Abstract
The echinoderm microtubule-associated protein-like 4 anaplastic lymphoma kinase (EML4-ALK) has emerged as the second most important driver oncogene in lung cancer and the first targetable fusion oncokinase to be identified in 4-6% of lung adenocarcinomas. Crizotinib, along with a diagnostic test—the Vysis ALK Break Apart fluorescence in situ hybridization (FISH) Probe Kit—is approved for the treatment of ALK positive advanced non-small cell lung cancer (NSCLC). However, the success of a targeted drug is critically dependent on a sensitive and specific screening assay to detect the molecular drug target. In our experience, reverse transcription polymerase chain reaction (RT-PCR)-based detection of EML4-ALK is a more sensitive and reliable approach compared to FISH and immunohistochemistry (IHC). Although ALK FISH is clinically validated, the assay can be technically challenging and other diagnostic modalities, including IHC and RT-PCR should be further explored.
Keywords: Anaplastic lymphoma kinase (ALK), fluorescence in situ hybridization (FISH), immunohistochemistry (IHC), non-small cell lung cancer (NSCLC), reverse transcription-polymerase chain reaction (RT-PCR)
Introduction
The echinoderm microtubule-associated protein-like 4 anaplastic lymphoma kinase (EML4-ALK) is a fusion gene comprising portions of the echinoderm EML4 and ALK genes and was first found in a subset of non-small cell lung cancer (NSCLC) cells (1). EML4-ALK is formed through disruption of EML4 at a position approximately 3.6-kb downstream of exon 13 and ligation to a position 297-bp upstream of exon 21 of ALK. EML4-ALK has transforming potential that is dependent on its kinase activity, while the coiledcoil domain of EML4 mediates the constitutive dimerization and cytoplasmic activation of EML4-ALK, which together are responsible for the oncogenic activity of the fusion protein (1).
Only four years after the discovery of EML4-ALK in NSCLC, crizotinib, a dual ALK-MET inhibitor, was approved by the Food and Drug Administration (FDA) in the United States for the treatment of advanced stage NSCLC harboring ALK rearrangements (2,3). One key issue in the detection of altered ALK is the method that best defines ALK status in a clinical setting.
The gold standard for detection of predictive ALK rearrangements is currently break-apart fluorescence in situ hybridization (FISH), as it is able to detect all known ALK-rearrangements and was clinically validated in crizotinib clinical trials (4-6). However, the ALK FISH assay is fraught with technical challenges, including FISH signal instability and scoring difficulties. The limitations of diagnosis with FISH have already been reported by Chihara & Suzuki in 2011, making clear that a reconsideration of the diagnostic method is needed for further studies of ALK inhibitors (7-9). An alternative method for determining ALK diagnostic status in NSCLC is to identify ALK protein overexpression using immunohistochemical methods. However, the sensitivity and reproducible detection of ALK by these methods present significant challenges because the differential expression of ALK protein occurs at a low level (10-12). Finally, a multiplex reverse transcription-polymerase chain reaction (RT-PCR)-based assay can sensitively detect certain ALK fusion-gene variants, although reproducible RT-PCR results are difficult to obtain in formalin-fixed paraffin-embedded (FFPE) tissue sections (12,13). Furthermore issues remain, such as the exact definition of ALK positivity, the best method of detection of ALK alterations that are clinically significant, the prognostic significance of these alterations, and whether there should be a preselection of patients for treatment based on the characteristics of molecular testing. Here, we summarize our current knowledge regarding the best methodology to detect EML4-ALK variant types in NSCLC.
ALK testing in non-small cell lung cancer: FISH, immunohistochemistry (IHC) or RT-PCR?
In the era of personalized cancer therapy the establishment of a rapid and cost effective diagnostic algorithm to identify an uncommon molecular subset of NSCLC, such as EML4-ALK is an unmet need (14). Although most patients with NSCLC positive for EML4-ALK derive benefit from treatment with ALK-TKIs, the clinical efficacy of these drugs varies greatly among such individuals and the molecular mechanism underlying this variability is unclear. For instance, crizotinib exerts marked antiproliferative and proapoptotic effects in H3122 cells harboring EML4-ALK v1 fusion, in contrast to H2228 cells that harbor the v3 fusion and are resistant to the effects of the drug (15). Interestingly, the longest variant (v2), which is the least stable, shows the highest degree of sensitivity to ALK inhibition (16). Thus, targeted therapy of ALK-positive lung cancer should take into account the precise ALK genotype. Although FISH has been used to enroll patients in clinical trials, the FDA-approved ALK probe does not identify the specific fusion partner or breakpoint variant. Furthermore, the optimal technique for large-scale screening in a clinical setting has not yet been determined.
In our experience in a cohort of 200 NSCLC patients, the EML4-ALK fusion gene was detected in 12.5% of cases by RT-PCR, more frequently than when compared to IHC (6.7%) or FISH analysis (4.5%). Among the RT-PCR EML4-ALK positive cases are included EML4-variant 1, 2 and 3 with intron 13, 20 or 6 of EML4 fused to intron 19 of ALK, respectively (Figure 1). Furthermore, in our routine cases examined by the three techniques, the EML4-ALK rearrangements can also be detected more frequently by RT-PCR. Among 139 NSCLC patients, EML4-ALK fusion gene was detected in 14 (10.1%) cases by IHC, 8 (5.8%) by FISH, and 24 (17.3%) by RT-PCR.
Figure 1.
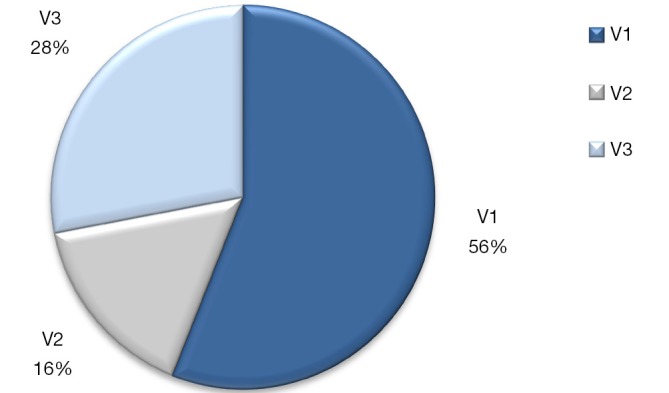
EML4-ALK positive variants in NSCLC tumors. Distribution of the most important EML4-ALK fusion variants.
Our frequency of detection compares favorably with other studies that enriched their study population based on associated clinical and histological characteristics (17-19). In a recent study, RT-PCR detected EML4-ALK variant 1 or 3a/b in 11 of 46 cases (24%) of lung adenocarcinomas that were enriched for wild-type EGFR status. Nine of these 46 cases (20%) were tested positive for variant 1 by RT-PCR; of those 9, only 1 (11%) was unanimously positive by FISH (20). Most similar is a study by Zhang et al., that detected EML4-ALK fusions in 43% (9 of 21) of adenocarcinomas which lacked EGFR and KRAS mutations using RACE-coupled PCR sequencing (19). Finally, in the study of Wallander et al., the FISH interpretation demonstrated great variability and the RT-PCR method was the most sensitive and least-subjective methodology for detection of EML4-ALK fusions (20). In contrast, a large scale screening of 7,344 archival FFPE NSCLC specimens was performed by David Gandara’s group. An EML4-ALK positive transcript was detected by quantitative RT-PCR only in 200 cases (2.7%) (14). A recent study by Tuononen et al., demonstrated that FISH, IHC, RT-PCR, and targeted resequencing can be performed successfully in FFPE tumor specimen material with significant concordance among the four methods (21).
Currently, both the National Comprehensive Cancer Network and the American Society of Clinical Oncology guidelines recommend EGFR mutation and ALK testing on all NSCLC that contain an adenocarcinoma component, regardless of histological grade or dominant histological subtype (22,23). It is worth emphasizing that our group was the first to detect by RT-PCR assay that the EML4-ALK rearrangement can be concomitant with EGFR mutations in a considerable number of EGFR mutant NSCLC patients, which may have a favorable effect on treatment outcome (24).
FISH is a relatively rapid, well-standardized, rather expensive method, which only requires a small amount of tumor biopsy. FISH could not, however, identify the fusion partner, nor can it distinguish between different fusion variants and may miss rare, complex rearrangements. The interpretation of FISH results is rather subjective, and requires specialized training because a considerable variability in interpretation of FISH results between readers has been reported (20). Furthermore, the cutoff point of 15% of positive cells to assess a sample as positive by FISH for EML4-ALK fusions significantly affects test specificity and sensitivity. This cutoff point was chosen, by the FDA-approved companion diagnostic, because it appeared to sit within a natural gap in the continuum of “percentage cells positive” that allows for distinguishing true-positive tumors from the technical background noise of the assay (18).
IHC is a rapid and relatively inexpensive method for diagnosing ALK-rearranged NSCLC that require only a small biopsy sample like FISH analysis. A great advantage of IHC is that tissue morphology is retained during analysis, and thus the detection of ALK expression can be focused on the tumor areas. In theory, IHC detects ALK expression for ALK fusion genes regardless of variant and fusion partner. However, variant and fusion partner may influence protein levels and location (25). Another advantage of IHC is that technical infrastructure is already available in many laboratories, in comparison with FISH, but there is no a FDA-approved ALK antibody. The reported sensitivity and specificity of IHC ALK testing has ranged from 67-100% and from 93-100%, respectively (using FISH as the standard procedure) (26,27); IHC has a very high negative predictive value (≥98.9%) (27). Two ALK IHC tests in particular, D5F3 (by Cell Signaling Technology, USA) and 5A4 (by Novocastra, UK), both of which are commercially available within Europe, are reported to be highly accurate and reliable in predicting ALK rearrangement in NSCLC patients (28,29). We have performed IHC by using ALK antibody clone 5A4, use of which has shown good sensitivity in ALK fusion detection in lung carcinomas (30). To a greater or lesser extent, IHC may eventually replace FISH for ALK testing (26). Several authors have proposed that a two-tiered approach may be appropriate, whereby all patients are initially screened using IHC (10,25). Those with faint, moderate or intense staining indicative of ALK gene expression are then tested by FISH for confirmation of ALK positivity (25). Alternatively, only patients with an IHC score of +1 or +2 are subjected to confirmatory testing by FISH, while those with an IHC score of 3+ are considered ALK-positive (25). In both algorithms, patients with no staining indicative of ALK gene expression are considered ALK-negative and are therefore not retested (25). Such approaches do, however, require clinical validation in large-scale studies. In our experience, the EML4-ALK rearrangement can be detected in 10.1% of cases by IHC, with easily interpreted results. In Europe, use of crizotinib does not specify that a particular diagnostic test for ALK rearrangement must be used, in contrast to the US prescribing information for the drug which requires an FDA-approved test to be used (31).
The third methodology that we evaluated, RT-PCR, may be highly sensitive and specific but requires multiple primer sets to detect all EML4-ALK variants when using FFPE tissue. The accuracy of RT-PCR-based diagnosis depends greatly on the RNA quality of specimens (32). Because variants 1, 2 and 3a/b account for approximately 80% of reported positive cases, testing for only those 3 variants does appear to be clinically practical (4). A significant advantage of RT-PCR is that it can be readily applied to specimens such as bronchial washing fluid, pleural effusion or blood that may not be suitable for preparation of FFPE samples (12). To the best of our knowledge, we are the first to perform longitudinal monitoring of EML4-ALK in platelets by RT-PCR on EML4-ALK positive patients treated with crizotinib and correlate with clinical response as measured by serial radiographic CT (33). Although the specificity of RT-PCR as a screening tool is likely to be extremely high, especially if the amplified regions of combinational DNA (cDNA) are sequenced, the dangers of cross-contamination, for example, from reuse of the same blade in sectioning, means that false-positive results may occur with this highly sensitive technique. Furthermore, some fusion partners or breakpoint variants may be missed if the primers contained in the assay are not specifically designed to evaluate these alterations. When ALK FISH-positive cases were retrospectively analyzed for EML4-ALK transcripts by RT-PCR in the initial phase I study of crizotinib, transcripts could not be detected in 31% of cases (4). Although some of these RT-PCR-negative cases may represent a failure of technique, they also may represent uncommon EML-ALK variants or non-EML4 fusion variants that were being missed by the primer sets used in the assay.
Conclusions
Thanks to the efforts of scientific researchers and more rational design of clinical trials, we are closer to being able to stratify patients on the basis of specific genomic features before treatment selection, which could lead to more precise tailoring of treatment. ALK inhibitors offer a new opportunity to treat cancer patients according to the genetic characteristics of their tumors and, ultimately, improve treatment outcomes. RT-PCR is a sensitive method for detecting EML4-ALK variant 1, 2 and 3a/b that warrants further evaluation. RT-PCR is free from subjectivity in analysis, unlike IHC and FISH. Identification of the specific variant can only be determined using RT-PCR, which may become important in the future for predicting patient response to therapy. Additional primer sets can easily be added to the RT-PCR methodology in the future to detect the remaining 20% of reported EML4-ALK variants. The development of other techniques like targeted resequencing for detection of ALK gene fusions may be further explored. The clinical utility of RT-PCR should be evaluated with regard to ALK expression by FISH and IHC in prospective clinical studies.
Acknowledgements
The authors thank Ms. Sonia Rodriguez, Erika Aldeguer and Zaira Yeste for technical assistance.
Disclosure: The authors declare no conflict of interest.
References
- 1.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [DOI] [PubMed] [Google Scholar]
- 2.Chabner BA. Early accelerated approval for highly targeted cancer drugs. N Engl J Med 2011;364:1087-9. [DOI] [PubMed] [Google Scholar]
- 3.Savic S, Bode B, Diebold J, et al. Detection of ALK-positive non-small-cell lung cancers on cytological specimens: high accuracy of immunocytochemistry with the 5A4 clone. J Thorac Oncol 2013;8:1004-11. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [DOI] [PubMed] [Google Scholar]
- 7.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 2009;15:5216-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chihara D, Suzuki R.More on crizotinib. N Engl J Med 2011;364:776-7; author reply 778. [DOI] [PubMed] [Google Scholar]
- 9.Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thunnissen E, Bubendorf L, Dietel M, et al. EML4-ALK testing in non-small cell carcinomas of the lung: a review with recommendations. Virchows Arch 2012;461:245-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park HS, Lee JK, Kim DW, et al. Immunohistochemical screening for anaplastic lymphoma kinase (ALK) rearrangement in advanced non-small cell lung cancer patients. Lung Cancer 2012;77:288-92. [DOI] [PubMed] [Google Scholar]
- 12.Karachaliou N, Rosell R.Optimal detection of ALK rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:255-6. [DOI] [PubMed] [Google Scholar]
- 13.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008;3:13-7. [DOI] [PubMed] [Google Scholar]
- 14.Li T, Maus MK, Desai SJ, et al. Large-scale screening and molecular characterization of EML4-ALK fusion variants in archival non-small-cell lung cancer tumor specimens using quantitative reverse transcription polymerase chain reaction assays. J Thorac Oncol 2014;9:18-25. [DOI] [PubMed] [Google Scholar]
- 15.Tanizaki J, Okamoto I, Takezawa K, et al. Combined effect of ALK and MEK inhibitors in EML4-ALK-positive non-small-cell lung cancer cells. Br J Cancer 2012;106:763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 2012;18:4682-90. [DOI] [PubMed] [Google Scholar]
- 17.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res 2010;16:5581-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 2010;9:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallander ML, Geiersbach KB, Tripp SR, et al. Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med 2012;136:796-803. [DOI] [PubMed] [Google Scholar]
- 21.Tuononen K, Sarhadi VK, Wirtanen A, et al. Targeted resequencing reveals ALK fusions in non-small cell lung carcinomas detected by FISH, immunohistochemistry, and real-time RT-PCR: a comparison of four methods. Biomed Res Int 2013;2013:757490. [DOI] [PMC free article] [PubMed]
- 22.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 2011;29:2121-7. [DOI] [PubMed] [Google Scholar]
- 23.Cancer NSCL. NCCN Clinical Practice Guidelines in Oncology. Available online: www.nccn.com. Accessed on May 25. Version 3.2012.
- 24.Karachaliou N, Costa C, Gimenez-Capitan A, et al. The concomitant presence of echinoderm microtubule associated protein like 4 - anaplastic lymphoma kinase (EML4-ALK) EML4-ALK fusion gene in EGFR-mutant non-small-cell lung cancer (NSCLC) patients treated with erlotinib or chemotherapy in the EURTAC trial. MO26.08 Topic: Pathology, Session: MO26-Anatomical Pathology II. WCLC 2013; Mini oral.
- 25.Weickhardt AJ, Aisner DL, Franklin WA, et al. Diagnostic assays for identification of anaplastic lymphoma kinase-positive non-small cell lung cancer. Cancer 2013;119:1467-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JA, Bubendorf L, Stahel R, et al. Testing for anaplastic lymphoma kinase rearrangement to target crizotinib therapy: oncology, pathology and health economic perspectives. Expert Rev Anticancer Ther 2013;13:625-36. [DOI] [PubMed] [Google Scholar]
- 27.Yi ES, Chung JH, Kulig K, et al. Detection of anaplastic lymphoma kinase (ALK) gene rearrangement in non-small cell lung cancer and related issues in ALK inhibitor therapy: a literature review. Mol Diagn Ther 2012;16:143-50. [DOI] [PubMed] [Google Scholar]
- 28.Conklin CM, Craddock KJ, Have C, et al. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013;8:45-51. [DOI] [PubMed] [Google Scholar]
- 29.Martinez P, Hernández-Losa J, Montero MÁ, et al. Fluorescence in situ hybridization and immunohistochemistry as diagnostic methods for ALK positive non-small cell lung cancer patients. PLoS One 2013;8:e52261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paik JH, Choi CM, Kim H, et al. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer 2012;76:403-9. [DOI] [PubMed] [Google Scholar]
- 31.Riely GJ, Chaft JE, Ladanyi M, et al. Incorporation of crizotinib into the NCCN guidelines. J Natl Compr Canc Netw 2011;9:1328-30. [DOI] [PubMed] [Google Scholar]
- 32.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013;13:685-700. [DOI] [PubMed] [Google Scholar]
- 33.EML4-ALK monitoring of crizotinib response in blood platelets and plasma of NSCLC patients. 15th World Conference on Lung Cancer October 27-30, 2013; Sydney, Australia. Poster Session 2: Prognostic and Predictive Biomarkers, P2.06-046.


