier from degradation by microbial enzymes, Semin. Cell Dev. Biol. 20, 1064–1073. 10.1016/j.semcdb.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Leckie F., Mattei B., Capodicasa C., Hemmings A., Nuss L., Aracri B., et al. (1999). The specificity of polygalacturonaseinhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed b-strand/b-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J. 18: 2352–2363 10.1093/emboj/18.9.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. D., Bae H., Kang I. K., Byun J. K., Kang S. G. (2006). Characterization of an apple polygalacturonase-inhibiting protein (PGIP) that specifically inhibits an endopolygalacturonase (PG) purified from apple fruits infected with Botryosphaeria dothidea. J. Microbio. Biotech. 16, 1192–1200. [Google Scholar]
- Lim J. M., Aoki K., Angel P., Garrison D., King D., Tiemeyer M., et al. (2009). Mapping glycans onto specific N-linked glycosylation sites of Pyrus communis PGIP redefines the interface for EPG-PGIP interactions. J. Proteome Res. 8, 673–680. 10.1021/pr800855f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Li Z. (2002). The partial structure of wheat polygalacturonase inhibiting protein. Chin. J. Biochem. Mol. Bio. 18, 197–201. [Google Scholar]
- Liu D., Li W., He X., Ding Y., Chen C., Ge F. (2013). Characterization and functional analysis of a novel PGIP gene from Pyrus pyrifolia Nakai cv Huobali. Acta Physiol. Plant. 35, 1247–1256 10.1007/s11738-012-1164-y [DOI] [Google Scholar]
- Lu L., Zhou F., Zhou Y., Fan X., Ye S., Wang L., et al. (2012). Expression profile analysis of the polygalacturonase-inhibiting protein genes in rice and their responses to phytohormones and fungal infection. Plant Cell Rep. 31, 1173–1187. 10.1007/s00299-012-1239-7 [DOI] [PubMed] [Google Scholar]
- Machinandiarena M. F., Olivieri F. P., Daleo G. R., Oliva C. R. (2001). Isolation and characterization of a polygalacturonase-inhibiting protein from potato leaves. Accumulation in response tosalicylic acid, wounding and infection. Plant Physiol. Biochem. 39, 129–136 10.1016/S0981-9428(00)01228-6 [DOI] [Google Scholar]
- Manfredini C., Sicilia F., Ferrari S., Pontiggia D., Salvi G., Caprari C., et al. (2005). Polygalacturonase-inhibiting protein 2 of Phaseolus vulgaris inhibits BcPG1, a polygalacturonase of Botrytis cinerea important for pathogenicity, and protects transgenic plants from infection. Physiol. Mol. Plant Pathol. 67, 108–115 10.1016/j.pmpp.2005.10.002 [DOI] [Google Scholar]
- Mariotti L., Casasoli M., Migheli Q., Balmas V., Caprari C., De Lorenzo G. (2008). Reclassification of Fusarium verticillioides (syn. F. moniliforme) strain FC-10 as F. phyllophilum. Mycol. Res. 112, 1010–1011. 10.1016/j.mycres.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Mattei B., Bernalda M. S., Federici L., Roepstorff P., Cervone F., Boffi A. (2001). Secondary structure and post-translational modifications of the leucine-rich repeat protein PGIP (polygalacturonase-inhibiting protein) from Phaseolus vulgaris. Biochemistry 40, 569–576. 10.1021/bi0017632 [DOI] [PubMed] [Google Scholar]
- Maulik A., Ghosh H., Basu S. (2009). Comparative study of protein-protein interaction observed in Polygalacturonase-inhibiting proteins from Phaseolus vulgaris and Glycine max and Polygalacturonase from Fusarium moniliforme. BMC Genomics 10:S19. 10.1186/1471-2164-10-S3-S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Meyers B. C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Misas-Villamil J. C., van der Hoorn R. A. (2008). Enzyme-inhibitor interactions at the plant-pathogen interface. Curr. Opin. Plant Biol. 11, 380–388. 10.1016/j.pbi.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Mohammadzadeh R., Zamani M., Motallebi M., Norouzi P., Jourabchi E., Benedetti M., et al. (2012). Agrobacterium tumefaciens-mediated introduction of polygalacturonase inhibiting protein 2 gene (PvPGIP2) from Phaseolus vulgaris into sugar beet (Beta vulgaris L.). Aust. J. Crop Sci. 6, 1290–1297. [Google Scholar]
- Nguema-Ona E., Moore J. P., Fagerström A. D., Fangel J. U., Willats W. G., Hugo A., et al. (2013). Overexpression of the grapevine PGIP1 in tobacco results in compositional changes in the leaf arabinoxyloglucan network in the absence of fungal infection. BMC Plant Biol. 13:46. 10.1186/1471-2229-13-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietlispach D., Mott H. R., Stott K. M., Nielsen P. R., Thiru A., Laue E. D. (2004). Structure determination of protein complexes by NMR. Methods Mol. Biol. 278, 255–288. 10.1385/1-59259-809-9:255 [DOI] [PubMed] [Google Scholar]
- Nuss L., Mahe A., Clark A. J., Grisvard J., Dron M., Cervone F., et al. (1996). Differential accumulation of PGIP (polygalacturonase inhibiting protein) mRNA in two near-isogenic lines of Phaseolus vulgaris L. upon infection with Colletotrichum lindemuthianum. Physiol. Mol. Plant Pathol. 48, 83–89. [Google Scholar]
- O'Kennedy M., Burger J., Berger D. (2001). Transformation of elite white maize using the particle inflow gun and detailed analysis of a low-copy integration event. Plant Cell Rep. 20, 721–730 10.1007/s002990100383 [DOI] [Google Scholar]
- O'Brien J. A., Daudi A., Finch P., Butt V. S., Whitelegge J. P., Souda P., et al. (2012). A peroxidase-dependent apoplastic oxidative burst in cultured arabidopsis cells functions in MAMP-elicited defence. Plant Physiol. 158, 2013–2027. 10.1104/pp.111.190140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelofse D., Dubery I. A., Meyer R., Arendse M. S., Gazendam I., Berger D. K. (2006). Apple polygalacturonase inhibiting protein1 expressed in transgenic tobacco inhibits polygalacturonases from fungal pathogens of apple and the anthracnose pathogen of lupins. Phytochemistry 67, 255–263. 10.1016/j.phytochem.2005.10.029 [DOI] [PubMed] [Google Scholar]
- Oh D.-H., Dassanayake M., Bohnert H. J., Cheeseman J. M. (2012). Life at the extreme: lessons from the genome. Genome Biol. 13, 241. 10.1186/gb-2012-13-3-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. B., Nascimento L. B., Junior M. L., Petrofeza S. (2010). Characterization of the dry bean polygalacturonase-inhibiting protein (PGIP) gene family during Sclerotinia sclerotiorum (Sclerotiniaceae) infection. Genet. Mol. Res. 9, 994–1004. 10.4238/vol9-2gmr776 [DOI] [PubMed] [Google Scholar]
- Pérez-Donoso A. G., Sun Q., Roper M. C., Greve L. C., Kirkpatrick B., Labavitch J. M. (2010). Cell wall-degrading enzymes enlarge the pore size of intervessel pit membranes in healthy and Xylella fastidiosa-infected grapevines. Plant Physiol. 152, 1748–1759. 10.1104/pp.109.148791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A. L. T., Stotz H. U., Labavitch J. M., Bennett A. B. (1994). Glycoprotein inhibitors of fungal polygalacturonases, in Advances in Molecular Genetics of Plant-Microbe Interactions, Current Plant Science and Biotechnology in Agriculture, eds Daniels M. J., Downie J. A., Osbourn A. E. (Dordrecht: Springer; ), 399–402. [Google Scholar]
- Powell A. L. T., van Kan J., ten Have A., Visser J., Greve L. C., Bennett A. B., et al. (2000). Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol. Plant Microbe Interact. 13, 942–950. 10.1094/MPMI.2000.13.9.942 [DOI] [PubMed] [Google Scholar]
- Prabhu S. A., Kini K. R., Raj S. N., Moerschbacher B. M., Shetty H. S. (2012). Polygalacturonase-inhibitor proteins in pearl millet: possible involvement in resistance against downy mildew. Acta Biochim. Biophys. Sinica. 44, 415–423. 10.1093/abbs/gms015 [DOI] [PubMed] [Google Scholar]
- Prabhu S. A., Singh R., Kolkenbrock S., Sujeeth N., El Gueddari N. E., Moerschbacher B. M., et al. (2014). Experimental and bioinformatic characterization of recombinant polygalacturonase-inhibitor protein from pearl millet and its interaction with fungal polygalacturonases. J. Exp. Bot. 65, 5033–5047. 10.1093/jxb/eru266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S. A., Wagenknecht M., Melvin P., Gnanesh Kumar B. S., Veena M., Shailasree S., et al. (2015). Immuno-affinity purification of PglPGIP1, a polygalacturonase-inhibitor protein from pearl millet: studies on its inhibition of fungal polygalacturonases and role in resistance against the downy mildew pathogen. Mol. Biol. Rep. [Epub ahead of print]. 10.1007/s11033-015-3850-5 [DOI] [PubMed] [Google Scholar]
- Protsenko M. A., Buza N. L., Krinitsyna A. A., Bulantseva E. A., Korableva N. P. (2008). Polygalacturonase-inhibiting protein is a structural component of plant cell wall. Biochem. (Moscow) 73, 1053–1062 10.1134/S0006297908100015 [DOI] [PubMed] [Google Scholar]
- Raiola A., Sella L., Castiglioni C., Balmas V., Favaron F. (2008). A single amino acid substitution in highly similar endo-PGs from Fusarium verticillioides and related Fusarium species affects PGIP inhibition. Fungal Genet. Biol. 45, 776–789. 10.1016/j.fgb.2007.11.003 [DOI] [PubMed] [Google Scholar]
- Reignault P., Valette-Collet O., Boccara M. (2008). The importance of fungal pectinolytic enzymes in plant invasion, host adaptability and symptom type. Eur. J. Plant Pathol. 120, 1–11 10.1007/s10658-007-9184-y [DOI] [Google Scholar]
- Richter A., Jacobsen H. J., De Kathen A., De Lorenzo G., Briviba K., Hain R., et al. (2006). Transgenic peas (Pisum sativum) expressing polygalacturonase inhibiting protein from raspberry (Rubus idaeus) and stilbene synthase from grape (Vitis vinifera). Plant Cell Rep. 25, 1166–1173. 10.1007/s00299-006-0172-z [DOI] [PubMed] [Google Scholar]
- Ridley B. L., O'Neill M. A., Mohnen D. (2001). Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967. 10.1016/S0031-9422(01)00113-3 [DOI] [PubMed] [Google Scholar]
- Roper M. C., Greve L. C., Warren J. G., Labavitch J. M., Kirkpatrick B. C. (2007). Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol. Plant Microbe Interact. 20, 411–419. 10.1094/MPMI-20-4-0411 [DOI] [PubMed] [Google Scholar]
- Sathiyaraj G., Srinivasan S., Subramanium S., Kim Y. J., Kim Y. J., Kwon W. S., et al. (2010). Polygalacturonase inhibiting protein: isolation, developmental regulation and pathogen related expression in Panax ginseng C.A. Meyer. Mol. Biol. Rep. 37, 3445–3454. 10.1007/s11033-009-9936-1 [DOI] [PubMed] [Google Scholar]
- Sato A., Miura K. (2011). Root architecture remodeling induced by phosphate starvation. Plant Signal. Behav. 6, 1122–1126. 10.4161/psb.6.8.15752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaki Y., Iuchi S., Kobayashi Y., Kobayashi Y., Ikka T., Sakurai N., et al. (2009). STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294. 10.1104/pp.108.134700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht T., Unger C., Pich A., Wydra K. (2011). Endo- and exopolygalacturonases of Ralstonia solanacearum are inhibited by polygalacturonase-inhibiting protein (PGIP) activity in tomato stem extracts. Plant Physiol. Biochem. 49, 377–387. 10.1016/j.plaphy.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Sella L., Castiglioni C., Roberti S., D'Ovidio R., Favaron F. (2004). An endo-polygalacturonase (PG) of Fusarium moniliforme escaping inhibition by plant polygalacturonase-inhibiting proteins (PGIPs) provides new insights into the PG-PGIP interaction. FEMS Microbiol. Lett. 240, 117–124. 10.1016/j.femsle.2004.09.019 [DOI] [PubMed] [Google Scholar]
- Senthil G., Williamson B., Dinkins R. D., Ramsay G. (2004). An efficient transformation system for chickpea (Cicer arietinum L.). Plant Cell Rep. 23, 297–303. 10.1007/s00299-004-0854-3 [DOI] [PubMed] [Google Scholar]
- Shanmugam V. (2005). Role of extracytoplasmic leucine rich repeat proteins in plant defence mechanisms. Microbiol. Res. 160, 83–94. 10.1016/j.micres.2004.09.014 [DOI] [PubMed] [Google Scholar]
- Shivashankar S., Thimmareddy C., Roy T. K. (2010). Polygalacturonase inhibitor protein from fruits of anthracnose resistant and susceptible varieties of Chilli (Capsicum annuum L). Indian J. Biochem. Biophys. 47, 243–248. [PubMed] [Google Scholar]
- Sicilia F., Fernandez-Recio J., Caprari C., De Lorenzo G., Tsernoglou D., Cervone F., et al. (2005). The polygalacturonase-inhibiting protein PGIP2 of Phaseolus vulgaris has evolved a mixed mode of inhibition of endopolygalacturonase PG1 of Botrytis cinerea. Plant Physiol. 139, 1380–1388. 10.1104/pp.105.067546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K. H., Nam Y. W. (2005). Genomic organization and differential expression of two polygalacturonase-inhibiting protein genes from Medicago truncatula. J. Plant Biol. 48, 467–478 10.1007/BF03030589 [DOI] [Google Scholar]
- Spadoni S., Zabotina O., Di Matteo A., Mikkelsen J. D., Cervone F., De Lorenzo G., et al. (2006). Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 141, 557–564. 10.1104/pp.106.076950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli F., Mariotti L., Mattei B., Salvi G., Cervone F., Caprari C. (2009). Three aspartic acid residues of polygalacturonase-inhibiting protein (PGIP) from Phaseolus vulgaris are critical for inhibition of Fusarium phyllophilum PG. Plant Biol. 11, 738–743. 10.1111/j.1438-8677.2008.00175.x [DOI] [PubMed] [Google Scholar]
- Szankowski I., Briviba K., Fleschhut J., Schönherr J., Jacobsen H. J., Kiesecker H. (2003). Transformation of apple (Malus domestica Borkh.) with the stilbene synthase gene from grapevine (Vitis vinifera L.) and a PGIP gene from kiwi (Actinidia deliciosa). Plant Cell Rep. 22, 141–149. 10.1007/s00299-003-0668-8 [DOI] [PubMed] [Google Scholar]
- Tamburino R., Chambery A., Parente A., Di Maro A. (2012). A Novel Polygalacturonase-Inhibiting Protein (PGIP) from Lathyrus sativus L. seeds. Protein Pept. Lett. 19, 820–825. 10.2174/092986612801619561 [DOI] [PubMed] [Google Scholar]
- Tamura M., Gao M., Tao R., Labavitch J. M., Dandekar A. M. (2004). Transformation of persimmon with a pear fruit polygalacturonase inhibiting protein (PGIP) gene. Sci. Hortic. 103, 19–30 10.1016/j.scienta.2004.04.006 [DOI] [Google Scholar]
- ten Have A., Mulder W., Visser J., van Kan J. A. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 11, 1009–1016. 10.1094/MPMI.1998.11.10.1009 [DOI] [PubMed] [Google Scholar]
- Thornburg R. W., Carter C., Powell A. L., Mittler R., Rizhsky R., Horner H. T. (2003). A major function of the tobacco floral nectary is defense against microbial attack. Plant Syst. Evol. 238, 211–218 10.1007/s00606-003-0282-9 [DOI] [Google Scholar]
- Toubart P., Desiderio A., Salvi G., Cervone F., Daroda L., De Lorenzo G., et al. (1992). Cloning and characterization of the gene encoding the endopolygalacturonase-inhibiting protein (PGIP) of Phaseolus vulgaris L. Plant J. 2, 367–373. 10.1046/j.1365-313X.1992.t01-35-00999.x [DOI] [PubMed] [Google Scholar]
- van Santen Y., Benen J. A., Schroter K. H., Kalk K. H., Armand S., Visser J. (1999). 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J. Biol. Chem. 274, 30474–30480. 10.1074/jbc.274.43.30474 [DOI] [PubMed] [Google Scholar]
- Volpi C., Raiola A., Janni M., Gordon A., O'Sullivan D. M., Favaron F., et al. (2013). Claviceps purpurea expressing polygalacturonases escaping PGIP inhibition fully infects PvPGIP2 wheat transgenic plants but its infection is delayed in wheat transgenic plants with increased level of pectin methyl esterification. Plant Physiol. Biochem. 73, 294–301. 10.1016/j.plaphy.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Wand A. J., Englander S. W. (1996). Protein complexes studied by NMR spectroscopy. Curr. Opin. Biotechnol. 7, 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Wei X., Rong W., Dang L., Du L. P., Qi L., et al. (2014a). GmPGIP3 enhanced resistance to both take-all and common root rot diseases in transgenic wheat. Funct. Integr. Genomics. [Epub ahead of print]. 10.1007/s10142-014-0428-6 [DOI] [PubMed] [Google Scholar]
- Wang R., Lu L., Pan X., Hu Z., Ling F., Yan Y., et al. (2014b). Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 87, 181–191. 10.1007/s11103-014-0269-7 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhu X., Tooley P., Zhang X. (2013). Cloning and functional analysis of three genes encoding polygalacturonase-inhibiting proteins from Capsicum annuum and transgenic CaPGIP1 in tobacco in relation to increased resistance to two fungal pathogens. Plant Mol. Biol. 81, 379–400. 10.1007/s11103-013-0007-6 [DOI] [PubMed] [Google Scholar]