Abstract
The backbone of first-line treatment for Epidermal Growth Factor (EGFR) wild-type (wt) advanced Non-small cell lung cancer (NSCLC) patients is the use of a platinum-based chemotherapy combination. The treatment is characterized by great inter-individual variability in outcome. Molecular predictive markers are extremely needed in order to identify patients most likely to benefit from platinum-based treatment and resistant ones, thus optimizing chemotherapy approach in NSCLC. Several components of DNA repair response (DRR) have been investigated as potential predictive markers. Among them, high levels of expression of ERCC1, both at protein and mRNA levels, have been associated with resistance to cisplatin in NSCLC. In addition, low levels of expression of RRM1, a target for gemcitabine, have been associated with improved OS in advanced NSCLC patients treated with cisplatin and gemcitabine. Preclinical data and retrospective analyses showed that BRCA1 is able to induce resistance to cisplatin and sensitivity to antimicrotubule agents. In addition, the mRNA levels of expression of RAP80, encoding for a protein cooperating with BRCA1 in homologous recombination (HR), have demonstrated to further sub-classify low BRCA1 NSCLC tumors, improving the predictive model. On the basis of biological knowledge on DNA repair pathway and recent controversial results from clinical validation of potential molecular markers, integrated analysis of multiple DNA repair components could improve predictive information and pave the way to a new approach to customized chemotherapy clinical trials.
Key Words: Platinum, ercc1, brca1, dna repair, predictive modeling
Introduction
The most important progresses in the field of advanced NSCLC disease are related to the capacity of individuating so-called driver-mutations, that is molecular alterations able to render the tumors specifically sensitive to targeted therapy. The first and best known example is the discovery of EGFR mutations, characterizing a subgroup of tumors in which the treatment with EGFR selective inhibitors (gefitinib, erlotinib, afatinib) significantly improves survival and quality of life (1-6). Currently, we also know that patients carrying ALK rearrangements could significantly benefit from crizotinib treatment (7) and that we can individuate several other subgroups of patients with lung adenocarcinoma characterized by dysregulation of main oncogenic pathways induced by a specific genetic alteration (8,9). Finally, a series of potentially targetable molecular alterations have been recently found also in squamous cell carcinoma (SCC) (10). Nevertheless, still about 80% of advanced NSCLC patients receive standard first-line chemotherapy treatment and their best therapeutic option is considered platinum-based chemotherapy, when clinically feasible. Clinical and radiological responses are obtained only in a subgroup of these patients and the median overall survival (OS) of the chemotherapy-treated population is still inferior to one year. Moreover, platinum-based chemotherapy is currently the standard second-line treatment after progression to an EGFR-inhibitor in EGFR-mutated patients.
In this clinical context, the aim of the research concerning molecular predictive markers of platinum sensitivity is to optimize chemotherapy approach and provide more precise information to patient at diagnosis.
Biological rationale for predictive models in NSCLC
Cisplatin and carboplatin act as DNA-damaging agents and have largely overlapping resistance mechanisms. For this reason, defective DNA repair capacity, one of the main factors responsible for carcinogenesis, may contribute to the cytotoxic effect of the drugs. On the other hand, DNA repair capacity, contributing to genome stability, is one of the most studied mechanisms of platinum resistance.
Cellular DNA repair capacity depends on complex inter-related mechanisms, also interacting with cell cycle control and apoptotic pathways. For this reason, considerable efforts have been made to validate predictive markers as surrogate of DNA repair capacity and, in particular, of the capacity of repairing the lesions induced by platinum on DNA.
Cisplatin and carboplatin inhibit DNA replication mainly acting as cross-links inducing agents. They bind DNA, and in prevalence nucleophilic N7-sites on purine bases, leading to the generation of protein-DNA and DNA-DNA intra- and, less commonly, interstrand adducts. Platinum-induced lesions cause distortions in DNA structure that are recognized by multiple DNA repair pathways. These DNA distortions are mainly repaired by the nucleotide excision repair (NER) system. NER is a pathway involved in DRR specifically targeting DNA helix-distorting lesions, including cisplatin- and ultraviolet-induced lesions. It functions as a so-called “cut-and-paste” mechanism including different sequential steps: DNA damage recognition, local opening of the DNA helix around the lesion, damage excision and gap filling. It consists of two sub-pathways: global genome NER (GG-NER) and transcription-coupled NER (TC-NER), sharing the same core mechanism but differing in the way that DNA lesions are recognized and in the target DNA sequences. TC-NER specifically recognizes actively transcribed DNA sequences (Figure 1).
Figure 1.
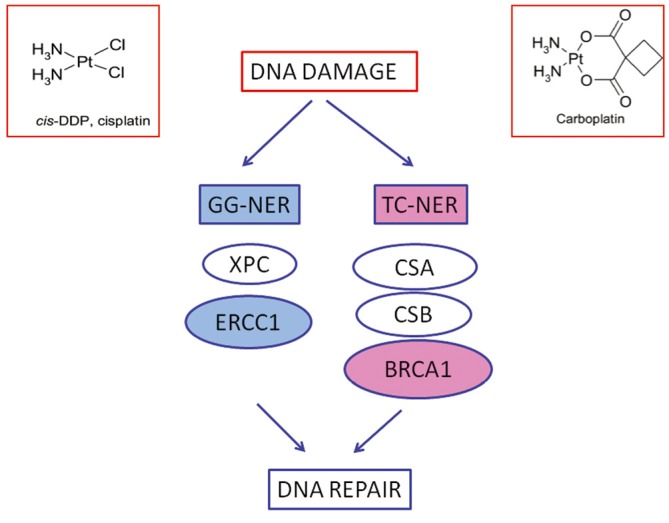
The figure shows the two simplified sub-pathways of nucleotide excision repair (NER): Global-genome (GG)-NER targets the whole genome sequences, while transcription-coupled (TC)-NER recognizes specifically lesions involving actively transcribed DNA. ERCC1 is a fundamental element in GG-NER, while BRCA1 is mainly involved in TC-NER.
The structure-specific endonuclease excision repair cross-complementing 1 (ERCC1) is a protein playing pivotal role in GG-NER. It is thus involved in the rate-limiting step of the pathway, that is incision process. Together with its XP group F (XPF) partner, it cuts the damaged DNA strand at the 5 site of the helix-distorting lesion. In addition, the ERCC1/XPF complex is also involved in the homologous recombination (HR) repair of platinum-induced DNA damage. In tumor experimental models cisplatin exposure is able to increase ERCC1 mRNA expression levels. The mRNA expression of ERCC1 correlates with the capacity of DNA adducts repair (11,12) while higher activation of ERCC1 is associated with platinum resistance in several tumor models (13).
RRM1 is the regulatory subunit of ribonucleotide reductase and controls the function of the enzyme involved in deoxynucleotide production. Deoxynucleotide availability is essential to conclude NER and this could explain a potential predictive role for ribonucleotide reductase subunit M1 (RRM1) in patients treated with platinum, in addition to known data about gemcitabine sensitivity. Gemcitabine is an inhibitor of ribonucleotide reductase and increased RRM1 expression has been associated with gemcitabine resistance (14,15). In clinical setting, low mRNA levels of RRM1 have been associated with improved outcome of patients treated with platinum and gemcitabine, showing a sort of synergism in the DNA-repair-linked resistance mechanisms of the two drugs (16-18).
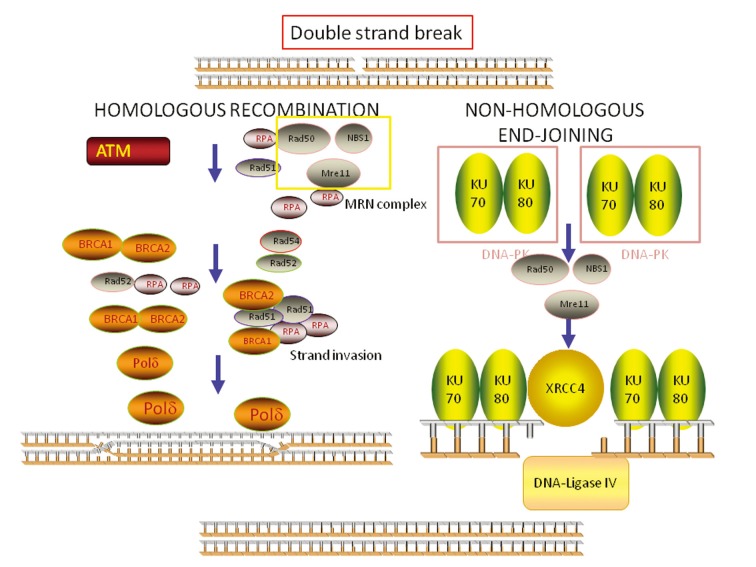
Replication blocks induced by cisplatin lead to activation of HR, creating the so-called “stalled replication forks” and, in this way, the sequential coordinated action of NER and HR is required for repairing the platinum-induced DNA damage. HR is one of the major pathways involved in DNA double strand breaks (DSBs) repair. It acts using the non-damaged strand as a template and so it is considered an “error-free” system (Figure 2). The role of HR in the repair of platinum-induced lesions is at the basis of the potential role of Breast cancer susceptibility gene 1 (BRCA1), one main component of HR, in predicting resistance to platinum.
Figure 2.
The figure shows the two main pathways involved in the repair of double strand breaks: homologous recombination, an error-free mechanism, using undamaged strand as template, and non-homologous end-joining, an error-prone mechanism.
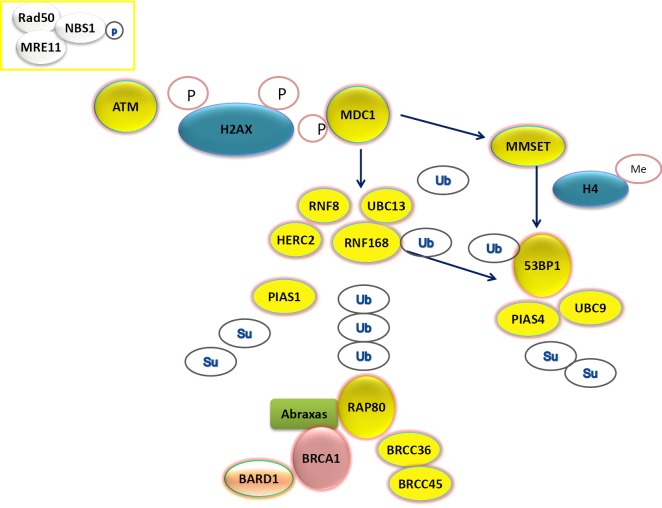
HR is a complex mechanism initiating with the recognition of DSBs by the multifunctional protein sensor complex (MRE11-RAD50-NBS1), the activation of the check-point phosphoinositide 3-kinase related ATM, ATR and DNA-PK and the subsequent phosphorylation of histone H2AX proteins (19). ATM phosphorylates the mediator of DNA checkpoint 1 (MDC1) at the region surrounding the DSB and in this way triggers the recruitment of DNA repair effectors (Figure 3). The assembly process requires a series of post-translational modifications of DNA repair components. In particular, the E3 ubiquitin-ligase RNF8 recognizes the phosphorylated MDC1 and creates a complex with the E3 ubiquitin-ligase RNF168 and the E2 ubiquitin-conjugating enzyme UBC13, leading to the recruitment of BRCA1 (20). The assembly of the RNF8-UBC13 complex is facilitated by the activity of the HECT type E3 ligase (HERC2) (21), an E3 ubiquitin-ligase which can also target BRCA1 for degradation (22). A large proportion of BRCA1 present at DSBs co-localizes with a group of proteins forming a complex called BRCA1-A complex, including also BARD1, BRCC36, ABRAXAS and RAP80 (Figure 3). In particular, RAP80, an ubiquitin-interaction motif (UIM) protein, recognizes the histones ubiquitinated by RNF8/UBC13 and triggers the formation of the complex. In experimental model, the presence of RAP80 is essential for the recruitment of the BRCA1-A complex at DNA damage sites, while its loss abrogates the repair response (23-27). On the other hand, BRCA1 forms also other complexes at DNA damage sites, increasing the level of complexity.
Figure 3.
The recruitment of BRCA1-A complex and of 53BP1 at DNA damage sites after double strand breaks. The detailed molecular processes are described in the text.
Notably, BRCA1 is considered to play a fundamental role also in TC-NER (28,29) (Figure 1). This point could be particularly relevant, since GG-NER could have low affinity for cisplatin-induced DNA adducts, while TC-NER is specifically initiated by cisplatin cross-links and an experimental model of TC-NER deficient cells demonstrated hypersensitivity to cisplatin (30-32). The effect of the BRCA1 in determining resistance to platinum has been also directly demonstrated in preclinical models and, in parallel, BRCA1-deficiency has been associated with platinum-resistance (33-35).
BRCA1 is also involved in Non homologous end-joining (NHEJ), an error-prone pathway involved in DSBs repair and in mismatch repair (MMR), which can recognize cisplatin-induced DNA lesions, while normally dealing with erroneous insertions, deletions and mis-incorporations of bases during DNA replication (36).
The role of BRCA1 in HR and NHEJ response to DSBs (Figure 2) implies inter-relation with other DNA repair components. In particular, recent findings show a complex functional interplay with 53BPB1, a protein acting as an activator of P53, but also involved in NHEJ and HR. The protein 53BP1 modulates the chromatin structure at DNA damage sites and contributes in maintaining genomic stability (37). In addition, it is able to negatively regulate HR repair, by inhibiting CTIP, a protein that creates a complex with BRCA1 (BRCA1-C complex), thus supporting HR (38). Interestingly, 53BP1 function could contribute to the expression of BRCA1-loss phenotype and, in the absence of 53BP1, HR capacity could be maintained independently on BRCA1 (38). Finally, BRCA1-independent HR capacity can be suppressed by abrogating RNF8, consistently with the complexity of the pathway (39). The protein 53BP1 is recruited at DNA damage sites through two different mechanisms, activated after MDC1 recruitment at DNA damage sites. The first mechanism of 53BP1 recruitment depends on RNF8/UBC13 complex formation and subsequent histone ubiquitination (20). The second mechanism of 53BP1 localization at DSBs is driven by histone methylation, regulated by the methyltransferase MMSET (40) (Figure 3).
In addition to ubiquitination process, the assembly of DNA repair effectors at DSB requires also another post-translational modification process called sumoylation. For this reason, specific small ubiquitin-related modifier (SUMO)-conjugating systems are required. In particular, the E3 ligase PIAS1 and PIAS4 are recruited at DSBs and their depletion reduces BRCA1 accumulation at DSBs (41). PIAS4 forms a complex with E2 ligase UBC9, positively modulating 53BP1 function (42) (Figure 3).
The complex and inter-related pathways involved in the repair of platinum-induced DNA lesions explain the difficulty in finding one single marker measurable in patients’ blood or tumor biopsies able to effectively predict resistance to platinum in clinical setting.
Strategies for building predictive models in lung cancer
The biological observation that tumor cells with defective capacity of removing cisplatin-DNA adducts are hypersensitive to platinum and the increasing knowledge about DNA repair pathways, forming a complex integrated network, carries great potential clinical application. In translational application of this knowledge the issue is to find a reliable marker able to measure the cellular capacity of repairing platinum-induced DNA damage and clinically performable in samples from patients’ blood or small biopsies.
The most direct method of quantifying DNA repair capacity would be to measure and to monitor the rate of unrepaired DNA-adducts in tumor cells following platinum exposure directly (Table 1). For practical application, it is possible to measure DNA repair capacity in vitro by culturing patients’ peripheral lymphocytes and measuring the unrepaired DNA adducts induced by a cross-links inducing agent. Recently, it has been suggested that the DNA repair capacity, quantified with this method, could predict the patients’ outcome to platinum-based chemotherapy. A retrospective analysis in a large but heterogeneous population of NSCLC showed a trend for improved overall survival (OS) in patients with the lowest rate of DNA repair capacity measured in peripheral lymphocytes. This trend was higher in the subgroup of patients with stage I-IIIA disease and in adenocarcinoma histology (43).
Table 1. Main clinical data depicted in the text about potential predictive biomarkers in platinum-treated NSCLC patients. In the table the data are summarized according to the methodology used.
| Methodology | Clincal study type | Clinical setting | Biomarker | Results | Reference |
|---|---|---|---|---|---|
| DNA repair capacity in peripheral lymphocytes | Retrospective evaluation | Platinum-treated NSCLC | DNA repair capacity | Trend for improved OS in patients with low DNA repair capacity | (43) |
| Single DNA repair component at protein level | Retrospective evaluation | Adjuvant | ERCC1 | OS benefit from platinum-treatment only in low/negative expressing ps | (44) |
| Retrospective evaluation | Adjuvant | ERCC1; BRCA1 | Improved DFS for SCC with low/negative expression of ERCC1 | (45) | |
| Single DNA repair component at mRNA level | Retrospective evaluation | Advanced disease | ERCC1; RRM1 | Improved OS for low-expressing ps | (18) |
| Retrospective evaluation | Advanced disease | ERCC1 | Improved OS for low-expressing ps | (46) | |
| Retrospective evaluation | Advanced disease (second-line) | ERCC1; BRCA1 | Higher RR, PFS, OS for low-expressing ps | (47) | |
| Retrospective evaluation | Advanced disease | RRM1 | Improved OS for low-expressing ps | (16) | |
| Retrospective evaluation | Advanced disease | RRM1 | Improved OS for low-expressing ps | (17) | |
| Individual patient analysis | Advanced disease | ERCC1; RRM1 | Improved RR and OS in ps treated with customized CT according to ERCC1 and RRM1 | (48) | |
| Phase III trial | Advanced disease | ERCC1 | Improved RR in ps treated with customized CT according to ERCC1 | (49) | |
| Retrospective evaluation | Neoadjuvant | BRCA1 | Improved OS for low-expressing ps | (50) | |
| Phase II | Advanced disease | BRCA1 | Feasibility of customized CT according to BRCA1 mRNA | (51) | |
| Retrospective evaluation | Advanced disease | ERCC1 | Negative | (52) | |
| Retrospective evaluation | Advanced disease | ERCC1; BRCA1 | Improved RR, PFS, OS for ps expressing low ERCC1 | (53) | |
| CTCs evaluation of a single DNA repair component | Retrospective evaluation | Advanced disease | ERCC1 | Improved PFS in ps expressing low ERCC1 | (54) |
| Integrated DNA repair components analysis | Retrospective evaluation | Advanced disease | BRCA1; RAP80 | Trend for Improved PFS and OS for ps expressing low BRCA1 and RAP80 | (51) |
Abbreviations: CTCs, circulating tumor cells; DFS, disease free survival; NSCLC, non-small cell lung cancer; OS, overall survival; PFS, progression free survival; ps, patients; RR, response rate; SCC, squamous cell carcinoma
Anyway, the most studied way to translate preclinical findings about DNA repair and platinum efficacy into clinical benefit is based on the idea that one single protein could lead the repair of DNA-induced damage and that its activity could mirror global capacity of repairing platinum-induced lesions and, consequently, predict resistance to platinum. Actually, most of the clinical data available and currently under prospective evaluation concerns the use of a molecular marker, considered the main protagonist of a DNA repair pathway, as potential predictive marker of platinum resistance (Table 1). In particular the most studied molecular markers are the aforementioned ERCC1, BRCA1 and RRM1. These markers have been studied both at protein levels, with immunoistochemistry (ICH), and at mRNA level, through quantitative reverse transcriptase PCR. From the technical point of view, quantitative reverse transcriptase shows high sensitivity and reproducibility. The use of mRNA measurement could be suitable for screening a series of markers in retrospective analyses, and could be successfully performed also in small biopsies and formalin-fixed paraffin-embedded tissues (51), even though it could have some limits in quantifying differences in the expression of very low expressing genes, using this kind of samples. In addition, it could require centralized evaluation in laboratories with long-term experience in the specific field. On the other hand, IHC is a really cost-effective method, although sometimes appearing less reproducible and objective. Once a marker is validated, IHC testing has the potentiality of being performed at each pathology centre providing rapid predictive information, if sufficient tumor material is available. Particularly interesting is also the possibility of studying DNA repair components expression in circulating tumor cells (CTCs), which are present in peripheral blood of patients with metastatic disease and can be analyzed quantitatively and qualitatively with several techniques. The most important point is the possibility to analyze the expression of specific biomarkers in CTCs, as surrogate of tumor samples, and to monitor functional changes of these biomarkers induced by treatment. Preliminary promising data are available from a small cohort of patients and relevant difference in progression free survival (PFS) to platinum-based chemotherapy was described favoring the group of patients lacking ERCC1 expression. Unfortunately, serial evaluation at multiple time points was missing for most of the patients (54) (Table 1).
On the basis of DNA repair complexity, some retrospective data are also already available about the possibility that integrated analysis of more than one component of a DNA repair pathway could provide more precise predictive information (51) (Table 1).
Clinical data available about DNA repair components as molecular predictive markers in NSCLC
Several potential molecular markers have been suggested for predicting the efficacy of platinum-based treatment and some of them are still under clinical evaluation.
ERCC1 and RRM1
In advanced NSCLC samples low ERCC1 mRNA expression levels have been associated with improved survival after first-line treatment with cisplatin and gemcitabine, thus seemingly confirming the biological resistance model (18,46). Recently, a predictive value for ERCC1 (and BRCA1) mRNA expression has been confirmed also in a series of patients treated with a platinum-based combination in second-line setting (47) (Table 1).
A meta-analysis including the results of 12 selected studies confirmed the potential predictive value of ERCC1, either at mRNA or at protein level, showing that negative/low ERCC1 expressing NSCLC patients treated with platinum-based combinations achieve higher response rate (RR) and improved median OS. In parallel, it indicated little difference favoring IHC in the results according to the technique used. The predictive effect seemed higher in the Asiatic population, compared to the Caucasian one and in patients treated with a combination including cisplatin, while it decreased when considering also carboplatin-based chemotherapy (55).
In advanced patients treated with this platinum and gemcitabine, also low RRM1 expression levels correlate with improved outcome (14,16-17). On the other hand, RRM1 is known to be associated with resistance to gemcitabine even when administered in non-platinum containing combinations (56-58). A meta-analysis, including the results of 18 selected trials, indicated an improved RR, PFS and OS for NSCLC expressing negative/low RRM1 treated with gemcitabine-containing chemotherapy (59).
A recently published individual patient analysis suggested that customizing chemotherapy according to ERCC1 and RRM1 expression could be translated into improved outcome for advanced NSCLC patients. The analysis included a comparison between two study populations, including four prospectively accrued cohorts of patients: the “personalized treatment” group and the “standard treatment” one (48). In the “personalized” group of treatment, ERCC1 and RRM1 were measured at mRNA level through real-time reverse transcriptase; patients whose tumors expressed low levels of both genes received carboplatin and gemcitabine, those with high levels of both genes received docetaxel and vinorelbine, patients with high ERCC1 and low RRM1 received gemcitabine and docetaxel, those with low ERCC1 but high RRM1 were treated with carboplatin and docetaxel (60). Even though the design of the study implies only hypothesis-generating results, it is encouraging to see that patients accrued in a phase II trial (NCT00215930) and thus treated according to the levels of expression of ERCC1 and RRM1 showed statistically significant improvement, both in terms of RR and of OS, when compared with patients receiving a “non-molecularly-driven” treatment (48) (Table 1).
In the adjuvant setting the predictive role of ERCC1 was suggested for the first time by an important retrospective analysis of tumor samples from patients enrolled in the International Adjuvant Lung Cancer Trial (IALT). The benefit of cisplatin-based adjuvant chemotherapy in terms of OS was limited to patients whose tumors did not express ERCC1. On the contrary, the expression of the marker was associated with improved prognosis in the group of patients who did not receive adjuvant chemotherapy. In this large study, ERCC1 was investigated at protein level using ICH (44) (Table 1).
Only the results of prospective phase III trials will let us draw definitive conclusions. Prospective results on the predictive role of ERCC1 are available in advanced disease setting. Standard chemotherapy with cisplatin and docetaxel was compared with customized treatment according to the mRNA levels of expression of ERCC1 measured in pretreatment biopsies. In the experimental arm, patients with low levels of ERCC1 received cisplatin and docetaxel, patients with high ERCC1 received docetaxel and gemcitabine. The patients allocated to the experimental arm demonstrated a significantly improved response rate (RR), not mirrored by increase of OS (49). This was the first prospective phase III clinical trial with available results in the field of customized chemotherapy and raised many questions about the methods to use to customize chemotherapy and the best chemotherapy combination for studying predictive models. Currently, several prospective trials are ongoing to validate ERCC1 predictive role both in early stage (phase III: TASTE, NCT00775385; ITACA) and in advanced stage disease (phase III: NCT00801736; NCT00499109; phase II: NCT01648517, NCT01356368; NCT00736814) (Table 2). In most of the studies the predictive value of ERCC1 is analyzed in conjuction with RRM1 evaluation, in particular when considering chemotherapy first-line combination including cis- or carboplatin and gemcitabine.
Table 2. Ongoing prospective clinical trial evaluating the predictive role of ERCC1, RRM1 and BRCA1.
| Trial number | Phase | Clinical setting | Biomarker | Methodology |
|---|---|---|---|---|
| NCT00775385 | Phase II-III | Adjuvant (II-IIIA) | ERCC1 | IHC |
| NCT00801736 | Phase III | Advanced disease | ERCC1 | NA |
| NCT00499109 | Phase III | Advanced disease | ERCC1; RRM1 | NA |
| NCT01648517 | Phase II | Advanced disease | ERCC1; RRM1 | mRNA |
| NCT01356368 | Phase II | Advanced disease | ERCC1; RRM1 | NA |
| NCT00736814 | Phase II | Advanced disease | ERCC1; RRM1 | mRNA |
| NCT00617656 | Phase III | Advanced disease | BRCA1; RAP80 | mRNA |
| NCT00478699 | Phase III | Adjuvant (II-IIIA) | BRCA1 | mRNA |
| Eudra-ct: 2008-001764-36 | Phase III | Adjuvant (II-IIIA) | ERCC1; TS | IHC and mRNA |
Abbreviations: IHC, immunohistochemistry; NA, not available
BRCA1
Clinical investigation concerning the predictive role of BRCA1 expression in NSCLC has retrospectively confirmed that low mRNA expression is correlated with good prognosis and increased sensitivity to cisplatin (50,61). A retrospective analysis in paraffin-embedded NSCLC samples, collected before neoadjuvant treatment with cisplatin and gemcitabine, demonstrated that the group of tumors with the lowest expression of BRCA1 obtained the greatest benefit from cisplatin-gemcitabine treatment (50) (Table 1). On the basis of this finding, a prospective phase II trial was planned. In this trial, the treatment of advanced non-squamous NCSLCs was customized according to mRNA expression levels of BRCA1 measured by reverse transcriptase PCR. Patients whose tumors expressed low levels of BRCA1 received cisplatin and gemcitabine in first line setting; patients with high levels of the two received docetaxel alone, while patients with intermediate expression of BRCA1 were treated with cisplatin and docetaxel. The study demonstrated the feasibility of BRCA1 expression analysis in clinical practice and the median OS achieved was similar in the three genotyped groups. The samples from patients enrolled in the prospective trial were retrospectively analyzed to measure mRNA expression of RAP80 and ABRAXAS, as main components of BRCA1-A complex (Figure 3). The mRNA expression of RAP80 resulted as potential new predictive marker, able to further subclassify the outcome of low-BRCA1 expressing patients. In the study population, patients with low levels of both BRCA1 and RAP80 obtained an impressive median PFS of 14 months (51).
Consequently, a multi-centric phase III trial has been coordinated in order to confirm the predictive value of integrated BRCA1-RAP80 analysis and their applicability in clinical practice (BREC, NTC00617656) (Table 2). In the trial, the outcome of advanced NSCLC patients treated with non-personalized chemotherapy (cisplatin-docetaxel) in first-line setting is compared to the one of patients receiving customized chemotherapy. The primary end-point is time to progression (TTP). All the samples of patients allocated to the experimental arm are analyzed for mRNA expression of BRCA1 and RAP80 through real-time PCR and the levels of expression of the two genes are categorized using tertiles as cut-off points. Patients in the experimental arm receive cisplatin and gemcitabine if RAP80 is low, independently on the levels of BRCA1, cisplatin and docetaxel if RAP80 is intermediate or high in the presence of low or intermediate BRCA1, docetaxel alone when BRCA1 is high and RAP80 intermediate or high. The accrual has been completed and interim analysis results will be soon available. Another phase III prospective trial is ongoing to test the predictive value of BRCA1 in the adjuvant setting (GEPC-SCAT, NCT00478699) (Table 2). Patients with stage II or IIIA NSCLCs, after complete surgical resection, are randomized to receive adjuvant chemotherapy with cisplatin and docetaxel or adjuvant chemotherapy customized according to BRCA1 mRNA levels. Patients allocated to the experimental arm receive cisplatin and gemcitabine if BRCA1 is low, cisplatin and docetaxel if BRCA1 is intermediate, docetaxel alone if BRCA1 is high.
New perspectives: integrated analysis of multiple DNA repair components and histology-driven analyses
While prospective trials are ongoing, controversial data about DNA repair components as predictive markers of platinum-based chemotherapy efficacy are available. In other words, not all the retrospective series confirmed the predictive role of ERCC1 and BRCA1 and several are the possible explanations. Among the most recent retrospective series, an analysis of mRNA expression levels of ERCC1 in formalin-fixed paraffin embedded tumor samples did not confirm the predictive role of ERCC1. The mRNA expression of ERCC1 was neither correlated to RR nor to OS in advanced NSCLC patients prospectively recruited in a phase III trial and treated with platinum-based chemotherapy (52). In a more recent retrospective evaluation, tumor tissues from patients prospectively enrolled in a phase III trial and treated with platinum and gemcitabine were analyzed for the expression of six DNA repair components. In this case, the authors found no predictive value for BRCA1 and RAP80 mRNA, whereas low ERCC1 and ABRAXAS levels were associated with increased RR and improved OS mRNA and PFS. Notably, in this study only 45 patients out of 137 had sufficient tumor material to perform planned analyses (53). These results are also in contrast with the data of the retrospective analysis published in 2009, showing potential predictive value for RAP80, but not for ABRAXAS (51). In a larger series of patients, treated with platinum-based chemotherapy in adjuvant setting, protein expression of seven DNA repair components has been analyzed using IHC. The number of cases with evaluable results was variable according to the different biomarkers, with a range of 550-716 cases. Despite the large study population, neither tested biomarker was able to predict the benefit from adjuvant chemotherapy with statistical significance. The analyzed DNA repair components were generally expressed at higher levels in squamous cell carcinomas, with respect to adenocarcinoma histology. In the analysis by histology, higher benefit from platinum-based adjuvant chemotherapy was demonstrated in squamous cell carcinoma patients with low expression of ERCC1 and ATM. This difference in outcome was measured in terms of disease free survival (DFS) (45). Actually, histology has gained increasing role in NSCLC definition and treatment decision making in the latest years and we know that different biology characterizes adenocarcinoma versus squamous cell carcinoma.
All mentioned controversial results open new questions and suggest a new approach to predictive modeling in lung cancer.
Probably analysis by histology will help in the interpretation of retrospective analyses and ongoing prospective validation of potential predictive markers. Squamous cell carcinoma should probably be analyzed separately for non-completely known biological reason. One possible explanation is that the pattern of expression of DRR genes is different according to histology. Recently we have confirmed higher levels of expression of BRCA1, but also 53BP1 and UBC9, in squamous histology in a retrospective series of 115 advanced NSCLCs (62). Also in our series the differential expression according to histology was not mirrored by differential sensitivity to platinum-based chemotherapy according to histology. Another point to take into account is the differential sensitivity to different platinum-based chemotherapy according to histology. It is possible that the chemotherapy combinations could influence the outcome of patients differently in different histological subtypes, increasing the level of complexity. We already know that the partner for platinum in chemotherapy combinations could influence predictive modeling interpretation. As a matter of fact, platinum and gemcitabine seem to show a sort of synergism in DNA repair-associated resistance mechanisms (16-18), whereas taxanes and vinca alkaloids could not be the best partner for platinum in customized chemotherapy approach. BRCA1 is modulator of cellular response to chemotherapy drugs, inducing resistance to platinum and sensitivity to antimicrotubule agents (63). Finally, when interpreting the results in retrospective series treated with platinum-based combinations also containing pemetrexed, the well-known differential sensitivity to the drugs should be taken into account according to histology (64) and the molecular modulators of pemetrexed sensitivity (65).
In addition to histology-driven analyses, predictive information in lung cancer could be improved by building predictive models able to integrate the influence of several DNA repair components, contributing to cellular response to DNA repair damage. Actually, cellular response to DNA damage includes redundant mechanisms normally contributing to genome stability and interacting in sometimes unexpected ways to chemotherapy-induced DNA damage. We know that RAP80 expression could improve BRCA1 predictive information, when analyzed at mRNA levels in low BRCA1 expressing NSCLCs (52). One of the most interesting inter-relation is the one between BRCA1 and 53BP1. We have recently explored the predictive value of integrated BRCA1 and 53BP1 analysis in advanced NSCLC patients treated with a platinum-based combination not including antimicrotubules agents in first-line setting. In this series, we measured BRCA1, 53BP1 and other six components of 53BP1 pathway at mRNA level using real-time PCR. The levels of mRNA expression were considered as categorical variables using median values as cut-off points. BRCA1 was not confirmed as predictive marker, when considered in isolation. Among patients expressing low levels of BRCA1 mRNA, patients with low levels of 53BP1 obtained an impressive median OS of more than 19 months and a PFS of 10 months, in sharp contrast with patients with low BRCA1 and high 53BP1 (median OS: 8.2 months, median PFS: 5.9 months; P=0.01 for OS, P<0.0001 for PFS). In patients with high levels of BRCA1 the median OS was 10 months, independently on 53BP1 (62). These results demonstrate the potentiality of integrated predictive modeling in lung cancer.
Conclusions
Current clinical data concerning the potential predictive role of DDR components require confirmation by large prospective randomized phase III trials but highlight the possibility of significant improvement in outcome of advanced NSCLC patients treated with chemotherapy. Available data suggest that different platinum-based doublets should be analyzed separately, considering the possible influences of different predictive markers and that histology-driven analysis could improve predictive modeling interpretation. In addition, only optimal clinical stratification of patients will permit correct interpretation of results concerning predictive biomarkers. Finally, current results demonstrate that it is difficult to identify a single marker able to predict response to a drug or a combination of drugs. Integrated analysis of several potential biomarkers based on the study of DNA repair pathways biology will probably provide more insight in predictive modeling in lung cancer.
Acknowledgements
The author thanks Martina Boscaro for reviewing the language of the article.
Disclosure: The author declares no conflict of interest.
References
- 1.Pao W, Chmielecki J.Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010;10:760-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [DOI] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [DOI] [PubMed] [Google Scholar]
- 6.Yang JC, Shih JY, Su WC, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): a phase 2 trial. Lancet Oncol 2012;13:539-48. [DOI] [PubMed] [Google Scholar]
- 7.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pao W, Girard N.New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [DOI] [PubMed] [Google Scholar]
- 10.Drilon A, Rekhtman N, Ladanyi M, et al. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol 2012;13:e418-26. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Gardner K, Zhang L, et al. Cisplatin induction of ERCC-1 mRNA expression in A2780/CP70 human ovarian cancer cells. J Biol Chem 1998;273:23419-25. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Yu JJ, Mu C, et al. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res 2000;20:645-52. [PubMed] [Google Scholar]
- 13.Arora S, Kothandapani A, Tillison K, et al. Downregulation of XPF-ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair (Amst) 2010;9:745-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol 2006;24:4731-7. [DOI] [PubMed] [Google Scholar]
- 15.Davidson JD, Ma L, Flagella M, et al. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res 2004;64:3761-6. [DOI] [PubMed] [Google Scholar]
- 16.Rosell R, Scagliotti G, Danenberg KD, et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene 2003;22:3548-53. [DOI] [PubMed] [Google Scholar]
- 17.Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res 2004;10:1318-25. [DOI] [PubMed] [Google Scholar]
- 18.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol 2006;17:1818-25. [DOI] [PubMed] [Google Scholar]
- 19.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 2007;85:509-20. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett 2008;271:179-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 2010;12:80-6;sup pp 1-12. [DOI] [PubMed] [Google Scholar]
- 22.Wu W, Sato K, Koike A, et al. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res 2010;70:6384-92. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 2007;316:1194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Chen J, Yu X.Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007;316:1202-5. [DOI] [PubMed] [Google Scholar]
- 25.Sobhian B, Shao G, Lilli DR, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007;316:1198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan J, Kim YS, Yang XP, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 2007;67:6647-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Liu C, Chen J, et al. RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo. J Biol Chem 2012;287:22919-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Page F, Randrianarison V, Marot D, et al. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res 2000;60:5548-52. [PubMed] [Google Scholar]
- 29.Abbott DW, Thompson ME, Robinson-Benion C, et al. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J Biol Chem 1999;274:18808-12. [DOI] [PubMed] [Google Scholar]
- 30.Lainé JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J 2006;25:387-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tremeau-Bravard A, Riedl T, Egly JM, et al. Fate of RNA polymerase II stalled at a cisplatin lesion. J Biol Chem 2004;279:7751-9. [DOI] [PubMed] [Google Scholar]
- 32.Furuta T, Ueda T, Aune G, et al. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res 2002;62:4899-902. [PubMed] [Google Scholar]
- 33.Bhattacharyya A, Ear US, Koller BH, et al. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 2000;275:23899-903. [DOI] [PubMed] [Google Scholar]
- 34.Husain A, He G, Venkatraman ES, et al. BRCA1 up-regulation is associated with repair-mediated resistance to cis-diamminedichloroplatinum(II). Cancer Res 1998;58:1120-3. [PubMed] [Google Scholar]
- 35.Tassone P, Di Martino MT, Ventura M, et al. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol Ther 2009;8:648-53. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy RD, Quinn JE, Johnston PG, et al. BRCA1: mechanisms of inactivation and implications for management of patients. Lancet 2002;360:1007-14. [DOI] [PubMed] [Google Scholar]
- 37.Bothmer A, Robbiani DF, Di Virgilio M, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 2011;42:319-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunting SF, Callén E, Wong N, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010;141:243-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakada S, Yonamine RM, Matsuo K. RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res 2012;72:4974-83. [DOI] [PubMed] [Google Scholar]
- 40.Pei H, Zhang L, Luo K, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011;470:124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris JR, Boutell C, Keppler M, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 2009;462:886-90. [DOI] [PubMed] [Google Scholar]
- 42.Galanty Y, Belotserkovskaya R, Coates J, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009;462:935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang LE, Yin M, Dong Q, et al. DNA repair capacity in peripheral lymphocytes predicts survival of patients with non-small-cell lung cancer treated with first-line platinum-based chemotherapy. J Clin Oncol 2011;29:4121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [DOI] [PubMed] [Google Scholar]
- 45.Pierceall WE, Olaussen KA, Rousseau V, et al. Cisplatin benefit is predicted by immunohistochemical analysis of DNA repair proteins in squamous cell carcinoma but not adenocarcinoma: theranostic modeling by NSCLC constituent histological subclasses. Ann Oncol 2012;23:2245-52. [DOI] [PubMed] [Google Scholar]
- 46.Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 2002;8:2286-91. [PubMed] [Google Scholar]
- 47.Papadaki C, Sfakianaki M, Ioannidis G, et al. ERCC1 and BRAC1 mRNA expression levels in the primary tumor could predict the effectiveness of the second-line cisplatin-based chemotherapy in pretreated patients with metastatic non-small cell lung cancer. J Thorac Oncol 2012;7:663-71. [DOI] [PubMed] [Google Scholar]
- 48.Simon GR, Schell MJ, Begum M, et al. Preliminary indication of survival benefit from ERCC1 and RRM1-tailored chemotherapy in patients with advanced nonsmall cell lung cancer: evidence from an individual patient analysis. Cancer 2012;118:2525-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol 2007;25:2747-54. [DOI] [PubMed] [Google Scholar]
- 50.Taron M, Rosell R, Felip E, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet 2004;13:2443-9. [DOI] [PubMed] [Google Scholar]
- 51.Rosell R, Perez-Roca L, Sanchez JJ, et al. Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One 2009;4:e5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Booton R, Ward T, Ashcroft L, et al. ERCC1 mRNA expression is not associated with response and survival after platinum-based chemotherapy regimens in advanced non-small cell lung cancer. J Thorac Oncol 2007;2:902-6. [DOI] [PubMed] [Google Scholar]
- 53.Joerger M, deJong D, Burylo A, et al. Tubulin, BRCA1, ERCC1, Abraxas, RAP80 mRNA expression, p53/p21 immunohistochemistry and clinical outcome in patients with advanced non small-cell lung cancer receiving first-line platinum-gemcitabine chemotherapy. Lung Cancer 2011;74:310-7. [DOI] [PubMed] [Google Scholar]
- 54.Das M, Riess JW, Frankel P, et al. ERCC1 expression in circulating tumor cells (CTCs) using a novel detection platform correlates with progression-free survival (PFS) in patients with metastatic non-small-cell lung cancer (NSCLC) receiving platinum chemotherapy. Lung Cancer 2012;77:421-6. [DOI] [PubMed] [Google Scholar]
- 55.Chen S, Zhang J, Wang R, et al. The platinum-based treatments for advanced non-small cell lung cancer, is low/negative ERCC1 expression better than high/positive ERCC1 expression? A meta-analysis. Lung Cancer 2010;70:63-70. [DOI] [PubMed] [Google Scholar]
- 56.Nakano Y, Tanno S, Koizumi K, et al. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer 2007;96:457-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bepler G, Sommers KE, Cantor A, et al. Clinical efficacy and predictive molecular markers of neoadjuvant gemcitabine and pemetrexed in resectable non-small cell lung cancer. J Thorac Oncol 2008;3:1112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Souglakos J, Boukovinas I, Taron M, et al. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br J Cancer 2008;98:1710-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong W, Zhang X, Wu J, et al. RRM1 expression and clinical outcome of gemcitabine-containing chemotherapy for advanced non-small-cell lung cancer: a meta-analysis. Lung Cancer 2012;75:374-80. [DOI] [PubMed] [Google Scholar]
- 60.Simon G, Sharma A, Li X, et al. Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. J Clin Oncol 2007;25:2741-6. [DOI] [PubMed] [Google Scholar]
- 61.Rosell R, Skrzypski M, Jassem E, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One 2007;2:e1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonanno L, Costa C, Majem M, et al. The predictive role of the 53BP1 pathway in advanced Non-small-cell lung cancer (NSCLC) patients (p) treated with first-line platinum-based chemotherapy. Ann Oncol 2012;23:abstr 1183.
- 63.Quinn JE, Kennedy RD, Mullan PB, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res 2003;63:6221-8. [PubMed] [Google Scholar]
- 64.Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [DOI] [PubMed] [Google Scholar]
- 65.Wu MF, Hsiao YM, Huang CF, et al. Genetic determinants of pemetrexed responsiveness and nonresponsiveness in non-small cell lung cancer cells. J Thorac Oncol 2010;5:1143-51. [DOI] [PubMed] [Google Scholar]