Abstract
Background
Several lines of evidence indicate mitochondrial impairment in the pathophysiology of autism. As one of the most common biomarkers for mitochondrial dysfunction, mitochondrial DNA (mtDNA) copy number has also been linked to autism, but the relationship between mtDNA copy number and autism was still obscured. In this study, we performed a case–control study to investigate whether mtDNA copy number in peripheral blood cells is related to patients with autism.
Methods
Relative mtDNA copy number in peripheral blood cells was measured by using real-time polymerase chain reaction method. The participants in this study included 78 patients with childhood autism and 83 typically developing children.
Results
We observed children with autism had significantly elevated relative mtDNA copy number than healthy controls (Beta = −0.173, P = 0.0003). However, there were no significant correlations between mtDNA copy number and clinical features (paternal age, maternal age, age of onset, illness of duration, CARS score and ABC score) in childhood autism.
Conclusion
We show that elevated mtDNA copy number in peripheral blood is associated with autism, indicating that there may be mitochondrial dysfunction in children with autism.
Keywords: Autism, Mitochondrial dysfunction, mtDNA copy number, CARS, ABC
Background
Autism is a neurodevelopmental disorder, characterized by social deficits, communication impairment and unusually restricted, repetitive behaviors with onset prior to three years of age. The worldwide prevalence of autism spectrum disorder (ASD) is estimated at about 0.7%, although the estimates vary with populations [1,2]. Because of the complex, multifactorial etiology and pathophysiology underlying this illness and the limited scientific advance, the pathogenesis of autism is still elusive. It is consensually believed that gene–environment interplay contributes to the development of autism [3,4]. Increasing evidence has shown that mitochondrial dysfunction is associated with autism and there is much higher prevalence of mitochondrial diseases in autism than that in general population of children [5-7].
Mitochondria are specialized cellular organelle generating adenosine triphosphate (ATP) through oxidative phosphorylation that is a series of chemical reactions in the electron transport chain (ETC). In addition, mitochondria play important roles in other biological activities of cells, such as apoptosis and calcium regulation, and are the major intracellular source and primary target of reactive oxygen species (ROS) that is toxic for mitochondria [8,9]. In normal cells, each mitochondrion carries about 2 to 10 copies of mitochondrial DNA (mtDNA). The mtDNA is a 16.5 kb circular double-stranded molecule containing 37 genes, which codes partial proteins of ECT enzyme complexes and partial components of the machinery of intramitochondrial protein synthesis. Comparing with nuclear DNA, mtDNA is lack of protective histones and has limited DNA repair capacity, and therefore is highly susceptible to intramitochondrial ROS [10].
Increased oxidative stress has been found in patients with autism [11-13]. This leads to high rates of mutation and deletion for mtDNA, and subsequently impairs the mitochondrial function [14]. Evidence of different lines supports a role of mitochondrial impairment in the pathophysiology of autism [5,15-21]. Studies of peripheral blood and lymphoblastoid cell lines have identified ETC complexes deficiency and disordered mitochondrial energy metabolism such as plasma lactate, pyruvate, carnitine and amino acids in individuals with autism [19]. Similarly, previous magnetic resonance spectroscopy study has showed increased lactate doublets, decreased synthesis of ATP and a disturbed energy metabolism in the brain of autism [21]. Postmortem human brain studies in autism have also found decreased protein expressions of ETC complexes in specific region of brain, reinforcing the mitochondrial dysfunction in this illness [20]. Abnormal mtDNA number, as one of the most common biomarkers, has been observed associated with mitochondrial dysfunction and increased oxidative stress [22-25].
Previous studies have suggested that mtDNA copy number may increase with mtDNA damage or mitochondrial dysfunction, and compensate for the mitochondrial energy metabolism in patients with ASD [20]. Other studies failed to find this abnormality in ASD [19,26]. Considering the ASD defined by a constellation of different classes of subtypes, the diversity of ASD and the limited sample size in these studies may account for the inconsistent results. In an attempt to elucidate this association, we performed a large case–control study to investigate the status of peripheral blood mtDNA copy number in subjects with childhood autism.
Methods
Subjects
The study included 78 children with autism and 83 typically developing children. All subjects age from 3 to 6 years and were Han Chinese ethnicity. Children with autism were consecutively recruited from the department of Children psychology, MCH Hospital of Shenzhen and Institute of Mental Health, Second Xiangya Hospital of Central South University from July 2011 to December 2012. All patients were diagnosed by two senior psychiatrists according to DSM-IV diagnostic criteria-based structured interview for autism. In addition, all cases were also assessed by using childhood autism rating scale (CARS) and autism behavior checklist (ABC) [27,28]. Exclusion criteria for children included: autistic patients with age above 6 years, patients with Rett syndrome, childhood schizophrenia, Asperger’s syndrome or pervasive developmental disorder-not otherwise specified (PDD-NOS).
All typically developing children were recruited from community volunteers. The current mental status and history of mental disorders of the control subjects were evaluated by a senior psychiatrist. Controls recruited when individuals met the criteria and did not have any history of mental disorders, neurological disorders, substance abuse and serious physical diseases. Unfortunately we failed to obtain a family history of mental disorders.
Written informed consents conforming to the principles expressed in the Declaration of Helsinki were obtained from the guardians of all participants. The study was approved by the Human Ethics Committee of the Second Xiangya Hospital.
Clinical assessment
The severity of autism was assessed by CARS, which rates the child on a scale from one to four in each of 15-items behavioral rating scale. The items are: relating to people; emotional response; imitation; body use; object use; listening response; fear or nervousness; verbal communication; non-verbal communication; activity level; level and consistency of intellectual response; adaptation to change; visual response; taste, smell and touch response and general impressions. Children with a total score above 30 were considered autistic and were included in this study [27].
The ABC consists in 57 items about the atypical behaviors and these behaviors are related to five areas (sensory stimuli sensorial; relating; body and object use; language social; self-help). Most items scored from 1 to 4 according to the impairment degree. Children with scores above 53 were considered autism and were included in this study [28].
Laboratory analysis
Peripheral blood samples were collected in EDTA tubes and kept at −20°C before use. Genomic DNA was isolated from 200 ul of each blood sample using a commercial DNA Isolation Kit NEP004-1 (Beijing Dingguo Changsheng Biotechnology Co., Ltd., China). The procedure for DNA extraction and purification was performed by using the silica-membrane-based spin column method. The quantity and purity of the DNA was assayed using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and all DNA samples all had OD260/OD280 values of 1.7–2.0. Then total DNA samples were stored at −70°C until use.
The relative mtDNA copy number was measured by quantitative real-time polymerase chain reaction (PCR) and normalized by simultaneous measurement of the nuclear DNA according the method described in previous studies [29-31]. In brief, the primer sequences L394, 5′-CACCAGCCTAACCAGATTTC-3′/H475, 5′-GGGTTGTATT-GATGAGATTAGT-3′ were used for measuring the mtDNA content, and primers HBG1F, 5′-GCTTCTGACACAACTGTGTTCACTAGC-3′/HBG1R 5′-CACCAACTTCATCCACGTTCACC-3′ were used for amplification the single-copy nuclear ß-globin gene [29]. Assay was performed by using the Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) supplied by CFX96 Real-Time Detection Systems (BioRad Laboratories, Hercules, CA, USA). The qPCR was performed under the following conditions: denaturation at 95°C for 10 minutes followed by 40 cycles of 10s at 95°C, 30 at 60°C and 30s at 72°C. All assays were carried out in triplicate using 10 ng DNA per 10 μl reaction. The acceptable standard deviation (SD) of the triplicate threshold cycle (Ct) values was set at 0.3. If the result was out of the acceptable range, then the run was repeated for the same sample. The relative mtDNA copy number was calculated by the equation 2−ΔΔCt (ΔCt = CtmtDNA-Ctβ-globin) [32,33].
Statistical analysis
All statistical analyses were performed with the Statistical Package for the Social Sciences (SPSS, version 20.0 for Windows). The difference in the distribution of characteristics between the cases and the controls was evaluated by chi-square test for categorical variables (gender) and Student’s t-test for continuous variables (age). Age and sex of the participants, as well as the batch used for mtDNA copy number measurement, were included as covariates in linear regression in order to rule out confounding effects. For comparisons between multiple groups, one-way ANOVA followed by LSD test for multiple comparisons between any of the two groups was carried out. Monotonic relationships between mtDNA copy number and clinic parameters of autism were examined by using linear regression analysis. All P-values were two-sided and considered statistically significant at <0.05.
Results
A total 78 patients with autism and 83 healthy controls were included in this study. The mean age of autism patients and healthy controls were 45.4 ± 12.3 and 47.3 ± 12.5 months, respectively (p = 0.413, Table 1). There was no sex difference between autism cases and healthy control samples (p = 0.175), although there were more female controls (n = 16) than cases (n = 9). In addition, the mean CARS and ABC scores in cases were 34.48 ± 3.01 and 105.44 ± 9.55, respectively. The relative mtDNA copy number was skewed distributed in both autism cases and healthy control. The mean of relative mtDNA copy numbers was 9.16 (SD = 11.6) in cases and 3.052 (SD = 4.252) in controls; both were far from its medians, 1.624 for autism cases and 1.257 for controls (Table 2). There was a quite large standard deviation in both groups. We performed eighth root transformation, which made the means and median of transformed mtDNA very close in both autism cases (1.135 ± 0.293 vs 1.062) and controls (0.960 ±0.288 vs 1.029), suggesting the transformed mtDNA are symmetric distribution, and variations were close in two groups.
Table 1.
Demographic and clinical characteristics of autism patients and healthycontrols
| Variables | Cases (n = 78) | Controls (n = 83) | P value |
|---|---|---|---|
| Age, months (mean±SD) | 45.4±12.3 | 47.3±12. 5 | 0.413 |
| Sex, n, (%) | 0.175 | ||
| Male | 69 (88.46) | 67 (80.72) | |
| Female | 9 (11.54) | 16 (19.23) | |
| Family training (yes/no)* | 27/35 | ||
| Paternal age, years (mean±SD)* | 30.18±4.64 | ||
| Maternal age, years (mean±SD)* | 27.34±4.68 | ||
| Age of onset, years (mean±SD)* | 2.05±0.71 | ||
| Illness of duration, years (mean±SD)* | 2.49±1.20 | ||
| CARS Score, (mean±SD)* | 34.48±3.01 | ||
| ABC Score, (mean±SD)* | 105.44±9.55 |
*Data from 16 of 78 cases (same subjects) were missing.
Table 2.
Summary statistics of relative mtDNA in autism cases and controls and by sexin autism cases and controls
| Status | N | Variable | Mean | Median | SD | Min | Max | 25th Pctl | 75th Pctl | |
|---|---|---|---|---|---|---|---|---|---|---|
| Overall | ||||||||||
| Control | 83 | mtDNA | 3.052 | 1.257 | 4.252 | 0.004 | 23.702 | 0.041 | 5.153 | |
| sqrtmtDNA | 0.960 | 1.029 | 0.288 | 0.495 | 1.485 | 0.671 | 1.227 | |||
| Autism | 78 | mtDNA | 9.165 | 1.624 | 11.596 | 0.015 | 35.585 | 0.807 | 19.004 | |
| sqrtmtDNA | 1.135 | 1.062 | 0.293 | 0.590 | 1.563 | 0.974 | 1.445 | |||
| Male | ||||||||||
| Control | 67 | mtDNA | 3.056 | 0.979 | 4.454 | 0.004 | 23.702 | 0.045 | 4.885 | |
| sqrtmtDNA | 0.955 | 0.997 | 0.289 | 0.495 | 1.485 | 0.679 | 1.219 | |||
| Autism | 69 | mtDNA | 8.994 | 1.617 | 11.369 | 0.015 | 35.585 | 0.762 | 18.724 | |
| sqrtmtDNA | 1.131 | 1.062 | 0.295 | 0.590 | 1.563 | 0.967 | 1.442 | |||
| Female | ||||||||||
| Control | 16 | mtDNA | 3.037 | 1.585 | 3.399 | 0.014 | 8.473 | 0.036 | 6.681 | |
| sqrtmtDNA | 0.979 | 1.057 | 0.291 | 0.589 | 1.306 | 0.660 | 1.267 | |||
| Autism | 9 | mtDNA | 10.478 | 2.570 | 13.908 | 0.038 | 32.939 | 1.030 | 20.978 | |
| sqrtmtDNA | 1.167 | 1.125 | 0.298 | 0.665 | 1.548 | 1.004 | 1.463 |
Note: sqrtmtDNA—Eighth-root transformation of mtDNA.
Linear regression analysis of both mtDNA and transformed mtDNA while controlling for age and sex showed an elevated mtDNA copy number in autism cases (Table 3). In the transformed mtDNA, mtDNA copy number is lower in controls compared to the autism cases (beta=−0.173, p=0.0003; Figure 1). Age and sex were not associated with mtDNA and transformed mtDNA. Stratification analysis by sex also showed the similar trend in males (beta=−0.172, p=0.0008). However, we did not observe significant association of mtDNA copy number with the disease in females (beta=−0.214, p=0.1056), although the effect size is slight larger. This may because the sample size is relative small and lack adequate statistical power to detect this association in females. We did not observe any significant interaction between the disease status and sex, and age.
Table 3.
Linear regression analysis of mtDNA and eighth-root transformed mtDNA and by sex in autism cases and controls
| mtDNA | Eighth-root transformed | |||||
|---|---|---|---|---|---|---|
| Parameter | Beta | SE | P | Beta | SE | P |
| Overall | ||||||
| Intercept | 12.296 | 3.050 | <.0001 | 1.248 | 0.103 | <.0001 |
| Control vs autism | −6.037 | 1.375 | <.0001 | −0.173 | 0.046 | 0.0003 |
| Female vs Male | −0.317 | 1.904 | 0.8679 | −0.0207 | 0.064 | 0.7469 |
| Age | −0.062 | 0.056 | 0.2636 | −0.0021 | 0.0019 | 0.2715 |
| Male | ||||||
| Intercept | 11.976 | 2.906 | <.0001 | 1.259 | 0.097 | <.0001 |
| Control vs autism | −5.849 | 1.490 | 0.0001 | −0.172 | 0.050 | 0.0008 |
| Age | −0.064 | 0.058 | 0.2735 | −0.0028 | 0.0019 | 0.16 |
| Female | ||||||
| Intercept | 11.763 | 8.332 | 0.172 | 0.9504 | 0.2782 | 0.0025 |
| Control vs autism | −7.284 | 3.803 | 0.0685 | −0.2144 | 0.1270 | 0.1056 |
| Age | −0.032 | 0.193 | 0.8705 | 0.00535 | 0.0064 | 0.4143 |
Figure 1.
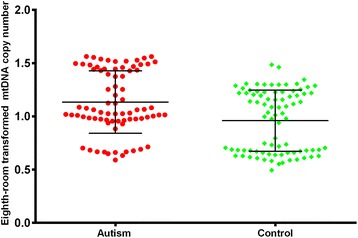
Relative mtDNAcopy number of autistic patients and health controls.
The associations between relative mtDNA copy number and clinical features in autism cases are shown in Table 4. No significant associations were observed between relative mtDNA copy number and clinical features (paternal age, maternal age, age of onset, illness of duration, CARS score and ABC score) in patients with childhood autism. Similarly, there was no difference of relative mtDNA copy number between patients with and without family training interaction (Figure 2).
Table 4.
Correlations between mtDNA copy number and clinical variables in childhood autism
| Variables | Beta | P value |
|---|---|---|
| Paternal age | 0.009 | 0.297 |
| Maternal age | 0.002 | 0.778 |
| Age of onset | −0.032 | 0.562 |
| Illness of duration | −0.030 | 0.361 |
| CARS Score | −0.002 | 0.873 |
| ABC Score | −0.001 | 0.844 |
Figure 2.
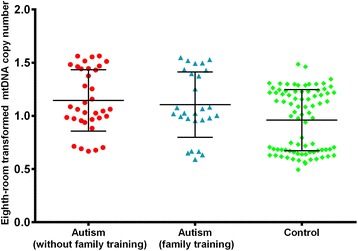
MtDNA copy number in autism with and without family training compared with controls. One-way analysis of variance (ANOVA) of the mean values was used to analyze the difference of mtDNA copy number among groups (F=6.042, P = 0.003) and the LSD test was used for multiple comparisons between any of the two groups. The differences of mtDNA copy number betweencontrol group and autism subgroup without family training, and autism subgroup withfamily trainingwere statistically significant (P = 0.002, 0.024, respectively). There is no significant difference in mtDNA copy number between patients with and without family training(p=0.592).
Discussion
In this study, we measured the relative mitochondrial DNA copy number in peripheral blood cells using quantitative PCR technique in autism patients and healthy controls. Our study found significant difference in the relative mtDNA copy number between childhood autism and healthy controls, and an elevated mtDNA content level was observed in patients with autism.
Unlike the findings reported in previous studies that the mtDNA copy number was negatively correlative with age in healthy populations [34-36], this study showed no association between mtDNA copy number and age in case and control groups. This may due to the narrow age span of subjects included in our study (3–6 years), and lack of adequate statistical power to detect this association. In addition, no significant associations were observed between relative mtDNA copy number and paternal age, maternal age, age of onset, illness of duration, CARS score, ABC score, and family training status in childhood autism. This indicates that the mtDNA copy number has no direct impact on clinical features of autism.
Our finding is consistent with previous brain tissue-based study that showed an aberrant elevated mtDNA copy number in autism patients [20]. Gu et al. analyzed mtDNA copy number with the brain tissues in 9 autistic children and 9 controls, and they found increased copy numbers of three mitochondrial genes (ND1, ND4, CYTB) in autistic patients, which indicated higher mtDNA copy numbers in autism. However, this finding was failed to be replicated by a following postmortem study [26]. Tang et al. examined the of brain mtDNA in childhood subjects, including 8 ASD children and 7 controls and found that no mtDNA copy number are associated with ASD patients [26]. Similarly, Giulivi et al. evaluated mtDNA copy number in peripheral lymphocytes from 10 children with autism and 10 controls. They found that the mean mtDNA copy number in lymphocytes was not significantly different in overall association. However, 5 of 10 children with autism presented mtDNA over-replication compared with 95% CI of the value in control children [19]. A small sample size and the different tissues cells may account for this inconsistency in previous studies. We measured mtDNA content in peripheral lymphocytes from 161 subjects and found autistic children had higher copy numbers of mtDNA when compared with healthy children group. The increased mtDNA copy number may be caused by compensatory mtDNA over-replication or a disruption of mtDNA degradation, which indicated a dysfunctional state in mitochondria. Therefore, these findings showed mitochondrial dysfunction in children with autism and proved a previous finding that mitochondrial dysfunction may be a biological subtype of autism spectrum disorder [21].
The major strength of our study is the unique and highly homogenous patient subjects. ASD is a heterogeneous group of common developmental disorders including autism, Rett syndrome, Asperger’s syndrome and PDD-NOS. Our study is just restricted to childhood autism, excluding the patients with atypical autism and adult autism. Thus, any potential confounding effects yielded by different sub-diseases aetiology were eliminated, which may be efficient in this association study to evaluate the mtDNA status in childhood autism. Furthermore, we analyzed 161 children subjects in this study, which is much larger than the sample size in previous studies.
We have to note that there are several limitations in the present study. First, because the inherent limitation of case–control design, the finding may not be used to make causal inference on the association of elevated mtDNA copy number with the risk of autism; and potential population stratification was not controlled. In addition, while we analyzed mtDNA copy number in peripheral blood cells, brain tissues are considered as the standard target tissue for studying autism or other brain disorders. However, peripheral blood cells can be obtained in noninvasive method; and the mtDNA copy number in peripheral blood has been associated with that in brain tissues [37,38]. The mtDNA copy number in peripheral blood might serve as an alternative indicator for the mitochondrial function in brain tissues. Furthermore, the incomplete data on characteristics of autism and controls did not allow controlling more potential confounding.
Conclusion
In this study we observed the mtDNA copy number in peripheral blood is significantly elevated in children with autism. Our finding supports the previous statement that there is mitochondrial dysfunction in patients with autism and indicates mitochondrial dysfunction may be a biological subtype of childhood autism. Additional research is needed to assess whether the association is replicable in the future.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (81271484 and 81471361 to XC, Grant No. 30900486 and 81371480 to JT, 81100996 to YL), the National Key Basic Research and Development Program (973) (Grant No. 2012CB517904 to XC). However, the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- mtDNA
mitochondrial DNA
- ASD
Autism spectrum disorder
- PDD-NOS
Pervasive developmental disorder-not otherwise specified
- ATP
Adenosine triphosphate
- ETC
Electron transport chain
- ROS
Reactive oxygen species
- DSM-IV
Diagnostic and statistical manual of mental disorders fourth edition
- CARS
Childhood autism rating scale
- ABC
Autism behavior checklist
- PCR
Polymerase chain reaction
- ND1
Nicotinamide Adenine Dinucleotide Hydrogen dehydrogenase gene 1
- ND4
Nicotinamide Adenine Dinucleotide Hydrogen dehydrogenase gene 4
- CYTB
Cytochrome-b encoded complex III subunit gene
Footnotes
Shan Chen and Zongchang Li contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SC, JT, KX and XC designed the study; SC, ZL, YH, HL, YL, GW, ZW, MH acquired the data, ZL, JT, XX, FZ performed the data analysis. SC, ZL, JT and XC wrote the article; FZ made critical comments and revision on the manuscript. All authors read and approved the final manuscript.
Contributor Information
Shan Chen, Email: chenshan0121@163.com.
Zongchang Li, Email: zongchanglicn@gmail.com.
Ying He, Email: heying@dr.com.
Fengyu Zhang, Email: fzhang20@jhmi.edu.
Hong Li, Email: lh3067@163.com.
Yanhui Liao, Email: tangliaoyanhui@gmail.com.
Zhen Wei, Email: weizhen14@163.com.
Guobin Wan, Email: wgb1978@yahoo.com.cn.
Xi Xiang, Email: xiangxihaha@163.com.
Maolin Hu, Email: humaolin@126.com.
Kun Xia, Email: xiakun48@163.com.
Xiaogang Chen, Email: chenxghn@gmail.com.
Jinsong Tang, Email: tangjinsonghn@gmail.com.
References
- 1.Elsabbagh M, Divan G, Koh YJ, Kim YS, Kauchali S, Marcín C, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5(3):160–79. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai M-C, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383(9920):896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 3.Tordjman S, Somogyi E, Coulon N, Kermarrec S, Cohen D, Bronsard G, et al. Gene x Environment interactions in autism spectrum disorders: role of epigenetic mechanisms. Front Psychiatry. 2014;5:53. doi: 10.3389/fpsyt.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C-S, Lee H-T, Lee S-Y, Shen Y-A, Wang L-S, Chen Y-J, et al. High mitochondrial DNA copy number and bioenergetic function are associated with tumor invasion of esophageal squamous cell carcinoma cell lines. Int J Mol Sci. 2012;13(9):11228–46. doi: 10.3390/ijms130911228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye RE, Rossignol DA. Mitochondrial dysfunction can connect the diverse medical symptoms associated with autism spectrum disorders. Pediatr Res. 2011;69:41R–7. doi: 10.1203/PDR.0b013e318212f16b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossignol D, Frye R. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legido A, Jethva R, Goldenthal MJ. Mitochondrial dysfunction in autism. Semin Pediatr Neurol. 2013;20(3):163–75. doi: 10.1016/j.spen.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Kowaltowski AJ, de Souza-Pinto NC, Castilho RF, Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med. 2009;47(4):333–43. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51(5):440–50. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 10.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–9. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80(6):1611–7. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13(3):171–81. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Zoroglu SS, Armutcu F, Ozen S, Gurel A, Sivasli E, Yetkin O, et al. Increased oxidative stress and altered activities of erythrocyte free radical scavenging enzymes in autism. Eur Arch Psychiatry Clin Neurosci. 2004;254(3):143–7. doi: 10.1007/s00406-004-0456-7. [DOI] [PubMed] [Google Scholar]
- 14.Napoli E, Wong S, Giulivi C. Evidence of reactive oxygen species-mediated damage to mitochondrial DNA in children with typical autism. Mol Autism. 2013;4(1):2. doi: 10.1186/2040-2392-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, Brown WT, et al. Brain region‐specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem. 2011;117(2):209–20. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filiano JJ, Goldenthal MJ, Rhodes CH, Marin-Garcia J. Mitochondrial dysfunction in patients with hypotonia, epilepsy, autism, and developmental delay: HEADD syndrome. J Child Neurol. 2002;17(6):435–9. doi: 10.1177/088307380201700607. [DOI] [PubMed] [Google Scholar]
- 17.Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. Am J Biochem Biotech. 2008;4(2):198–207. doi: 10.3844/ajbbsp.2008.198.207. [DOI] [Google Scholar]
- 18.Weissman JR, Kelley RI, Bauman ML, Cohen BH, Murray KF, Mitchell RL, et al. Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PLoS One. 2008;3(11):e3815. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giulivi C, Zhang Y-F, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389–96. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu F, Chauhan V, Kaur K, Brown W, Lafauci G, Wegiel J, et al. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl Psychiatry. 2013;3(9):e299. doi: 10.1038/tp.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh S, Dong Z, Zhang Y, DiMauro S, Peterson BS. Mitochondrial dysfunction as a neurobiological subtype of autism spectrum disorder: evidence from brain imaging. JAMA Psychiatry. 2014;71(6):665–71. doi: 10.1001/jamapsychiatry.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H-C, Wei Y-H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37(4):822–34. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13(5):481–92. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100(15):1104–12. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeng JY, Yeh TS, Lee JW, Lin SH, Fong TH, Hsieh RH. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103(2):347–57. doi: 10.1002/jcb.21625. [DOI] [PubMed] [Google Scholar]
- 26.Tang G, Gutierrez Rios P, Kuo S-H, Akman HO, Rosoklija G, Tanji K, et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 2013;54:349–61. doi: 10.1016/j.nbd.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schopler E, Reichler RJ, DeVellis RF, Daly K. Toward objective classification of childhood autism: Childhood Autism Rating Scale (CARS) J Autism Dev Disord. 1980;10(1):91–103. doi: 10.1007/BF02408436. [DOI] [PubMed] [Google Scholar]
- 28.Krug DA, Arick J, Almond P. Behavior checklist for identifying severely handicapped individuals with high levels of autistic behavior. J Child Psychol Psychiatry. 1980;21(3):221–9. doi: 10.1111/j.1469-7610.1980.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 29.Bi R, Zhang AM, Zhang W, Kong QP, Wu BL, Yang XH, et al. The acquisition of an inheritable 50‐bp deletion in the human mtDNA control region does not affect the mtDNA copy number in peripheral blood cells. Hum Mutat. 2010;31(5):538–43. doi: 10.1002/humu.21220. [DOI] [PubMed] [Google Scholar]
- 30.He Y, Tang J, Li Z, Li H, Liao Y, Tang Y, et al. Leukocyte mitochondrial DNA copy number in blood is not associated with major depressive disorder in young adults. PLoS One. 2014;9(5):e96869. doi: 10.1371/journal.pone.0096869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Su L-Y, Zhang A-M, Li Y-Y, Li X-A, Chen L-L, et al. Mitochondrial DNA copy number, but not haplogroup, confers a genetic susceptibility to leprosy in Han Chinese from Southwest China. PLoS One. 2012;7(6):e38848. doi: 10.1371/journal.pone.0038848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Liu CS, Tsai CS, Kuo CL, Chen HW, Lii CK, Ma YS, et al. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic Res. 2003;37(12):1307–17. doi: 10.1080/10715760310001621342. [DOI] [PubMed] [Google Scholar]
- 35.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10(1):62–8. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mengel-From J, Thinggaard M, Dalgard C, Kyvik KO, Christensen K, Christiansen L. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum Genet. 2014;133(9):1149–59. doi: 10.1007/s00439-014-1458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng YM, Jia YF, Su LY, Wang D, Lv L, Xu L, et al. Decreased mitochondrial DNA copy number in the hippocampus and peripheral blood during opiate addiction is mediated by autophagy and can be salvaged by melatonin. Autophagy. 2013;9(9):1395–406. doi: 10.4161/auto.25468. [DOI] [PubMed] [Google Scholar]
- 38.Kazachkova N, Raposo M, Montiel R, Cymbron T, Bettencourt C, Silva-Fernandes A, et al. Patterns of mitochondrial DNA damage in blood and brain tissues of a transgenic mouse model of Machado-Joseph disease. Neurodegener Dis. 2013;11(4):206–14. doi: 10.1159/000339207. [DOI] [PubMed] [Google Scholar]


