Abstract
Proteins that cap the ends of the actin filament are essential regulators of cytoskeleton dynamics. While several proteins cap the rapidly-growing barbed end, tropomodulin (Tmod) is the only protein known to cap the slowly-growing pointed end. The lack of structural information severely limits our understanding of Tmod’s capping mechanism. We describe crystal structures of actin complexes with the unstructured N-terminal and the leucine-rich repeat C-terminal domains of Tmod. The structures and biochemical analysis of structure-inspired mutants showed that one Tmod molecule interacts with three actin subunits at the pointed end, while also contacting two tropomyosin molecules on each side of the filament. We found Tmod achieves high affinity binding through several discrete low-affinity interactions, which suggests a mechanism for controlled subunit exchange at the pointed end.
The proteins that cap the ends of the actin filament play important roles in actin-driven processes such as cell migration and organelle trafficking by controlling the addition and dissociation of actin subunits at filament ends. Several proteins cap the barbed end of the filament, including capping protein (CP) and some gelsolin-family members (1, 2). In contrast, Tmod is the only protein known to cap the pointed end of tropomyosin (TM)-coated actin filaments (3). Four Tmod isoforms work in conjunction with one of several TM isoforms to stabilize actin structures characterized by a uniform distribution of the lengths of actin filaments. These structures include the sarcomere of cardiac and skeletal muscle cells and the spectrin-based membrane skeleton (3, 4).
The mechanism by which Tmod caps the pointed end is poorly understood. Quantification in skeletal muscle and erythrocytes led to the proposal that two Tmod molecules cap the pointed end (5, 6). In vitro, however, one Tmod molecule is sufficient to block pointed end elongation of TM-coated filaments (7), consistent with the unique domain architecture of Tmod, which harbors two actin- and two TM-binding sites. Thus, the N-terminal ~160-aa region is mostly unstructured in isolation (8), but contains three predicted helical segments that bind TM, actin, and TM in that order (9, 10). This region displays TM-dependent capping activity (9). Most of the C-terminal region (human Tmod1 residues 161–359) consists of a leucine-rich repeat (LRR) domain (11). This region displays limited capping activity on its own (9). While Tmod binds with nanomolar affinity to the pointed end (12), and even greater affinity in the presence of TM (13), it does not form an absolute cap. Instead, Tmod functions as a “leaky” cap, determining the length of the actin filaments while allowing for the controlled addition/dissociation of actin subunits at the pointed end (14). In the absence of high-resolution structures of the pointed end, rationalization of the existing data is difficult, and several models exist, featuring either one (10, 11) or two (5, 6) Tmod molecules at the pointed end.
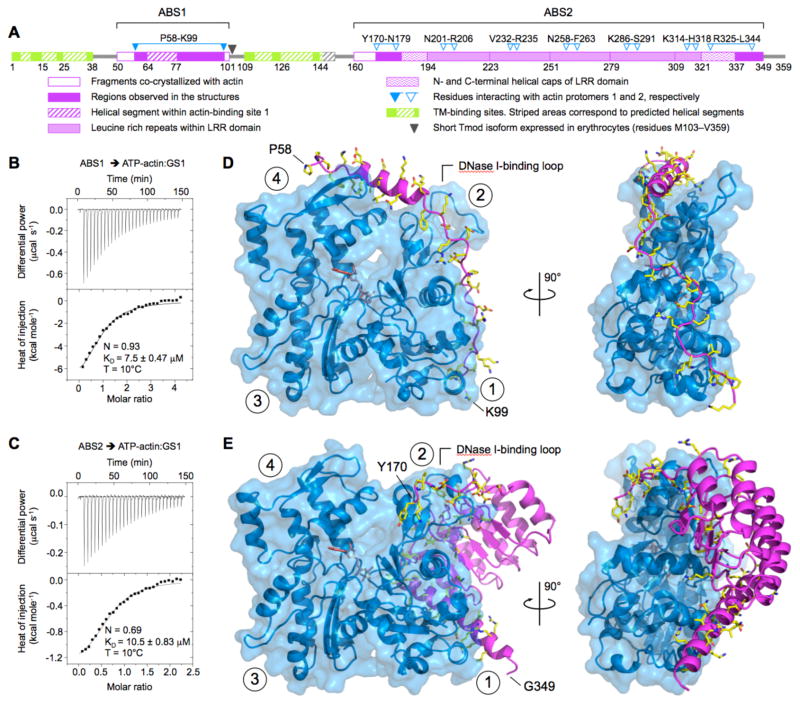
Actin polymerization prevents crystallization of capping complexes. We thus attempted crystallization of the N- and C-terminal actin-binding sites (ABS1 and ABS2) of Tmod in complex with monomeric actin. However, both sites bound with weak affinity to monomeric actin (see below), and polymerization persisted during crystallization. A solution was found by fusing ABS1 and ABS2 C-terminally to gelsolin segment 1 (GS1) via a 9-aa flexible linker (crystallization strategies are described in (15)). The Tmod fragments extended beyond the actin-binding sites defined previously (9, 16–18), with ABS1 and ABS2 comprising human Tmod1 residues 50–101 and 160–349, respectively (Fig. 1A). Importantly, ABS1 and ABS2 both bound actin:GS1 with 1:1 stoichiometry and with similar affinities (KD 7.5 μM and 10.5 μM for ABS1 and ABS2, respectively) when not connected by a linker (Fig. 1, B and C). Similar binding affinities were obtained at two different temperatures, 10°C and 20°C, and with ATP- or ADP-actin (fig. S1).
Fig. 1. Structures of Tmod’s ABS1 and ABS2 bound to actin.
(A) Domain diagram of human Tmod1, showing the interactions of ABS1 and ABS2 with actin. (B–C) ITC titrations of ABS1 (400 μM) (B) and ABS2 (632 μM) (C) into 20 μM and 60 μM of ATP-bound actin:GS1. For both titrations the best fit of the data corresponds to a one-site binding isotherm. The reported errors correspond to the s.d. of the fits. (D–E) Two perpendicular views of the structures of ABS1 (D) and ABS2 (E) bound to actin (blue). GS1, fused N-terminally to the two Tmod domains, is omitted from this figure, but shown in fig. S2. The side chains of Tmod residues that fall near actin are shown (colored by atom type: carbon, yellow; oxygen, red; nitrogen, blue). Circled numbers indicate actin subdomains 1 to 4. Note that, in the actin filament, subdomains 2 and 4 are exposed at the pointed end.
The complexes of ATP-actin with GS1-ABS1 and GS1-ABS2 crystallized under slightly different conditions, and with different unit cell parameters (15) (table S1). The structures were determined to 1.8 Å and 2.3 Å resolution for ABS1 and ABS2, respectively (Fig. 1, D and E, and fig. S2, A and B). Both structures were well defined in the electron density maps (fig. S2, C and D). The flexible linkers between GS1 and the Tmod fragments, and residues 50–57 and 100–101 of ABS1 and 160–169 of ABS2 were not visualized. These residues likely do not interact with actin, because the C-termini of ABS1 and ABS2 projected away from actin, and weak electron density that could not be modeled was also observed projecting away from actin at their N-termini. A structure of ABS1 was also obtained with ADP-actin at 2.15 Å resolution, and showed a similar conformation as that observed with ATP-actin (15). Crystals of ABS2 could not be obtained with ADP-actin.
The Tmod structures differ substantially from previously characterized actin complexes (19); while most actin-binding proteins bind in the cleft between actin subdomains 1 and 3, ABS1 and ABS2 bound at the pointed end of the actin monomer, and on the side of subdomains 2 and 1 (Fig. 1, D and E, and movies S1 and S2). An unusual feature of the structures was an interaction with the DNase I-binding loop in actin. As a result, this loop, which is disordered in most actin structures, was ordered in the Tmod complexes, albeit with different conformations in the two structures.
ABS1 extended beyond the α-helix suggested previously, with a total of 42 residues (P58-K99) mediating the interaction (Fig. 1, A and D). ABS1 adopted a rather extended conformation and contacted three out of the four actin subdomains. The α-helix was longer than anticipated, comprising residues 64–77. This helix played a crucial role in the interaction, by forming a bridge across the nucleotide-binding cleft on top of actin subdomains 4 and 2. As a result, the nucleotide cleft was slightly more closed in this structure than in that of ABS2.
The interactions of ABS2 were also more extensive than anticipated, and involved contacts along the entire region Y170-L344. Residues Y170-N179 formed a loop that interacted with the DNase I-binding loop in actin. Several additional contacts were mediated by the LRR domain, whose boundaries differ compared to the description of the structure of uncomplexed chicken Tmod1 residues 179–344 (11). The LRR motif is defined as a β-strand-loop-α-helix module (20). The loop of the motif is called the “ascending loop”, whereas the loop connecting one motif to the next is called the “descending loop”. By this criterion, Tmod contains not five but four and a half LRR motifs (Fig. 1A). The ascending loops typically mediate protein-protein interactions (20), and this principle was conserved in Tmod, where the ascending loops interacted on the back side (according to the classical view) of actin subdomains 2 and 1 (Fig. 1A and E). Most LRR domains have N- and C-terminal caps, which shield the hydrophobic core of the domain from solvent exposure (20). In Tmod, these caps consisted of α-helices that ran diagonally to the first and last LRR motifs: D182-N193 and Q321-N336. The C-terminal cap continued as an uninterrupted α-helix that ended with residue T348. This helix interacted along most of its length (R325-L344) with actin subdomain 1.
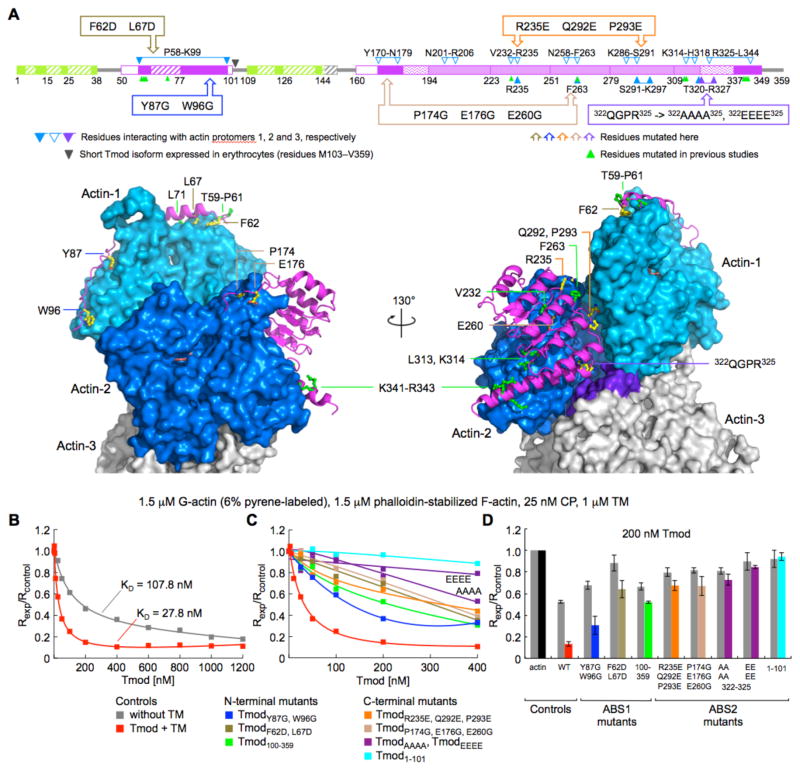
The structures of ABS1 and ABS2 were unequivocally overlaid onto the pointed end of the actin filament (21), producing a model of the pointed end that was then tested by mutagenesis (15). First, the structures showed that Tmod could only bind at the pointed end, because both ABS1 and ABS2 covered surfaces that are buried in the filament. Second, ABS1 and ABS2 had to bind to two different protomers at the pointed end, because their binding surfaces partially overlapped (fig. S3A). Third, the binding surfaces of ABS1 and ABS2 also overlapped when they were superimposed onto the second and first protomers of the filament, respectively (fig. S3B). This arrangement also placed the two actin-binding sites asymmetrically on one side of the filament, increasing the distance that must be covered by the intervening sequence 100–170 that contains the second TM-binding site. In contrast, no overlap was observed when ABS1 and ABS2 were superimposed onto the first and second protomers of the filament, respectively (Fig. 2A). This arrangement naturally positioned ABS2 at the interface between the first three subunits of the filament, where Tmod residues in contact with actin were highly conserved (fig. S4A). This model, and especially the location of the N-termini of ABS1 and ABS2, was further consistent with the azimuthal sliding of TM on the surface of the filament (fig. S5), which occurs upon Ca2+ binding to troponin and myosin binding to the filament (22). The two extreme positions of TM on the filament, blocked and open, are represented by EM structures of actin-TM (23) and actin-TM-myosin (24). The full extent of TM’s sliding is estimated at ~35° (22), which greatly restricts the available surface for Tmod binding at the pointed end, and strongly supports the model proposed here.
Fig. 2. Testing the interactions of Tmod at the pointed end by mutagenesis.
(A) Representation of the mutants studied here on a domain diagram of Tmod and on the structures of ABS1 and ABS2 superimposed onto the first two protomers (marine and blue) at the pointed end of the filament model (21) (see also movie S3). Residues mutated here are colored by atom type (carbon, yellow; oxygen, red; nitrogen, blue). Also shown are residues mutated in previous studies (green). Note that ABS1 only interacts with the first protomer, whereas ABS2 contacts the first three protomers of the filament. The third protomer is colored gray, except for the area that contacts ABS2, which is colored purple. (B) Normalized pointed end elongation rates of filament seeds as a function of Tmod concentration, with or without TM (conditions given on top). (C) Normalized pointed end elongation rates of filament seeds as a function of mutant Tmod concentration, and in the presence of TM (the individual titrations are shown in fig. S7). Each mutant is represented by a different color (keys are given on the bottom). (D) Comparison of the normalized pointed end elongation rates at 200 nM Tmod with or without (gray bars) TM.
Key interactions of Tmod with the first three protomers of the filament (Fig. 2A and fig. S4) were tested by mutagenesis and pointed end-capping assays. In these assays, the barbed end must be tightly capped, because faster dynamics at this end can mask subunit exchange at the pointed end. Traditionally, gelsolin has been used to cap the barbed end in Tmod studies (7, 10, 12, 16–18). We found, however, that CP and not gelsolin should be used in these assays (fig. S6, A to D), because monomer sequestration and filament severing by gelsolin interfered with its capping activity (15).
The optimal concentration of TM for our experiments was 1 μM (15) (fig. S6, E to H). Thus, the concentration dependence of Tmod capping was determined in the absence or the presence of 1 μM TM (Fig. 2B, and fig. S7, A and B). TM enhanced Tmod’s capping efficiency ~4-fold, resulting in a KD of 28 nM compared to 108 nM in the absence of TM. These affinity values could be directly compared, because the same stock of filament seeds was used in the experiments. Generally, however, the affinity values depend on the number of pointed ends, which varies from experiment to experiment. While substantial, a ~4-fold increase in affinity was far below the 1000-fold increase and picomolar affinity reported previously (13), which prompted several repetitions of the experiments with identical results. A nanomolar rather than picomolar affinity of Tmod at the pointed end seems more consistent with the observation of pointed end monomer exchange in sarcomeres (14).
We analyzed eight Tmod mutants in elongation assays (Fig. 2, and figs. S4 and S7, and movie S3). Erythrocytes express a short Tmod isoform (residues 103–359) (25), which reinforces an idea suggested by the structures; Tmod is better described as consisting of two TM-actin-binding modules. Thus, we expressed constructs 1–101 and 100–359 to test the importance of each module. Other mutations were introduced within full-length Tmod, and targeted the α-helix and tail region of ABS1, and interactions of ABS2 with the first three protomers of the filament. While TM increased the capping efficiency of the mutants, they all displayed reduced activity compared to wild type Tmod, supporting the importance of these residues for Tmod-actin interactions (Fig 2, C and D, and fig. S7, C to J). The most severe capping defects were observed with the deletion or mutations of the C-terminal TM-actin binding module. Particularly, replacing residues 322–325 at the beginning of the C-terminal α-helix with AAAA or EEEE had the most dramatic effect, consistent with the key location of this region at the interface between the first three protomers of the filament. Mutations reported to affect actin binding and capping (9, 16–18) also tended to localize to the contact surface with subunits of the filament (Fig. 2A, fig. S4, and movie S3).
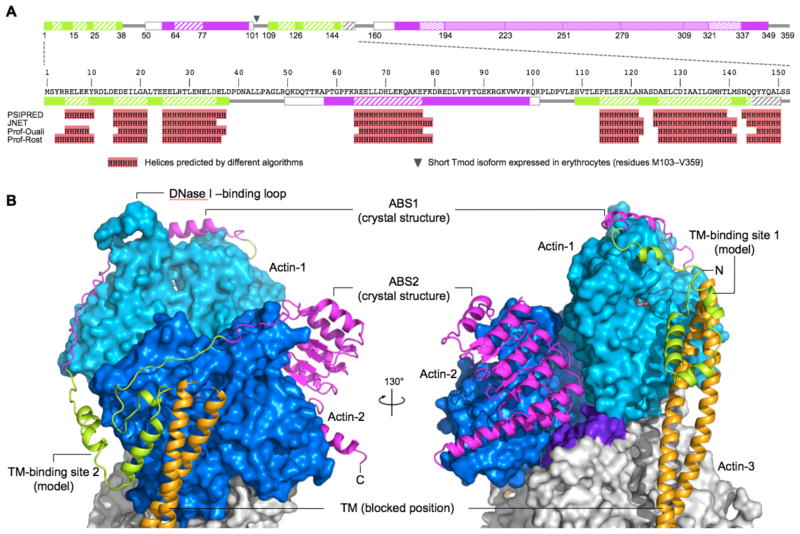
These results are consistent with a model in which a single Tmod molecule caps actin filaments at the pointed end (Fig. 3, and movie S4). ABS1 interacts only with the first protomer of the filament (interface area: 1,549 Å2) and ABS2 interacts mainly with the second protomer (interface area: 1,770 Å2), while it also contacts protomers 1 and 3 (estimated interface areas: 198 Å2 and 125 Å2). Secondary structure prediction using several algorithms further suggests that the two TM-binding sites have a related fold consisting of three helices (15). The model of the pointed end proposed here is supported by analysis of several Tmod mutants, and steric constraints imposed by the structure of the filament (21) and the blocked (23) and open (24) positions of TM on the filament.
Fig. 3. Model of the pointed end.
(A) Domain diagram of Tmod (according to Fig. 1A), showing a secondary structure prediction of the N-terminal region by several algorithms (15). The prediction suggests that the two TM-binding sites, which are similar in size, also share a similar fold, consisting of a three-helix bundle. (B) Model of the pointed end, with the structures of the complexes of actin with ABS1 and ABS2 (magenta) superimposed onto the first two protomers of the filament (marine and blue). ABS2 also contacts the third protomer (purple-colored area). TM is shown in the stable “blocked” position, which it assumes when bound alone to the filament (23). A tentative model of the two TM-binding sites of Tmod (green), based on the results of the secondary structure prediction and energy minimization (15), is shown for reference (see also movie S4).
The structure of the pointed end-binding VCD domain of Vibrio VopL was recently determined in complex with an actin trimer (26). One of the subunits of the VCD dimer binds in the same location as ABS2, i.e. at the interface between the first three subunits of the filament (fig. S8). VopL is a powerful nucleator, and this activity is enhanced many-fold by the presence of actin monomer-binding WH2 domains N-terminal to VCD (27, 28). Similarly, the Tmod related protein leiomodin is a powerful nucleator (29), and this activity is strongly enhanced by the presence of a C-terminal tail that contains a WH2 domain.
Separately, ABS1 and ABS2 interact with monomeric actin with KDs of ~10 μM, independent of the nucleotide state on actin, whereas full-length Tmod binds to the pointed end with KD 108 nM, and 28 nM in the presence of TM. Thus, the increased affinity of Tmod at the pointed end results from multiple, relatively weak interactions, involving three actin protomers and two TM molecules on each side of the filament. We suggest that Tmod’s interactions at the pointed end are better described in the form of two TM-actin-binding modules. Because of their relative independence and weaker individual affinities, one module can detach from the filament while the other remains bound, which could explain pointed end subunit exchange in sarcomeres (14). Newly incorporated monomers likely consist of ATP-actin, for which the two actin-binding sites of Tmod appear to have similar affinity as for ADP-actin, thought to be the predominant species at the pointed end.
Supplementary Material
Acknowledgments
Supported by National Institutes of Health grant R01 GM073791. X-ray data collection at beamline X6A of the National Synchrotron Light Source was supported by NIH grant GM-0080 and DOE contract DE-AC02–98CH10886. We thank Grzegorz Rebowski for the preparation of actin and TM for this study, and William Lehman and Marek Orzechowski for providing the TM models. The structures have been deposited in the Protein Data Bank under codes 4PKG, 4PKH and 4PKI.
Footnotes
REFERENCES AND NOTES
- 1.Cooper JA, Sept D. Int Rev Cell Mol Biol. 2008;267:183. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nag S, Larsson M, Robinson RC, Burtnick LD. Cytoskeleton. 2013;70:360. doi: 10.1002/cm.21117. [DOI] [PubMed] [Google Scholar]
- 3.Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM. Cytoskeleton. 2012;69:337. doi: 10.1002/cm.21031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett V, Baines AJ. Physiol Rev. 2001;81:1353. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 5.Fowler VM, Sussmann MA, Miller PG, Flucher BE, Daniels MP. J Cell Biol. 1993;120:411. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyer JD, et al. Blood. 2010;116:2590. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber A, Pennise CR, Fowler VM. J Biol Chem. 1999;274:34637. doi: 10.1074/jbc.274.49.34637. [DOI] [PubMed] [Google Scholar]
- 8.Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Biophys J. 2005;88:372. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler VM, Greenfield NJ, Moyer J. J Biol Chem. 2003;278:40000. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- 10.Kostyukova AS, Choy A, Rapp BA. Biochemistry. 2006;45:12068. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Biophys J. 2002;83:2716. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber A, Pennise CR, Babcock GG, Fowler VM. J Cell Biol. 1994;127:1627. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber A, Pennise CR, Fowler VM. J Biol Chem. 1999;274:34637. doi: 10.1074/jbc.274.49.34637. [DOI] [PubMed] [Google Scholar]
- 14.Littlefield R, Almenar-Queralt A, Fowler VM. Nat Cell Biol. 2001;3:544. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supplementary materials on Science Online.
- 16.Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Biochemistry. 2005;44:4905. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- 17.Yamashiro S, Speicher KD, Speicher DW, Fowler VM. J Biol Chem. 2010;285:33265. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukada T, et al. J Biol Chem. 2011;286:2194. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez R, Holmes KC. Annu Rev Biophys. 2011;40:169. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bella J, Hindle KL, McEwan PA, Lovell SC. Cell Mol Life Sci. 2008;65:2307. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii T, Iwane AH, Yanagida T, Namba K. Nature. 2010;467:724. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 22.Vibert P, Craig R, Lehman W. J Mol Biol. 1997;266:8. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 23.Li XE, et al. Biophys J. 2011;100:1005. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrmann E, et al. Cell. 2012;150:327. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao W, Sung LA. J Biol Chem. 2010;285:31408. doi: 10.1074/jbc.M110.130278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahm JA, et al. Cell. 2013;155:423. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namgoong S, et al. Nat Struct Mol Biol. 2011;18:1060. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu B, Cheng HC, Brautigam CA, Tomchick DR, Rosen MK. Nat Struct Mol Biol. 2011;18:1068. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chereau D, et al. Science. 2008;320:239. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.