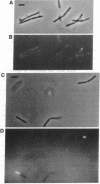
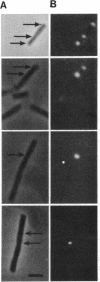
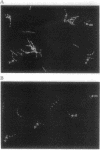
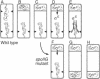
Abstract
Soon after the initiation of sporulation, Bacillus subtilis divides asymmetrically to produce sister cells that have very different developmental fates. Recently, it has been proposed that the differential gene expression which begins soon after this division is due to cell-specific activation of the transcription factors sigma F and sigma E in the prespore and the mother cell, respectively. We describe the use of a method for the localization of gene expression in individual sporulating cells that lends strong support to the cell-specific localization of sigma F and sigma E activities. The dependence of sigma E activity on integrity of the gene encoding sigma F has led to the suggestion that activation of sigma F in the prespore leads to a directional signal that triggers activation of sigma E only in the mother cell. Here we show that sigma E actually specifies the fate of the mother cell; in the absence of sigma E, two prespore-like cells are made. The appearance of sigma F activity at both poles of a sigma E-deficient mutant supports the idea that sigma F normally remains latent in the mother cell and that its activation depends on some morphological or physiological feature of the prespore. We present a model for the generation of asymmetry and the establishment of cell fate in B. subtilis.
Full text
PDF
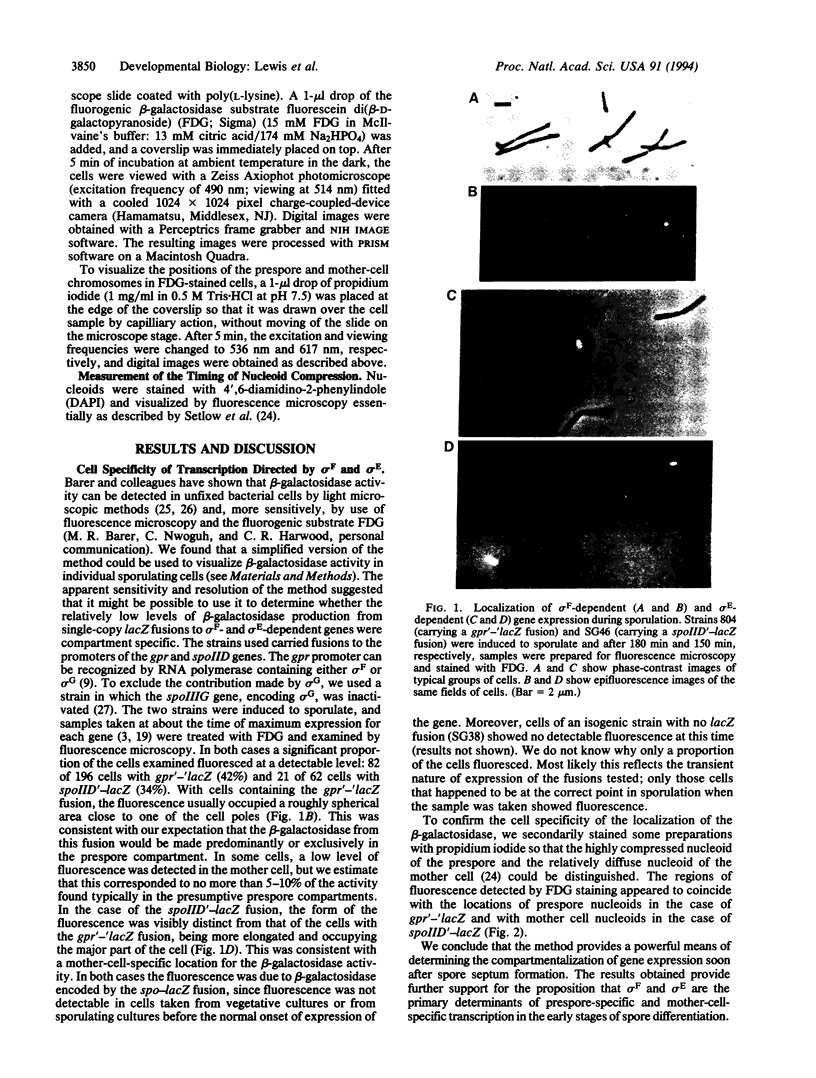
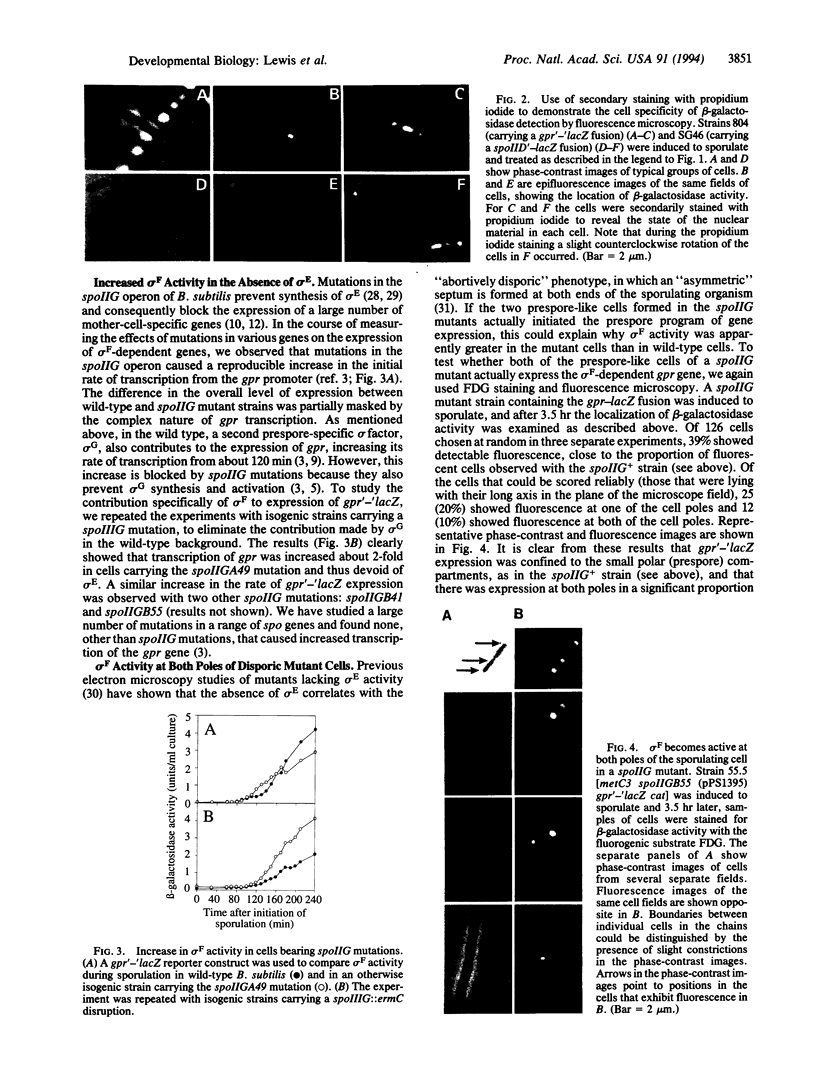
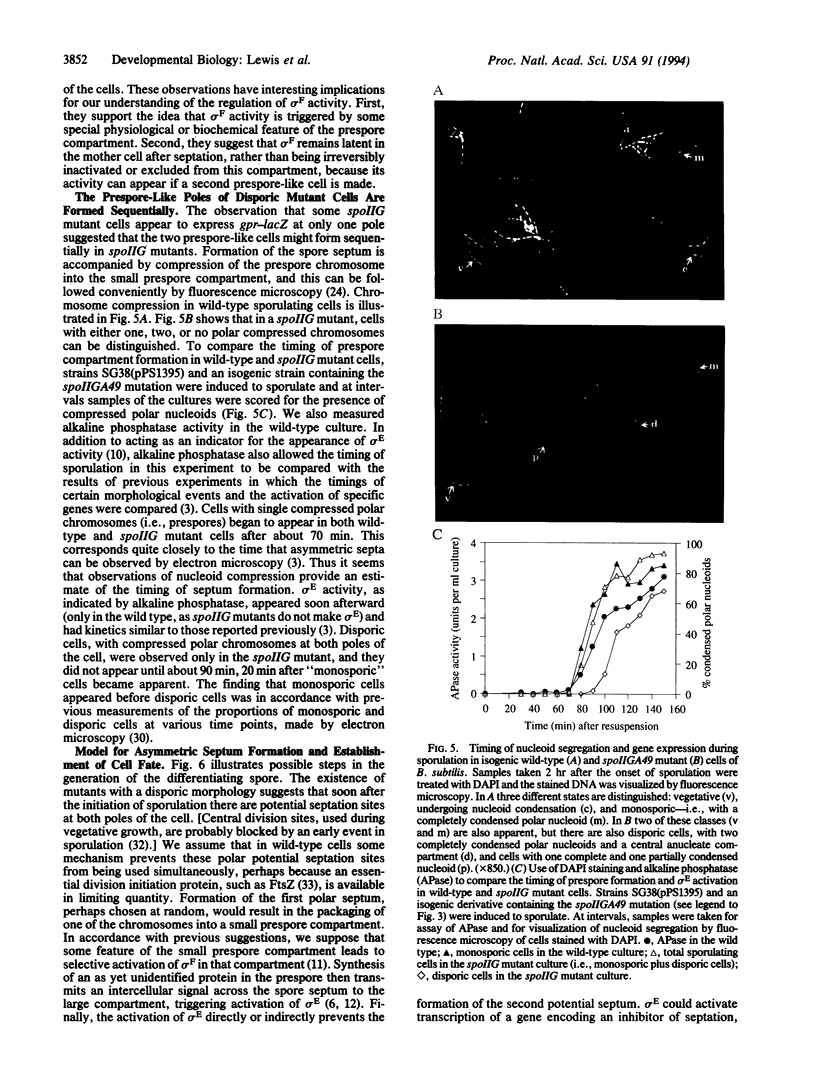

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barer M. R. New possibilities for bacterial cytochemistry: light microscopical demonstration of beta-galactosidase in unfixed immobilized bacteria. Histochem J. 1991 Nov-Dec;23(11-12):529–533. doi: 10.1007/BF01041179. [DOI] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991 Mar;5(3):447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Clarke S., Lopez-Diaz I., Mandelstam J. Use of lacZ gene fusions to determine the dependence pattern of the sporulation gene spoIID in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):2987–2994. doi: 10.1099/00221287-132-11-2987. [DOI] [PubMed] [Google Scholar]
- Driks A., Losick R. Compartmentalized expression of a gene under the control of sporulation transcription factor sigma E in Bacillus subtilis. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9934–9938. doi: 10.1073/pnas.88.22.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A., Roels S., Beall B., Moran C. P., Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994 Jan;8(2):234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- Dunn G., Torgersen D. M., Mandelstam J. Order of expression of genes affecting septum location during sporulation of Bacillus subtilis. J Bacteriol. 1976 Mar;125(3):776–779. doi: 10.1128/jb.125.3.776-779.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993 Mar;57(1):1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J., Illing N. Establishment of cell-specific transcription during sporulation in Bacillus subtilis. Mol Microbiol. 1992 Mar;6(6):689–695. doi: 10.1111/j.1365-2958.1992.tb01517.x. [DOI] [PubMed] [Google Scholar]
- Errington J., Mandelstam J. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- Errington J., Mandelstam J. Variety of sporulation phenotypes resulting from mutations in a single regulatory locus, spoIIA, in Bacillus subtilis. J Gen Microbiol. 1983 Jul;129(7):2091–2101. doi: 10.1099/00221287-129-7-2091. [DOI] [PubMed] [Google Scholar]
- Foulger D., Errington J. Effects of new mutations in the spoIIAB gene of Bacillus subtilis on the regulation of sigma F and sigma G activities. J Gen Microbiol. 1993 Dec;139(12):3197–3203. doi: 10.1099/00221287-139-12-3197. [DOI] [PubMed] [Google Scholar]
- Gholamhoseinian A., Piggot P. J. Timing of spoII gene expression relative to septum formation during sporulation of Bacillus subtilis. J Bacteriol. 1989 Oct;171(10):5747–5749. doi: 10.1128/jb.171.10.5747-5749.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N., Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J Bacteriol. 1991 May;173(10):3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H. F. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J Gen Microbiol. 1983 Jun;129(6):1945–1958. doi: 10.1099/00221287-129-6-1945. [DOI] [PubMed] [Google Scholar]
- Karmazyn-Campelli C., Bonamy C., Savelli B., Stragier P. Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 1989 Feb;3(2):150–157. doi: 10.1101/gad.3.2.150. [DOI] [PubMed] [Google Scholar]
- Losick R., Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992 Feb 13;355(6361):601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Margolis P., Driks A., Losick R. Establishment of cell type by compartmentalized activation of a transcription factor. Science. 1991 Oct 25;254(5031):562–565. doi: 10.1126/science.1948031. [DOI] [PubMed] [Google Scholar]
- Mason J. M., Hackett R. H., Setlow P. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J Bacteriol. 1988 Jan;170(1):239–244. doi: 10.1128/jb.170.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Errington J. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993 May;8(5):945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Foulger D., Errington J. The role of sigma F in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991 Mar;5(3):757–767. doi: 10.1111/j.1365-2958.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J. Mapping of asporogenous mutations of Bacillus subtilis: a minimum estimate of the number of sporeulation operons. J Bacteriol. 1973 Jun;114(3):1241–1253. doi: 10.1128/jb.114.3.1241-1253.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow B., Magill N., Febbroriello P., Nakhimovsky L., Koppel D. E., Setlow P. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol. 1991 Oct;173(19):6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini J. M., Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969 Jun;113(1):29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988 Mar 11;52(5):697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Sun D. X., Stragier P., Setlow P. Identification of a new sigma-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 1989 Feb;3(2):141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- Sun D., Fajardo-Cavazos P., Sussman M. D., Tovar-Rojo F., Cabrera-Martinez R. M., Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E sigma F: identification of features of good E sigma F-dependent promoters. J Bacteriol. 1991 Dec;173(24):7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. D., Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J Bacteriol. 1991 Jan;173(1):291–300. doi: 10.1128/jb.173.1.291-300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempy J. E., Bonamy C., Szulmajster J., Haldenwang W. G. Bacillus subtilis sigma factor sigma 29 is the product of the sporulation-essential gene spoIIG. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4189–4192. doi: 10.1073/pnas.82.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. R., Nwoguh C. E., Barer M. R. A microchamber system for the rapid cytochemical demonstration of beta-galactosidase and other properties in pathogenic microbes. Lett Appl Microbiol. 1994 Feb;18(2):102–104. doi: 10.1111/j.1472-765x.1994.tb00816.x. [DOI] [PubMed] [Google Scholar]