Abstract
Background and objectives
To investigate coagulase gene polymorphism of MRSA and MSSA isolates from Shiraz teaching hospitals from 2011 to 2012.
Materials and Methods
A total of 302 isolates of Staphylococcus aureus were collected from clinical specimens in three major teaching hospitals and confirmed on the basis of morphological characteristics and biochemical tests. The isolates were subjected to molecular typing on the basis of coagulase enzyme gene polymorphism by PCR-RFLP.
Results
There were 27 and 28 different RFLP patterns for AluI and HaeIII restriction enzymes respectively. This study showed that the discriminatory power of coagulase gene typing by Hae III enzyme was more than that of Alu I enzyme.
Conclusion
PCR-RFLP method is rapid, reproducible, simple and efficient for typing Staphylococcus aureus isolated from clinical specimens. This study showed that Hae III discriminatory power is better than Alu I for typing Staphylococcus aureus isolates.
Keywords: Antibiotic resistance, MRSA, PCR, RFLP, Coagulase gene
INTRODUCTION
Staphylococcus aureus is the most common cause of nosocomial infections that causes skin and soft tissue infections such as boils, carbuncle, cellulitis and abscesses (1, 2). Drug-resistant strains of the bacteria are rapidly being developed; thus, the treatment of this organism is difficult. Methicillin Resistant Staphylococcus aureus (MRSA) infection is a major cause of increasing morbidity and mortality (1).
The prevention and control of Staphylococcus aureus infections depends initially on detection of the risk factors in individuals exposed to S. aureus, analysis of the isolates by discriminatory DNA typing methods, and understanding the transmission of the bacterial infection (1). Molecular typing can play an important role in the epidemiological study of nosocomial infections, such as methicillin-resistant Staphylococcus aureus (MRSA) infection (3). In many countries, Staphylococcus aureus genotyping methods have become a part of the upcoming health care system and also the study of the strain origin, clonal relatedness and the epidemiology of the infection (2).
In the past two decades, a variety of molecular typing methods including pulsed-field gel electrophoresis (PFGE)(4), multilocus enzyme electrophoresis (MLEE), multilocus sequence typing (MLST)(5), spa typing(6), and coagulase genotyping(7) have developed to differentiate the related strains from unrelated ones (8) and to identify and compare the genotypes of S. aureus (9). Genotyping methods are relatively stable in natural conditions and produce reproducible results. These methods are rapid, do not require laboratory culture. The ability for typing by these methods is nearly 100% (2). Pulsed-field gel electrophoresis and multilocus sequence typing are the most reliable and the highest discriminatory typing methods, but are very hard, difficult, time-consuming and expensive to be used in a clinical microbiology laboratory (1). Coagulase gene (coa) typing is a simple, accurate, reproducible enough, easy to interpret and discriminatory method for typing Staphylococcus aureus isolates from various sources (2, 9-11).
It is well-known that coagulase enzyme is produced by most of the strains of S. aureus. Today, the ability to produce the coagulase in the clinical microbiology laboratory is used to detect S. aureus in human infections. It has been demonstrated that coagulase is an important virulence factor during the infection process (1,11,12). There are many different allelic forms of the S. aureus coa gene, each isolate produces one or more than one of these forms (11, 13, 14).
The discriminatory power of coagulase gene typing depends on the variability of the region containing the 81 bp tandem repeats at the 3’ coding region of the coagulase gene that differs both in the number of tandem repeats and the location of AluI and HaeIII restriction sites among different isolates (1-3,11,12,15-17). So, different S. aureus isolates could be discriminated using the coagulase gene typing method (11, 18).
In Iran, there is a little information about the genetic differences between aureus isolates from various hospitals and in particular, no information about coagulase gene polymorphisms. In present study, we have used coagulase gene typing method on the basis of the PCR-RFLP to discriminate S. aureus strains obtained from different specimens in teaching hospitals in Shiraz, Iran. Also, the ability of AluI and HaeIII restriction enzymes, for differentiating S. aureus isolates in PCR-RFLP method was also evaluated (1).
MATERIALS AND METHODS
Cultivation and distinction of bacteria
From August 2011 to July 2012, 302 clinical isolates of Staphylococcus aureus were collected from three major teaching hospitals in Shiraz, Iran. S. aureus strains were identified based on morphological characteristics and biochemical tests. Of all bacterial isolates, 39% were from female and 61% from male subjects. The percentage of the isolates from different wards of hospitals and various samples are presented in Tables 1 and 2.
Table 1.
The percentages of the studied isolates from various wards of the hospitals.
| Ward | Percentage | ward | Percentage |
|---|---|---|---|
| Surgery | 6.76% | Internal | 9.47 % |
| PER | 1.01 % | CCU | 0.72 % |
| OR | 2.74 % | Internal ICU | 21.28 % |
| Out Patient | 16.93 % | Pediatric Internal ICU | 3.1% |
| Neurology | 0.28% | Skin | 14.8 % |
| Neonatal Emergency | 3.32 % | Internal Neonatal | 0.73 % |
| Surgery ICU | 5.6 % | Emergency | 12.54 |
| KT | 0.72 % |
PER: Pediatric Emergency, OR: Operation Room, ICU: Intensive Care Unit, KT: Kidney Transplantation, CCU: Cardiac Care Unite.
Table 2.
The percentages of the studied bacterial isolates from various samples
| Sample | Percentage | Sample | Percentage |
|---|---|---|---|
| Inguinal | 0.3% | Wound | 9.4% |
| Sterile Fluid | 2.68% | Urine | 14.7% |
| Eye | 3.13% | Throat | 4% |
| Ear | 0.74% | Sputum | 26.2% |
| Lesion | 0.74% | Skin | 11.5% |
| CSF | 1.9% | Peritoneal | 0.35% |
| Blood | 15% | Plural | 0.3% |
| Axillary | 0.85% | Nasal | 7.81% |
| Abdominal | 0.4% |
Antibiotic susceptibility test
Disks containing cefoxitin (30 μg, Mast, UK) was used to discriminate the MRSA from MSSA isolates according to CLSI guideline recommendations. MHA (Muller Hinton Agar) plates were cultured with 0.5 McFarland standard of the bacterial broth culture and antibiotic disks were placed on the plates. Then this plate was incubated for 18 h at 37°C (1). There are several studies reporting that cefoxitin disk is preferable to oxacillin for detection of MRSA strains (19-23).
Extraction of Genomic DNA
Bacterial whole DNA was extracted from isolates by using the small-scale phenol-chloroform extraction method and used as template PCR(24).
Polymerase chain reaction
This method was carried out with a little difference from the method described by Himabindu et al. The forward primer sequence for coa gene was 5’CGAGACCAAGATTCAACAAG and the reverse primer sequence was 5’AAAGAAAACCACTCACATCA-3’ (1). PCR conditions were as follows: 94°C for 5 min, followed by 30 cycles of 95°C for 30 sec, 55°C for 45 sec and 72°C for 1.5 min, followed by a final extension of 72°C for 7 min. The PCR products were separated by 1.5% agarose gel electrophoresis.
Restriction Fragment Length Polymorphism (RFLP) analysis
Alu I (Fermentas, Lithuania) digestion of the products was performed by adding 15 μL of PCR product to 15 μL of the mixture that contained 2 U of Alu I, 3 μL of buffer enzyme and 8.11 μL of distilled injection water. Then, the reaction mixture was incubated at 37°C for 16 h. Hae III (Fermentas, Lithuania) digestion of the product was performed by adding 15 μL of PCR product to 15 μL of the mixture that contained 5 U of Hae III, 3 μL of buffer enzyme and 5.11 μL of distilled injection water. Then, the reaction mixture was incubated at 37°C for 16 h. To prevent evaporation of reaction mixture, 20 μL of sterile PCR oil was added. The restriction digest fragments were detected by 3% agarose gel electrophoresis. In this study, S. aureus strain 25923 was used as positive control.
Discriminatory power
The ability of a typing method to discriminate different types of the unrelated strains sampled from the hospital was assessed according to the Hunter-Gaston formula (1, 25). This is also called discriminatory index (D).
D= Numerical index of discrimination, N= The total number of isolates in the sample Population, s= The total number of types obtained, nj= The number of isolates belonging to the jth type contains (1, 25).
RESULTS
Coagulase gene amplification
The size of PCR products produced after coagulase gene amplification was separated into 6 different banding patterns in electrophoresis (Fig. 1, Table 3). The majority of isolates grouped in pattern 3 (729bp). We found that 2.4% of the isolates with the negative tube coagulase test, was positive for coagulase by PCR method, indicating the superiority of the molecular methods on phenotypic methods for typing of the Staphylococcus aureus. However, because the discriminatory power on the basis of the PCR alone was 0.68, which was very low, this typing method could not be used for S. aureus typing.
Fig. 1.
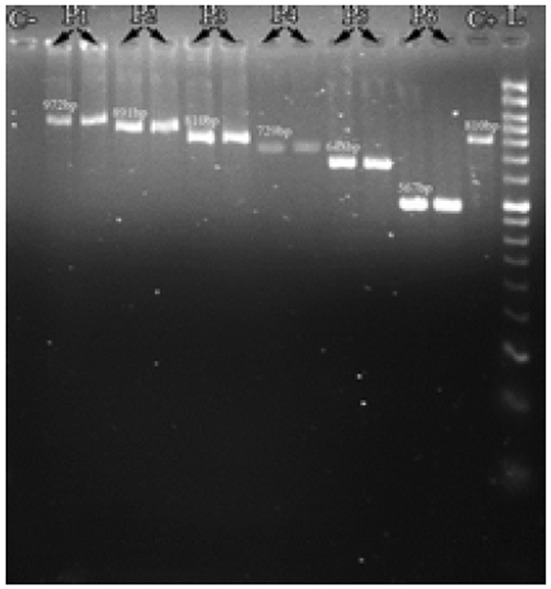
Representative agarose gel electrophoresis image of coagulase gene PCR products. L: 50bp DNA ladder, C-: Control negative, C+: Control positive, P1: 972, P2:891, P3:810, P4:729, P5: 648, P6: 567 bp bands
Table 3.
Band classes of PCR coagulase gene products in 3 hospitals.
| The size of PCR products (bp) | Hospital | Total | ||
|---|---|---|---|---|
| Faghihi | Ghotbodin | Namazi | ||
| 567 | 4(1.3%) | 0(0.0%) | 1(0.3%) | 5(1.7%) |
| 648 | 34(11.3%) | 0(0.0%) | 14(4.6%) | 48(15.9%) |
| 729 | 77(25.5%) | 4(1.3%) | 56(18.5%) | 137(45.4%) |
| 810 | 32(10.6%) | 1(0.3%) | 27(8.9%) | 60(19.9%) |
| 891 | 6(2.0%) | 1(0.3%) | 3(1.0%) | 10(3.3%) |
| 972 | 8(2.6%) | 0(0.0%) | 5(1.7%) | 13(4.3%) |
| Total | 161(58.9%) | 6(2.1%) | 106(38.8%) | 273(100.0%) |
PCR product digestion by AluI enzyme
After digesting of PCR products with Alu I and analysis of digestants on 3% agarose gel, 1 to 4 bands for each isolate were observed. The size of the band produced by enzyme digestion was divided into 12 different band classes (81, 162, 243, 324, 405, 486, 567, 648, 729, 810, 891 and 972 base pairs) each containing multiples of 81bp tandem repeat units.
After RFLP analysis of all the isolates, based on the number and size of the produced bands, 27 different patterns were observed (Table 4). The pattern 4 “324-405” was found to be the predominant type. The isolates grouped in pattern 27, produced only 1 band, indicating that this PCR product is not digested by Alu I. This probably indicates the lack of Alu I restriction sites amongst them. Discriminatory power was detected as 0.82 that indicated a good discriminatory power of this method for typing of the aureus. As a result, this method can be used for typing aureus.
Table 4.
Band patterns by AluI restriction enzyme digestion of PCR products
| HospitalM | Total | |||
|---|---|---|---|---|
| Faghihi | Ghotbodin | Namazi | ||
| 81-162-486 | 4(1.5%) | 0(0.0%) | 0(0.0%) | 4(1.5%) |
| 162-324-486 | 5(1.8%) | 0(0.0%) | 4(1.5%) | 9(3.3%) |
| 81-162-405 | 23(8.4%) | 0(0.0%) | 12(4.4%) | 35(12.8%) |
| 324-405 | 55(20.1%) | 3(1.1%) | 45(16.5%) | 103(37.7%) |
| 162-243-405 | 11(4.0%) | 0(0.0%) | 6(2.2%) | 17(6.2%) |
| 162-567 | 2(0.7%) | 0(0.0%) | 3(1.1%) | 5(1.8%) |
| 243-405 | 3(1.1%) | 0(0.0%) | 0(0.0%) | 3(1.1%) |
| 81-324-405 | 12(4.4%) | 0(0.0%) | 11(4.0%) | 23(8.4%) |
| 81-162-567 | 2(0.7%) | 0(0.0%) | 1(0.4%) | 3(1.1%) |
| 81-243-324 | 4(1.5%) | 0(0.0%) | 3(1.1%) | 7(2.6%) |
| 324-486 | 1(0.4%) | 1(0.4%) | 0(0.0%) | 2(0.7%) |
| 405-486 | 3(1.1%) | 0(0.0%) | 1(0.4%) | 4(1.5%) |
| 81-162-324 | 2(0.7%) | 0(0.0%) | 0(0.0%) | 2(0.7%) |
| 162-243-324 | 1(0.4%) | 0(0.0%) | 3(1.1%) | 4(1.5%) |
| 243-567 | 1(0.4%) | 0(0.0%) | 1(0.4%) | 2(0.7%) |
| 162-405 | 1(0.4%) | 0(0.0%) | 1(0.4%) | 2(0.7%) |
| 162-486 | 3(1.1%) | 0(0.0%) | 0(0.0%) | 3(1.1%) |
| 162-648 | 2(0.7%) | 0(0.0%) | 2(0.7%) | 4(1.5%) |
| Others* | 4(1.6%) | 1(0.4%) | 3(1.2%) | 8(3.2%) |
| Undigested | 22(8.1%) | 1(0.4%) | 10(3.7%) | 33(12.1%) |
| Total | 161(59.0%) | 6(2.2%) | 106(38.8%) | 273(100.0%) |
Others are the patterns that produced only by one sample including 81-162-324-405, 243-486, 81-243-567, 81-243-405, 81-405-486, 81-162-243-405, 162-729, 162-324-405
PCR product digestion by HaeIII enzyme
After digestion of PCR products with Hae III enzymes and analysis of amplicons in 3% agarose gel, 1 to 4 bands were observed for every isolate. The digestants were divided into 12 different band classes (81, 162, 243, 324, 405, 486, 567, 648, 729, 810, 891 and 972 base pairs) consisting of multiples of 81 bp tandem repeat units.
After RFLP analysis of all the isolates, based on the number and size of produced bands, 26 different patterns were observed (Table 5). The pattern 5 “324-405” was found to be the predominant pattern. The isolates that belonged to pattern 26 produced only 1 band which indicates that this PCR product is not digested by HaeIII. This probably indicates the lack of HaeIII restriction sites amongst them. The discriminatory power was detected as 0.90.
Table 5.
Band patterns by HaeIII restriction enzyme digestion of PCR products
| HaeIII | Hospital | Total | ||
|---|---|---|---|---|
| Faghihi | Ghotbodin | Namazi | ||
| 162-567 | 30(11.0%) | 1(0.4%) | 19(7.0%) | 50(18.3%) |
| 81-162-243-324 | 5(1.8%) | 0(0.0%) | 8(2.9%) | 13(4.8%) |
| 162-324-486 | 2(0.7%) | 0(0.0%) | 1(0.4%) | 3(1.1%) |
| 162-486 | 24(8.8%) | 0(0.0%) | 11(4.0%) | 35(12.8%) |
| 324-405 | 28(10.3%) | 1(0.4%) | 22(8.1%) | 51(18.7%) |
| 81-324-405 | 13(4.8%) | 0(0.0%) | 6(2.2%) | 19(7.0%) |
| 81-243-324 | 6(2.2%) | 0(0.0%) | 1(0.4%) | 7(2.6%) |
| 81-162-243-486 | 3(1.1%) | 0(0.0%) | 3(1.1%) | 6(2.2%) |
| 81-162-567 | 10(3.7%) | 0(0.0%) | 9(3.3%) | 19(7.0%) |
| 243-486 | 1(0.4%) | 0(0.0%) | 1(0.4%) | 2(0.7%) |
| 81-162-486 | 6(2.2%) | 2(0.7%) | 12(4.4%) | 20(7.3%) |
| 162-243-324 | 12(4.4%) | 0(0.0%) | 4(1.5%) | 16(5.9%) |
| 81-162-405 | 3(1.1%) | 0(0.0%) | 2(0.7%) | 5(1.8%) |
| 243-324-405 | 3(1.1%) | 0(0.0%) | 0(0.0%) | 3(1.1%) |
| 324-486 | 3(1.1%) | 1(0.4%) | 0(0.0%) | 4(1.5%) |
| 162-324-405 | 1(0.4%) | 1(0.4%) | 1(0.4%) | 3(1.1%) |
| 324-567 | 2(0.7%) | 0(0.0%) | 0(0.0%) | 2(0.7%) |
| 162-405 | 3(1.1%) | 0(0.0%) | 1(0.4%) | 4(1.5%) |
| 81-243-486 | 1(0.4%) | 0(0.0%) | 1(0.4%) | 2(0.7%) |
| 405-486 | 2(0.7%) | 0(0.0%) | 0(0.0%) | 2(0.7%) |
| 81-162-648 | 0(0.0%) | 0(0.0%) | 2(0.7%) | 2(0.7%) |
| Others* | 2(0.8%) | 0(0.0%) | 2(0.8%) | 4(1.6%) |
| Undigested | 1(0.4%) | 0(0.0%) | 0(0.0%) | 1(0.4%) |
| Total | 161 | 6 | 106 | 273 |
| 59.0% | 2.2% | 38.8% | 100.0% | |
Others are the patterns that produced only by one sample including 162-243-567, 81-243-405, 162-648, 81-162-243-405
DISCUSSION
Typing can be used to prevent or reduce an epidemic infection, reducing costs and hospital infection rates in hospitals (1). In this study, the polymorphism of coagulase gene among MRSA and MSSA isolates were investigated using PCR-RFLP analysis. The results of typing can be used to discriminate the strains within a given species (26). If strains from two patients have the same fingerprint, it indicates that both of them are infected from the one source. Discriminatory power of coagulase gene typing is high. It is an appropriate method for epidemiologic investigation of Staphylococcus aureus infection since it is applicable for typing of large number of strains in a short time.
he discriminatory power of this method is lower than PFGE and MLST but it is quicker and less costly (27). RFLP method can help to trace the source of infection and transmission; thus, this typing method can be used to prevent and control the spread of infection (1).
The purpose of this study was to investigate genetic variation in coagulase gene of aureus strains isolated from teaching hospitals in Shiraz, Iran. Although, MRSA strains are important cause of the nosocomial infections, no data are available on molecular typing of MRSA or even MSSA isolates by PCR-RFLP of coagulase gene in hospital isolates from Iran. There are few studies on this subject in Iran and one of them was in veterinary practice (9). However, in a study in Urmia, Iran, 26 S. aureus isolates from urine and skin in two hospitals were analyzed by PCR-RFLP using HaeIII enzyme. PCR products ranged from 490-790 bp in size and 6 distinct RFLP banding patterns were observed after the digestion of PCR products (2).
The discriminatory index of coagulase gene typing by PCR-RFLP on the basis of HaeIII enzyme is more than of this method based AluI. Thus, HaeIII enzyme is better than AluI enzyme for typing aureus.
In conclusion, our study proved that, of PCR-RFLP of coagulase gene is a rapid, simple and efficient method for typing S. aureus strains isolated from different clinical specimens in Shiraz teaching hospitals. This typing method can be used for tracing the source and transmission route of S. aureus infections and helps to prevent and control those related infections in our hospitals.
Fig. 2.
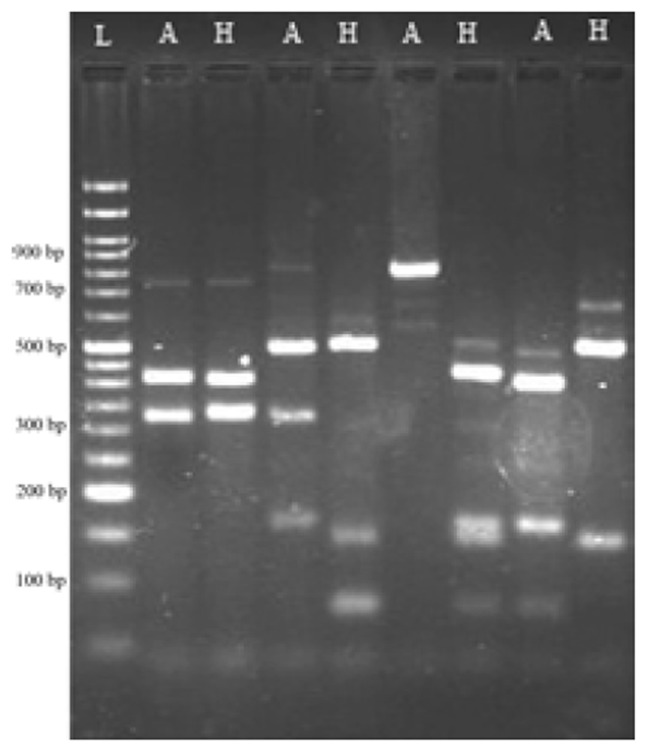
Representative agarose gel electrophoresis image of AluI and HaeIII restriction enzyme digestion products. L: 50bp DNA ladder, A: AluI restriction digested products, H: HaeIII restriction enzyme digestion products
Acknowledgments
This study was financially supported by Shiraz University of Medical Sciences, Grant No.90-5994. This paper is extracted from Mr Hossein Khoshkharam Roodmajani MSc thesis supervised by Dr. Mohammad Motamedifar.
References
- 1.Himabindu M, Muthamilselvan DS, Bishi DK, Verma RS. Molecular analysis of coagulase gene polymorphism in clinical isolates of methicillin resistant Staphylococcus aureus by restriction fragment length polymorphism based genotyping. Am J Infectious Diseases. 2009;5:170–176. [Google Scholar]
- 2.Talebi-Satlou R, Ahmadi M, Dastmalchi Saei H. Restriction fragment length polymorphism genotyping of human Staphylococcus aureus isolates from two hospitals in urmia region of iran using the coa gene. Jundishapur J Microbiol. 2012;5:416–420. [Google Scholar]
- 3.Ishino K, Tsuchizaki N, Ishikawa J, Hotta Usefulness of PCR-restriction fragment length polymorphism typing of the coagulase gene to discriminate Arbekacin-resistant methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2007;45:607–609. doi: 10.1128/JCM.02099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zadoks RN, Van Leeuwen WB, Kreft D, Fox LK, Barkema HW, Schukken HY, et al. Comparison of Staphylococcus aureus isolates from bovine and human skin, milking equipment, and bovine milk by phage typing, pulsed-field gel electrophoresis, and binary typing. J Clin Microbiol. 2002;40:3894–38902. doi: 10.1128/JCM.40.11.3894-3902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright MC, Day N, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nubel U, et al. spa Typing of Staphylococcus aureus as a Frontline Tool in Epidemiological Typing. J Clin Microbiol. 2008;46:574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinoso EB, El-Sayed A, Lämmler C, Bogni C, Zschöck M. Genotyping of Staphylococcus aureus isolated from humans, bovine subclinical mastitis and food samples in Argentina. Microbiol Res. 2008;163:314–322. doi: 10.1016/j.micres.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Janwithayanuchit I, Ngam-ululert S, Paungmoung P, Rangsipanuratn W. Epidemiologic Study of methicillin-resistant Staphylococcus aureus by coagulase gene polymorphism. Science Asia. 2006;32:127–132. [Google Scholar]
- 9.Dastmalchi Saei H, Ahmadi M, Mardani K, Batavani RA. Molecular typing of Staphylococcus aureus isolated from bovine mastitis based on polymorphism of the coagulase gene in the north west of Iran. Vet Microbiol. 2009;137:202–206. doi: 10.1016/j.vetmic.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Ciftci A, Emek Onuk E, Fındık A, Yıldırım T, Sogut MU. Molecular typing of Staphylococcus aureus strains from ovine mastitis by pulsed-field gel electrophoresis and polymerase chain reaction based on coagulase and protein A gene polymorphisms. J Vet Diagn Invest. 2009;21:849–853. doi: 10.1177/104063870902100614. [DOI] [PubMed] [Google Scholar]
- 11.Goh SH, Byrne SK, Zhang JL, Chow AW. Molecular Typing of Staphylococcus aureus on the Basis of Coagulase Gene Polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hookey JV, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on pcr restriction fragment length polymorphism and dna sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson A, Brodie J. Investigations on Staphylococcal coagulase. Br J Exp Pathol. 1963;44:524–528. [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves MW, Drummond MC, Tager M. Partial purification and characterization of the multiple molecular forms of Staphylococcal clotting activity (coagulase) J Bacteriol. 1981;148:861–868. doi: 10.1128/jb.148.3.861-868.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayaship N, Taniguchip K, Kojima K, Urasawa S, Uehara N, Omizu Y, et al. Analysis of methicillin-resistant and methicillin-susceptible Staphylococcus aureus by a molecular typing method based on coagulase gene polymorphisms. Epidemtilol Inifect. 1995;115:419–426. doi: 10.1017/s095026880005857x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belkum A, Scherer S, Alphen L, Verbrugh H. Short-sequence dna repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence C, Cosseron M, Mimoz O, Brun-Buisson C, Costa Y, Samii K, et al. Use of the coagnlase gene typing method for detection of carriers of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1996;37:687–696. doi: 10.1093/jac/37.4.687. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzkopf A, Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994;32:2407–2412. doi: 10.1128/jcm.32.10.2407-2412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skov R, Smyth R, Clausen M, Larsen AR, Frimodt-Møller N, Olsson-Liljequist B, et al. Evaluation of a cefoxitin 30 μg disc on Iso-Sensitest agar for detection of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2003;52:204–7. doi: 10.1093/jac/dkg325. [DOI] [PubMed] [Google Scholar]
- 20.Kahlmeter RWSaG. Mannitol Salt Agar-Cefoxitin Combination as a Screening Medium for Methicillin-Resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:3797–9. doi: 10.1128/JCM.43.8.3797-3799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Annie Felten BG. Philippe Henri Lagrange, and Isabelle Casin. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalactam, the Vitek 2 System, and the MRSA-Screen latex agglutination test. J Clin Microbiol. 2002;40:2766–71. doi: 10.1128/JCM.40.8.2766-2771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao Venkatakrishna I BKG, Kugaji Manohar S, Shantaram Manjula Pai Vidya. Detection of Methicillin Resistance in Staphylococcus aureus: Comparison of Disc diffusion and MIC with mecA gene detection by PCR. Int J Pharm Bio Sci. 2011;1:518–21. [Google Scholar]
- 23.Madhusudhan NSDS, Shoba DN. Correlation of cefoxitin disc diffusion test and oxacillin disc diffusion test for detecting mec a mediated oxacillin resistant staphylococcus aureus. J Pharm Bio Sci. 2011;10 [Google Scholar]
- 24.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006;1(1) doi: 10.1101/pdb.prot4455. [DOI] [PubMed] [Google Scholar]
- 25.Belkum A, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 26.Editorial. Molecular typing of micro-organisms: at the center of diagnosic, genomic and pathogenesisof infection disease? J Med Microbial. 2002;51:7–10. [Google Scholar]
- 27.Weller TM. Methicillin Resistant Staphylococcus aureus typing methods: which should be the international standard? J Hosp Infect. 2000;44:160–1. doi: 10.1053/jhin.1999.0701. [DOI] [PubMed] [Google Scholar]


