Abstract
Notwithstanding the encouraging results of the National Lung Screening Trial (NLST) the scientific community still debates on the cost-benefit profile of low dose computed tomography (LDCT) lung cancer screening. Several major concerns regard how to identify subjects at high risk of developing lung cancer, the optimal diagnostic algorithm, the management of lung nodules and the high false positive rates. The use of complementary biomarkers would be a useful strategy for dealing with most of these issues. This short review will focus on candidates’ biomarkers circulating in serum or plasma that already reached an advanced validation phase also in LDCT lung cancer screening series. The biomarkers presented below are examples of the value of searching candidates by looking not only to the tumor itself but also to the interplay between the tumor and the host in order to identify early changes related to the biological reactivity of the host to a developing cancer.
Keywords: Lung cancer, risk prediction, screening, diagnosis, prognosis, biomarkers
Background
Lung cancer is the second most frequent cancer both in man and women and continues to be the leading cause of death from cancer, accounting for over 20% of all cancer deaths in 2012 in Europe (http://globocan.iarc.fr).
The overall 5 years survival rate for lung cancer has risen from only 12% to 16% in the past 4 decades, due largely to the late stage at which most patients are diagnosed. This rate is very small if compared to that observed for the other big killers, colon and breast cancer, where survival exceeds 70% and 50%, respectively. In contrast survival of patients undergoing lung resection for small intrapulmonary cancers is greater than 80%. Thus in lung cancer, more than in any other cancer, early detection is essential to improve survivability through identification and therefore treatment of patients before their cancers become inoperable and lethal.
Imaging modalities and biomarkers
Great enthusiasm was raised by the publication in 2011 of the results of the National Lung Screening Trial (NLST), a randomized clinical screening trial enrolling 53,454 persons with three rounds of low dose computed tomography (LDCT) annual screening versus chest radiographs (1). It demonstrated a 20% reduction of lung cancer mortality and 7% reduction of all cause mortality in favor of LDCT. However, after three rounds of screening, 24.2% of subjects were classified as positive with 96.4% of these being a false positive with the need to screen 320 subjects to prevent 1 lung cancer death.
In a recent paper from the same team the issue of overdiagnosis in the trial was estimated (2). The authors reported an overdiagnosis global rate of >18% and that the number of cases of overdiagnosis in the 320 subjects needed to be screened to prevent 1 lung cancer death is 1.38. Thus reduction of false positive rate after initial screen, as well as reduction of overdiagnosis by more efficient prediction of tumor aggressiveness, represents critical and still unmet clinical needs.
Recently the results of three smaller European LDCT screening randomized trials were published and have reported non-significant mortality reductions (3-5). Two studies, the Multicentric Italian Lung Detection (MILD) (3) and the Danish Lung Cancer Screening Trial (DLCST) (5) showed a higher mortality in the screened LDCT arm and a meta-analysis of the four published studies demonstrated a small benefit in lung cancer mortality reduction (3).
In a systematic review of all randomized clinical trials that examined the benefits and harms of LDCT screening, the average nodule detection rate was around 25%, with 96% of nodules being benign. These high false positive rates of LDCT lead to multiple screening rounds and related radiation exposure, the use of unnecessary and sometimes harmful diagnostic follow-up and increased time and costs. The development of non-invasive complementary biomarkers could thus be very helpful for the reduction of subjects needed to be followed up and potentially to decrease false positive rate of CT scans and the over-diagnosis rate (Figure 1).
Figure 1.
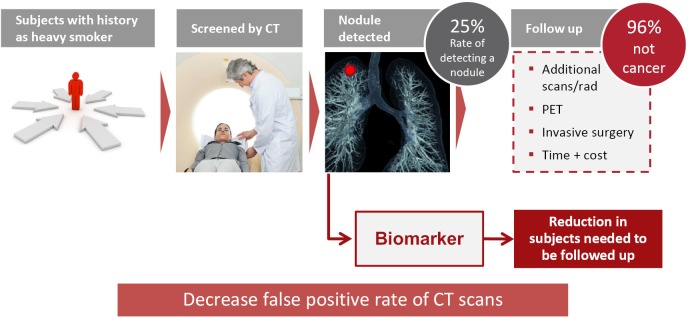
Clinical utility of biomarkers.
Biomarkers circulating in plasma or serum, if properly validated, could constitute the gold standard for a non invasive cancer diagnostics. In fact blood thanks to its rich content of different cellular and molecular elements that provide information on the health status of an individual, constitutes the ideal compartment to be tested for developing biomarkers. Moreover, blood samples can be easily and inexpensively collected by non invasive procedures throughout large clinical trials.
Several authors have based their biomarkers discovery strategy starting from the assumption that novel promising biomarkers are generated not only by cancer cells but also from the tumor microenvironment, the host response and their dynamic interaction. The cross talk among these components can be reflected in peripheral circulation and generate diagnostic and prognostic biomarkers and potentially, also biomarkers predicting the risk of disease development.
Table 1 reports the most promising candidate biomarkers for early lung cancer diagnostics detected in blood and their respective development phases according to the guide-lines published in JNCI (6) and taking also into account the workflow for biomarkers validation described by other authors (7,8).
Table 1. Circulating biomarkers for early lung cancer.
| Phase 1 | Phase 2 | Phase 3 | Phase 4 | Phase 5 | |
|---|---|---|---|---|---|
| Candidates | Discovery, prediction | Assay validation | Retro-longitudinal | Prospective screening | Cancer Control |
| Autoantibodies (earlyCDT-test) | × | × | × | × | |
| C4d protein | × | × | × | ||
| Serum microRNA | × | × | × | ||
| Plasma microRNA (MSC test) | × | × | × | × |
Several biomarkers have reached phase 3 which evaluates, as a function of time before clinical diagnosis, the capacity of the biomarker to detect preclinical disease. However, only few of them reached phase 4, prospective screening, which studies screen people and lead to diagnosis and treatment. None of them has reached so far phase 5, the final phase that will address whether screening with selected biomarkers will result in an overall benefit for the screened population by impacting on survival. A good biomarker should reduce the burden of cancer and would be not useful if it does not lead to change in treatments or outcomes and if it is only efficient in picking up indolent cancers.
However, concerning biomarkers, it must be recognized that there is a disconnection between promise and product and several reasons could be evoked:
Discovery methods are often neither reliable nor efficient. This is in part related to the rapidly changing technology;
Selection of candidates: the choice of tumor-specific or high-throughput approaches. In particular genetic heterogeneity of tumors has limited the success of these initiatives;
Reproducibility of the laboratory assays: several studies have to deal with over fitting, and lack of cross-validation and external validation;
Most studies have poor design, just rely on case-control comparison and are not in the clinical context;
The low concentration of analytes to be measured influences the reproducibility of the results;
The availability of very few prospective collections of biological samples and in particular of bio-repositories related to screening trials.
Blood-based biomarkers
This review will focus on candidates’ biomarkers circulating in serum or plasma since they are so far those that reached the more advanced validation phase.
All the studies selected in this review have validated their biomarkers in the context of LDCT lung cancer screening trials, by studying high risk subjects, and showed to be of value to predict the risk of lung cancer in asymptomatic individuals.
The biomarkers presented below are also examples of the value of searching candidates by looking not only to the tumor but also to the interplay between the tumor and the host in order to identify early changes related to the biological reactivity of the host to an incipient cancer.
Immune response biomarkers
C4d complement split product (9)—Phase of development: phase 2
These authors used an alternative approach not looking for cancer but for the immune response to cancer. In fact, immune activation may generate host-derived markers that are more homogeneous than cancer-derived markers. Immune responses against intracellular and surface tumor antigens are well documented in patients with lung cancer (10). In particular, the complement system is activated in lung tumor cells (11-14). Complement is a central component of innate immunity that plays an essential role in immune surveillance and homeostasis (15).
In their study these authors showed that lung tumors activate the classical complement pathway and generate C4d, a degradation product of this pathway and they evaluated if C4d may be of value for the diagnosis and prognosis of lung cancer.
They first examined plasma samples from 50 patients with early (stage I-II), clinically detected lung cancer and showed statistically significantly higher levels of C4d than those from 50 matched control subjects. The area under the ROC curve was 0.782 (P<0.001). Patients with higher levels of C4d (>3 µg/mL) had a statistically significantly shorter overall survival than those with low C4d levels (P=0.002). They also measured the levels of C4d in paired plasma samples (pre- and post-surgery) from 25 lung cancer patients with high (>2 µg/mL) C4d levels in the pre-surgery plasma. In all but one case, C4d levels were reduced after surgical removal of the tumor (P<0.001). As expected, in 19 patients with low plasma C4d levels (<2 µg/mL), the concentration of the marker did not change after resection of the tumor. These results provided evidence that plasma C4d levels depend on the presence of the tumor.
Plasma C4d levels were further evaluated in plasma samples from 190 asymptomatic individuals enrolled in a LDCT screening program. Thirty-two of them were diagnosed with lung cancer in the context of the program while the remaining 158 individuals had no evidence of cancer after LDCT screening. Both groups were matched by sex, age, and smoking history. Plasma C4d levels were statistically significantly higher in individuals with lung cancer than in individuals without the disease.
This result suggests that C4d levels may be of value to predict the risk of lung cancer in asymptomatic individuals. Additional validation sets are required to establish reliable cutoff values of this biomarker and it would be also critical to evaluate the performance of the test in specific clinical applications (e.g., in the context of a screening program) or in a cohort of prospectively collected patients presenting with one or more lung nodules discovered by chest LDCT.
Autoantibody signature (16)—Phase of development: phase 4
A more advanced and validated biomarker is the Autoantibody (AAB) signature developed by the group of Richardson JF in United Kingdom and now released by Oncimmune USA LLC.
It is well established that cancer patients produce autoantibodies to tumor proteins that are mutated, misfolded, ectopically presented, over-expressed, aberrantly degraded or anomalously glycosylated.
These authors discovered a 7 AAB signatures, previously 6 AAB, against oncogenes and TSG involved in lung cancer and also in other tumors: CAGE, GBU 4–5, HER2, p53, c-myc, NY-ES0-1 and MUC1. The strength of this AAB signature, called EarlyCDT-Lung test, is that it was validated in large series of patients and controls including either early and late stages tumors, NSCLC and SCLC. Across the various series, the signature showed high specificity, around 93%, but quite low sensitivity ranging around 40% in NSCLC and 55% in SCLC (Table 2) (16-20). However the test has the advantage to rely in an Elisa assay that is easily accomplished in a clinical laboratory.
Table 2. Performances of the autoantibody EarlyCDT®-Lung test.
| Cases | Controls | Sensitivity | Specificity | |
|---|---|---|---|---|
| Case-control studies | 235 | 266 | 41% | 91% |
| Clinical audit dataset | 61 | 1,538 | 41% | 87% |
| CT-detected lung nodules | 43 | 146 | 44% | 88% |
In a recent paper (21) the test’s performance characteristics in routine clinical practice were evaluated by auditing clinical outcomes of 1,600 US patients deemed at high risk for lung cancer by their physician, who ordered the EarlyCDT-Lung test for their patient.
The results obtained mirrored that of the extensive case–control training and validation studies previously reported (17-19,22). This audit has confirmed that EarlyCDT-Lung detects all types of lung cancer, all stages of the disease, and performs in clinical practice with the same sensitivity and specificity measured in the case–control studies. This is, therefore, the first autoantibody test that detects early stage lung cancer as shown with prospective validation data on a large number of individuals from a routine clinical practice setting (Table 2).
Recently Massion et al. evaluated the performance of the 7 AAB test in 189 lung nodules detected by LDCT, of which 43 malignant and 146 benign, and reported that EarlyCDT- Lung Oncimmune can provide significant discrimination between malignant and non-malignant lung nodules with sensitivity 44.2%, specificity 88.4%, PPV 52.8%, NPV 84.3%, with even better performance for nodules between 8-20 mm of diameter (Table 2) (unpublished data).
A prospective study is ongoing in Scotland (ECLS study) with the purpose to assess the value of the EarlyCDT-Lung test as a pre-CT screening tool. The study will enroll 10,000 people (50-75 yrs, smokers or ex-smokers) from Glasgow and the surrounding areas. Half of those taking part will be offered the EarlyCDT-Lung test (lung cancer test group). The other half (non-test group) will also have their blood taken, but it will not be tested as part of this study. People who have a positive lung cancer blood test will get a chest X-ray and a lung scan and 6 monthly scans for 2 years. However, only 1 in 9 people with a positive test is expected to develop LC within 2 years. People with a negative lung cancer blood test and those in the non-test group will not get any X-rays or scans will be monitored by their GP as normal: 98-99/100 people with a negative test are expected to not have LC at that time.
This study will potentially give insights on the utility of this biomarker as a first-line test to select subjects at increased risk for lung cancer development who need to undertake regular LDCT, potentially avoiding radiological exposure to low risk individuals with a negative test.
Blood circulating miRNAs
Circulating microRNA in plasma and serum are promising biomarkers for a non invasive cancer diagnostics. After being transcribed in the nucleus, pre-miRNA molecules can be processed further by Dicer in the cytoplasm. In addition, based on recent findings there are at least two ways that pre-miRNAs can be packaged and transported using exosomes and MVBs or other (not fully explored) pathways together with RNA-binding proteins. After fusion with the plasma membrane, MVBs release exosomes into the circulating compartments and bloodstream. Likewise, pre-miRNA inside the donor cell can be stably exported in conjunction with RNA-binding proteins, such as NPM1 and Ago2, or by HDL (23). Circulating miRNAs enter the bloodstream and are taken up by the recipient cells by endocytosis or, hypothetically, binding to receptors present at the recipient cellular membrane capable of recognizing RNA-binding proteins. More studies are necessary to elucidate how miRNAs are loaded into exosomes and how they can be internalized by recipient cells. Exosomal miRNAs are processed by the same machinery used in miRNA biogenesis and thus have widespread consequences within the cell by inhibiting the expression of target protein-coding genes.
Thus, for their nature and biogenesis, miRNAs seem to remain rather intact and stable in biological fluids and, importantly, they are detectable quantitatively with simple assays (i.e., RT-qPCR) that are suitable also in a clinical context.
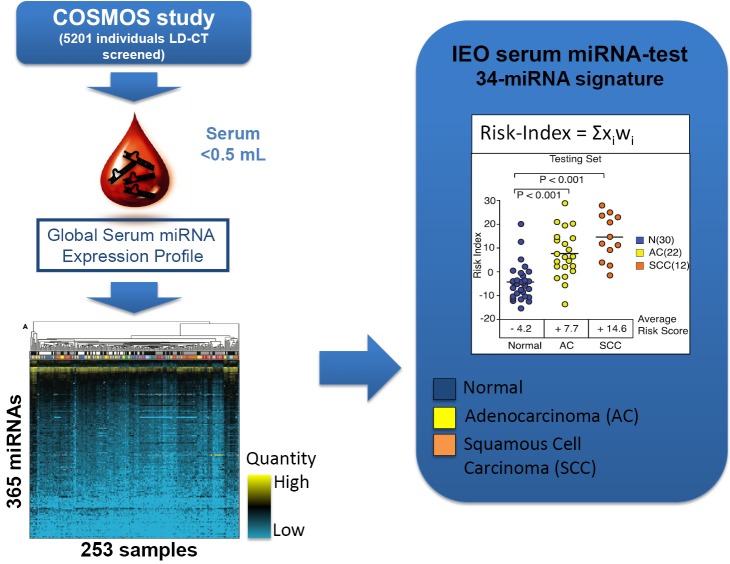
Serum-based 34 miRNA signature (24)—Phase of development: phase 4
The group of F. Bianchi at European Institution of Oncology (Milan, Italy) has developed a blood test for lung cancer diagnosis in asymptomatic high-risk individuals (heavy smokers, aged over 50) based on the detection of miRNAs from serum. Sera were collected from high-risk subjects enrolled in a large prospective early detection trial (the COSMOS study) for lung cancer by annual LD-CT. Starting from a total of 365 miRNA assay (microfluidic cards) the authors selected a pool of 147 miRNAs that were informative in a total of serum 253 samples from lung cancer screening patients and controls (COSMOS), symptomatic lung cancer patients and as a control group, a breast cancer and benign nodules series (Figure 2).
Figure 2.
Flowchart of the COSMOS study.
They used the training set to derive a diagnostic 34-miRNA signature capable of separating tumor from normal sera. As discriminant predictor a risk index was calculated based on the inner sum of the weights (wi) and expression (xi) of the 34 miRNAs greater than the threshold determined in the training set (S wi xi>3.235).
The performance of the IEO test in the validation set was 71% sensitivity, 90% specificity and 80% accuracy with better performance in stage II-IV only (30 normal/12 tumors) with 82% sensitivity, 90% specificity and 90% accuracy.
An analysis of the 34-miRNA model prediction strength in the testing set (all, 30 normal and 34 tumors) stratified by available clinical-pathological parameters showed odds ratio higher in Stage II-IV disease, in squamous carcinoma and in women.
When the 34-miRNA predictor was applied to evaluate the risk in a symptomatic set of 36 NSCLC patients and in 15 pulmonary hamartomas, it performed remarkably well.
By comparing the performance of the predictor in the normal sera of the testing set and in the sera of patients with the LDCT-detected benign nodules no significant differences in the average risk of the normal and nodule categories were found.
The authors also analyzed a group of sera collected before the onset of NSCLC (i.e., from patients who were negative at the screening round but who developed lung cancer >1 year after). For 13 of such cases, both the sera harvested before disease onset (BDO) and the tumor sera that were already included in the training or testing sets were available. When the risk predictor algorithm was applied, it indicated a significantly increased average risk index for sera collected after the onset of the disease (average risk BDO, 7.1; tumor, 10.4; P<0.001, paired t-test). Thus, at least in the cases analyzed, the 34-miRNA model was capable of detecting the conversion from a normal to a malignant state.
Finally, they tackled the question of the specificity of the 34-miRNA predictor for NSCLC detection, as opposed to other types of cancer, by screening sera from a cohort of 18 patients with invasive ductal breast carcinoma and 10 with breast benign nodules. When the 34-miRNA risk predictor algorithm was applied, it could not discriminate between breast tumors and benign breast nodules.
Plasma-based miRNA signature (25)—Phase of development: phase 4
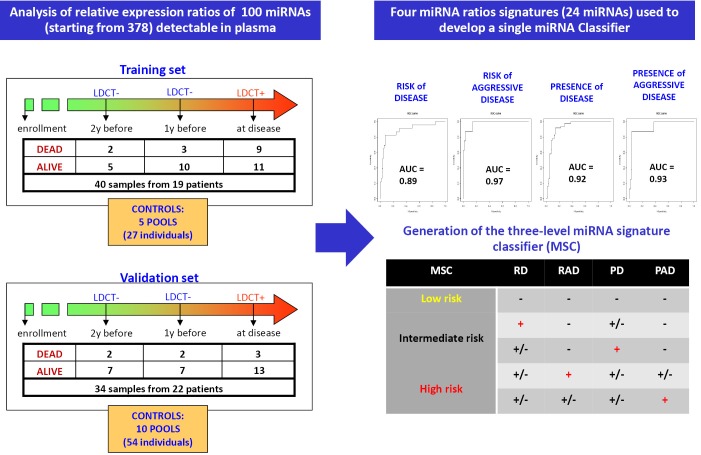
In our first exploratory study we investigated miRNA profiles in plasma samples collected before and at time of disease detection in subjects enrolled in the first observational trial and we validated selected miRNAs signatures in an independent series of subjects belonging to the randomized MILD trial (25). High-throughput miRNA expression profiles of plasma samples using TaqMan microfluidic cards and single assays for validation studies were performed and, importantly, we generated an original method to analyze data by looking at reciprocal miRNA ratios, an approach that allowed us to bypass the controversial issue of data normalization of miRNA in plasma. In this way, we identified 24 miRNAs whose reciprocal ratios were able to discriminate patients at risk of developing lung cancer and at risk for aggressive disease development in samples collected before disease detection, as well as diagnostic and prognostic signatures in plasma collected at the time of disease detection (Figure 3).
Figure 3.
miRNA signatures discovery and initial validation.
In order to have a more friendly and useful tool to classify plasma samples in clinical trials we recently generated a three-level risk categorization for disease: low, intermediate and high miRNA signature classifier (MSC) by combining the different signatures (Figure 4) and we used this pre-specified classifier to test diagnostic and prognostic performance in a Clinical Validation Study using the Multicentric Italian Lung Detection (MILD) Trial [2005-2012] cohort.
Figure 4.
Comparison between serum- (IEO) and plasma-based (INT) miRNA tests.
For this study, 1,000 consecutive plasma samples collected from June 2009 to July 2010 among lung cancer-free individuals enrolled in the trial were used to determine the specificity of the MSC. Plasma samples were first assayed for hemolysis to remove samples from patients that were potentially contaminated by red blood cells miRNAs (26,27).
Of the 1,000 samples, 130 were not evaluable because of hemolysis. Of the remaining 870 subjects, 594 (68%) belonged to the LDCT arms and 276 (32%) to the observational arm. To obtain a cohort for determining the sensitivity performance of MSC, plasma samples from almost all patients with lung cancer diagnosed by September 2012 were obtained (N=85). For 69 of these 85 patients, at least one evaluable sample was collected. For all patients we considered the sample closest to LDCT examination resulting in cancer diagnosis. Specifically, a sample at-diagnosis was available for 50 patients and a pre-disease sample for 19 patients. The pre-disease samples were collected from 8 to 35 months before lung cancer detection with a median lag time of 18 months.
Diagnostic and prognostic performance of MSC
MSC risk groups were examined for all 939 subjects according to lung cancer occurrence, lung cancer death, and tumor stage. MSC Intermediate and High correctly classified 60 of 69 lung cancer patients with 87% SE, 81% SP, 27% PPV and 99% NPV (Table 3). MSC risk groups were not significantly associated (P=0.40) with varying tumor stage (I, II-III or IV). No significant differences were observed between MSC risk groups and histological subtypes (χ12=1.60, p=0.4485), and between adenocarcinoma and squamous cell carcinoma (χ12=0.55, P=0.759).
Table 3. Overall diagnostic performance of MSC.
| Total | MSC (risk of lung cancer) |
|||
|---|---|---|---|---|
| High (%) | Intermediate (%) | Low (%) | ||
| All subjects | 939 | 63 (6.7) | 159 (16.9) | 717 (76.4) |
| No lung cancer | 870 | 32 (3.7) | 130 (14.9) | 708 (81.4) |
| Lung cancer | 69 | 31 (44.9) | 29 (42.0) | 9 (13.0) |
MSC, miRNA signature classifier.
Time dependency analysis of diagnostic performance of MSC, showed similar values of SE, SP, PPV and NPV at 6-, 12-, 18- and 24-month intervals between blood sampling and lung cancer diagnosis supporting a strong diagnostic performance of MSC to predict LC development up to 24 months before disease detection.
Complementary diagnostic performance of LDCT and MSC
Restricting the analysis to the total of 652 subjects in the LDCT arm, LDCT identified 46 of 58 lung cancer subjects missing three patients with no pulmonary nodule detected and nine patients because of an interval cancer for a SE of 79%. Pre-specified binary risk groups of MSC (considering High and Intermediate versus Low) identified 40 of 46 LDCT-detected cancers, 8 of 9 interval cancers and all three subjects with “no pulmonary nodule”.
LDCT had a SP of 81% for the clinically actionable subgroup of non-calcified nodules >5 mm and an associated false positive rate of 19.4% (115/594). When double-positive (LDCT and MSC) subjects were considered, the false positive rate decreased to 3.7% (22/594), with a decrease in SE (40/58, 69%). On the other hand, MSC detected 9 of 11 (82%) lung cancers that occurred in the observational arm.
The 5-fold reduction in false positives obtained by combining the MSC Lung Cancer assay to the results of the LDCT scan is of great clinical relevance in the context of reducing the false positive rate and the potential side effects associated with repeated LDCT scans or other unnecessary invasive diagnostic follow-ups.
Association of MSC risk groups with survival
The prognostic performance of the three pre-defined MSC risk groups to predict overall survival from plasma samples collected for all subjects with 3-year follow-up (N=939) was also evaluated. Three-year survival was 100%, 97% and 77% for Low, Intermediate and High respectively. The difference in survival between High/Intermediate and Low MSC was statistically significant (χ12=49.53, P<0.0001) also after adjustment for age and gender (χ12=12.57, P=0.0004).
This correlative study in lung cancer is the first of its kind, validating a biomarker using prospectively collected blood samples from a large randomized lung cancer screening trial. In addition to a significant reduction in the rate of false positive results, the performance of the MSC Lung Cancer assay was independent of the stage of lung cancer, as well as the time prior to detection of cancer with LDCT. This suggests additional potential utility for diagnosis and early detection with the MSC Lung Cancer assay.
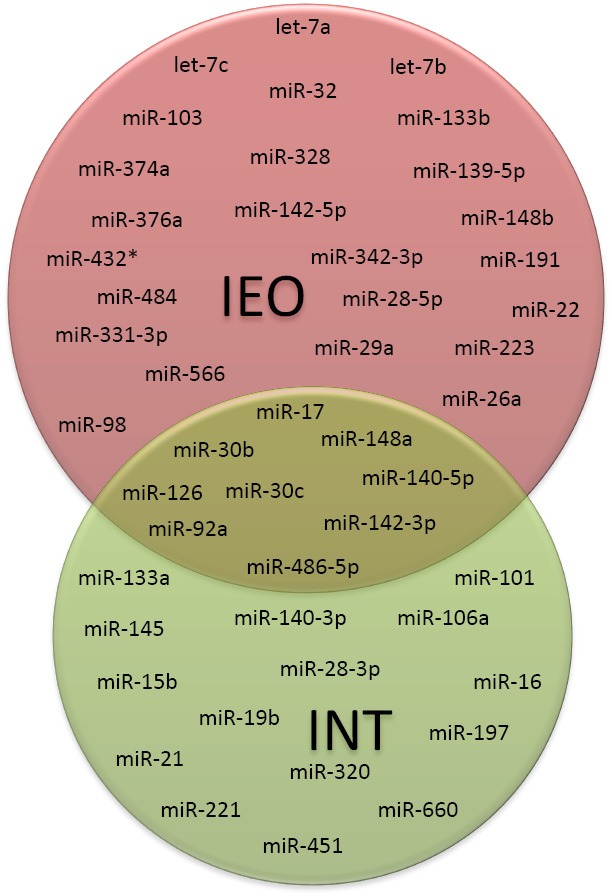
Comparison between serum and plasma-based miRNA tests
Between the two miRNA signatures developed in serum and plasma, only nine miRNAs were overlapping, suggesting the relevance of this core of miRNAs for early lung cancer diagnosis (Figure 4).
The differences in the remaining miRNAs composing the signatures may be likely related to the type of biological samples used (i.e., serum vs. plasma) and the study design. In fact, our findings and those reported in literature suggest that miRNAs not released in physiological process, as during the cell lysis that occur during clot formation in serum samples, have a different physical state than miRNAs physiologically released and protected by lipoproteic complex or microvesicles (28,29). Moreover, the plasma signature was trained in samples of patients collected also before (and at the time of) disease detection, thus reflecting earlier, microenvironment-related changes whereas the serum-based signature was trained in serum samples of patients at the time of lung cancer diagnosis likely detecting more advanced tumor-specific changes.
A large validation phases in two different prospective screening trials in ongoing for both miRNA tests.
Conclusions
Early detection candidate biomarkers exist but only few of them are validated or tested in screening settings. The priority is now to validate existing candidates.
Biomarkers should provide knowledge about added value and therefore should be integrated to clinical, laboratory and imaging (LDCT) routine data.
To demonstrate clinical utility requires significant investment in effort and resources towards prospective biomarkers driven clinical trial.
Acknowledgements
Supported by Investigator Grants No. 14318 and 12162 (Special Program “Innovative Tools for Cancer Risk Assessment and early Diagnosis,” 5×1,000) from the Italian Association for Cancer Research, Grant No. RF-2010 from the Italian Ministry of Health, Grant EDRN UO1 CA166905 from the National Cancer Institute, and by Gensignia inc.
Disclosure: Gabriella Sozzi and Mattia Boeri are coinventors for two patent applications regarding the MSC.
References
- 1.National Lung Screening Trial Research Team , Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patz EF, Jr, Pinsky P, Gatsonis C, et al. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern Med 2014;174:269-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastorino U, Rossi M, Rosato V, et al. Annual or biennial CT screening versus observation in heavy smokers: 5-year results of the MILD trial. Eur J Cancer Prev 2012;21:308-15. [DOI] [PubMed] [Google Scholar]
- 4.Infante M, Cavuto S, Lutman FR, et al. A randomized study of lung cancer screening with spiral computed tomography: three-year results from the DANTE trial. Am J Respir Crit Care Med 2009;180:445-53. [DOI] [PubMed] [Google Scholar]
- 5.Saghir Z, Dirksen A, Ashraf H, et al. CT screening for lung cancer brings forward early disease. The randomised Danish Lung Cancer Screening Trial: status after five annual screening rounds with low-dose CT. Thorax 2012;67:296-301. [DOI] [PubMed] [Google Scholar]
- 6.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001;93:1054-61. [DOI] [PubMed] [Google Scholar]
- 7.Baker SG, Kramer BS, McIntosh M, et al. Evaluating markers for the early detection of cancer: overview of study designs and methods. Clin Trials 2006;3:43-56. [DOI] [PubMed] [Google Scholar]
- 8.Pass HI, Beer DG, Joseph S, et al. Biomarkers and molecular testing for early detection, diagnosis, and therapeutic prediction of lung cancer. Thorac Surg Clin 2013;23:211-24. [DOI] [PubMed] [Google Scholar]
- 9.Ajona D, Pajares MJ, Corrales L, et al. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst 2013;105:1385-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd FA, Douillard JY, Blumenschein GR, Jr. Immunotherapy for non-small cell lung cancer: novel approaches to improve patient outcome. J Thorac Oncol 2011;6:1763-73. [DOI] [PubMed] [Google Scholar]
- 11.Kay AB, Smith AF, McGavin CR, et al. Immunoglobulins and complement in pleural effusions associated with bronchogenic carcinoma. J Clin Pathol 1976;29:887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gmiński J, Mykała-Cieśla J, Machalski M, et al. Immunoglobulins and complement components levels in patients with lung cancer. Rom J Intern Med 1992;30:39-44. [PubMed] [Google Scholar]
- 13.Varsano S, Rashkovsky L, Shapiro H, et al. Human lung cancer cell lines express cell membrane complement inhibitory proteins and are extremely resistant to complement-mediated lysis; a comparison with normal human respiratory epithelium in vitro, and an insight into mechanism(s) of resistance. Clin Exp Immunol 1998;113:173-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ajona D, Castaño Z, Garayoa M, et al. Expression of complement factor H by lung cancer cells: effects on the activation of the alternative pathway of complement. Cancer Res 2004;64:6310-8. [DOI] [PubMed] [Google Scholar]
- 15.Ricklin D, Hajishengallis G, Yang K, et al. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 2010;11:785-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman CJ, Murray A, McElveen JE, et al. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax 2008;63:228-33. [DOI] [PubMed] [Google Scholar]
- 17.Chapman CJ, Healey GF, Murray A, et al. EarlyCDT®-Lung test: improved clinical utility through additional autoantibody assays. Tumour Biol 2012;33:1319-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam S, Boyle P, Healey GF, et al. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 2011;4:1126-34. [DOI] [PubMed] [Google Scholar]
- 19.Boyle P, Chapman CJ, Holdenrieder S, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol 2011;22:383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Healey GF, Lam S, Boyle P, et al. Signal stratification of autoantibody levels in serum samples and its application to the early detection of lung cancer. J Thorac Dis 2013;5:618-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jett JR, Peek LJ, Fredericks L, et al. Audit of the autoantibody test, EarlyCDT®-lung, in 1600 patients: an evaluation of its performance in routine clinical practice. Lung Cancer 2014;83:51-5. [DOI] [PubMed] [Google Scholar]
- 22.Chapman CJ, Thorpe AJ, Murray A, et al. Immunobiomarkers in small cell lung cancer: potential early cancer signals. Clin Cancer Res 2011;17:1474-80. [DOI] [PubMed] [Google Scholar]
- 23.Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011;8:467-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirschner MB, Edelman JJ, Kao SC, et al. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet 2013;4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 2011;57:833-40. [DOI] [PubMed] [Google Scholar]
- 29.Wang K, Yuan Y, Cho JH, et al. Comparing the MicroRNA spectrum between serum and plasma. PLoS One 2012;7:e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]