Abstract
Mouse butyrylcholinesterase (mBChE) and an mBChE-based cocaine hydrolase (mCocH, i.e. the A199S/S227A/S287G/A328W/Y332G mutant) have been characterized for their catalytic activities against cocaine, i.e. naturally occurring (−)-cocaine, in comparison with the corresponding human BChE (hBChE) and an hBChE-based cocaine hydrolase (hCocH, i.e. the A199S/F227A/S287G/A328W/Y332G mutant). It has been demonstrated that mCocH and hCocH have improved the catalytic efficiency of mBChE and hBChE against (−)-cocaine by ~8- and ~2000-fold, respectively, although the catalytic efficiencies of mCocH and hCocH against other substrates, including acetylcholine (ACh) and butyrylthiocholine (BTC), are close to those of the corresponding wild-type enzymes mBChE and hBChE. According to the kinetic data, the catalytic efficiency (kcat/KM) of mBChE against (−)-cocaine is comparable to that of hBChE, but the catalytic efficiency of mCocH against (−)-cocaine is remarkably lower than that of hCocH by ~250-fold. The remarkable difference in the catalytic activity between mCocH and hCocH is consistent with the difference between the enzyme-(−)-cocaine binding modes obtained from molecular modeling. Further, both mBChE and hBChE demonstrated substrate activation for all of the examined substrates ((−)-cocaine, ACh, and BTC) at high concentrations, whereas both mCocH and hCocH showed substrate inhibition for all three substrates at high concentrations. The amino-acid mutations have remarkably converted substrate activation of the enzymes into substrate inhibition, implying that the rate-determining step of the reaction in mCocH and hCocH might be different from that in mBChE and hBChE.
Keywords: Cholinesterase, hydrolysis, kinetics, drug abuse, enzyme
INTRODUCTION
Cocaine is a widely abused drug[1] with no FDA (U.S. Food and Drug Administration)-approved medication available for treatment-seeking users. A promising concept for anti-cocaine medication is to accelerate cocaine metabolism by hydrolysis at the benzoyl ester, producing biologically inactive metabolites.[2–7] In humans, butyrylcholinesterase (BChE) is the primary endogenous cocaine-metabolizing enzyme capable of catalyzing this reaction in plasma. However, wild-type human BChE (hBChE) has a low catalytic activity against naturally occurring (−)-cocaine (kcat = 4.1 min−1 and KM = 4.5 µM).[8–12] Our previous efforts were focused on improving the catalytic activity of hBChE against (−)-cocaine, leading to discovery of various hBChE mutants[8, 9, 13–18] with a considerably improved catalytic efficiency towards that drug. The highly efficient hBChE mutants, such as A199S/S287G/A328W/Y332G (kcat = 3060 min−1 and KM = 3.1 µM)[14, 19] or A199S/F227A/S287G/A328W/Y332G (kcat = 5700 min−1 and KM = 3.1 µM),[16] can be described as cocaine hydrolases (CocH). Initial experiments in rats and mice[16, 17, 20–29] showed that CocH is likely to be effective as an enzyme therapy or gene therapy for treating cocaine abuse by greatly reducing the reward value of a given drug dosage. In addition, we have not seen any acute toxicity of hCocH in mice or rats nor have other investigators found that wild-type hBChE elicited adverse effects in experimental animals.[30–32] Two clinical trials (NCT00333515 and NCT00333528) of hBChE have been performed by Baxter Healthcare Corporation, although the clinical data have not been made available.
Not surprisingly, some mice and rats eventually develop antibodies against hBChE and hCocH, accelerating the clearance of these enzymes and lowering their plasma levels,[25] although no immune response is noted when mouse BChE (mBChE) is injected into mice.[33] This outcome was expected because hBChE shares only ~80% sequence identity with its rodent counterparts.[34] We deemed it unlikely that human beings would generate antibodies to hCocH as the mutated residues are not exposed on the surface but occupy a deep and narrow catalytic gorge. Nonetheless, the mouse response called for further experiments to test the hypothesis that mutations in the catalytic site are not antigenic. Therefore we developed a conspecific cocaine hydrolase with equivalent mutations in mBChE: A199S/S227A/S287G/A328W/Y332G, designated mouse CocH or “mCocH”. The catalytic properties of mCocH were compared with those of mBChE, hBChE, and hCocH, and it was incorporated into viral gene transfer vector for in vivo studies with the aim of avoiding complications from an immune response in the animals.
In fact, initial gene transfer experiments with mCocH showed that very high levels of enzyme protein could be generated, on the order of 1000-fold above the native mBChE background level.[25] However, the levels of cocaine hydrolysis did not increase to the extent achieved with hCocH. This outcome suggested that although mCocH and hCocH contain similar mutations, their catalytic efficiencies with (−)-cocaine are different, and results from the gene transfer study were consistent with this interpretation.[25] To explore the reason, in the present study, mBChE and mCocH proteins were compared with hBChE and hCocH in regard to catalytic properties against (−)-cocaine and various other substrates. In addition, homology modeling and molecular dynamics (MD) simulations were used to compare structural features of the two mutated enzymes. As will be shown here, the catalytic efficiency of mCocH against (−)-cocaine is indeed lower than that of hCocH, and computational modeling of the detailed three-dimensional (3D) structures provides some insight into the reasons for this conclusion, which in turn may facilitate future attempts at re-engineering enzymes for therapeutic purposes.
MATERIALS AND METHODS
Materials
The cDNA for mBChE containing N-terminal signal was kindly provided by Dr. Palmer Taylor (Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, CA). Cloned pfu DNA polymerase and Dpn I endonuclease were obtained from Stratagene (La Jolla, CA). Restriction enzyme, alkaline phosphatase (CIP), and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). All oligonucleotides were synthesized by Eurofins MWG Operon (Huntsville, AL). Vector pCMV-MCS was obtained from Agilent Technologies (Santa Clara, CA). The QIAprep Spin Plasmid Miniprep Kit and QIAquick Gel Extraction Kit were obtained from QIAGEN (Valencia, CA). Chinese hamster ovary (CHO)-S cells and FreeStyle™ CHO Expression Medium were ordered from Invitrogen (Grand Island, NY). [3H](−)-Cocaine (50 Ci/mmol) and [3H]acetylcholine (ACh) were purchased from PerkinElmer Life and Analytical Sciences (Waltham, MA). Butyrylthiocholine (BTC) was obtained from Sigma-Aldrich (St Louis, MO).
Construction of eukaryotic expression plasmids
Site-directed mutagenesis for obtaining the mCocH cDNA was carried out using the QuikChange method.[35] The cDNA for full-length mBChE or mCocH was constructed in a pCMV-MCS expression plasmid by using restriction enzyme EcoR I to digest the original vector and cDNA. Before the ligation, alkaline phosphatase (CIP) was used to dephosphorylate the 5' end of vector. Gel-purified cDNA was ligated with pCMV-MCS vector using T4 DNA ligase. Plasmids encoding hBChE and hCocH were obtained as previously described.[16]
Protein expression and purification
All proteins (m/hBChE and m/hCocH) were expressed in CHO-S cells separately. Cells were incubated at 37°C in a humidified atmosphere containing 8% CO2, and transfected with plasmids encoding various proteins using TransIT-PRO® transfection kit once cells had grown to a density of ~1.0 × 106 cells/ml. The culture medium (Gibco® FreeStyle™ CHO expression medium with 8 mM glutamine) was harvested 7 days after transfection. Secreted enzyme in the culture medium was purified by a two-step approach described previously,[27] including ion exchange chromatography using QFF anion exchanger and affinity chromatography using procainamide-sepharose. Pre-equilibrated procainamide-sepharose was added into protein sample purified by ion exchange chromatography, and incubated for 3 h with occasional stirring. After washing the column with 20 mM potassium phosphate, 1 mM EDTA, pH 7.0 until OD280 < 0.02, enzyme was eluted by buffer containing 0.3 M NaCl and 0.1 M procainamide-HCl. The eluate was dialyzed in phosphate buffer, pH 7.4 by Millipore centrifugal filter device. The entire purification process was carried out in a cold-room at 4°C. Concentration of the active enzyme was determined through active site titration with diisopropylfluorophosphate (DFP) as described previously.[25] Purified enzymes were stored at 4°C before enzyme activity assays.
Enzyme activity assays
The catalytic activities of enzymes against (−)-cocaine were determined with a radiometric assay based on toluene extraction of [3H](−)-cocaine labeled on its benzene ring. 150 µl enzyme solution (100 mM phosphate buffer, pH 7.4) was added to 50 µl [3H](−)-cocaine solution with varying concentration. The reactions were stopped by adding 200 µl of 0.1 M HCl which neutralized the liberated benzoic acid while ensuring a positive charge on the residual (−)-cocaine. [3H]Benzoic acid was extracted by 1 ml of toluene and measured by scintillation counting. The assays to determine catalytic activity with [3H]ACh differed only in that the reaction was stopped with 200 µl of 0.2 M HCl containing 2 M NaCl. To determine catalytic activities of enzymes against BTC, UV-Vis spectrophotometric assays were carried out in a GENios Pro Microplate Reader (TECAN, Research Triangle Park, NC) with XFluor software. 100 µl enzyme solution was mixed with 50 µl of 25 mM dithiobisnitrobenzoic acid and 50 µl of BTC in varying concentrations. Reaction rates were measured by recording the time-dependent absorption at 450 nm. All measurements were performed at 25°C. Kinetic data were analyzed by performing non-linear, least-squares fitting to Eq.(1) (which accounts for the potential secondary binding site of the enzyme, i.e. a peripheral anionic binding site around D70).[36, 37]
| (1) |
In Eq.(1), S represents the concentration of the substrate, Vmax = kcat[E] in which [E] is the enzyme concentration, Kss is a binding constant for substrate at the secondary binding site, and b is a factor reflecting whether or not there is a substrate activation/inhibition. When b = 1, there is no substrate activation or inhibition, and the enzymatic reaction follows Michaelis-Menten kinetics. There is substrate activation when b > 1, and substrate inhibition when b < 1. Kinetic data were analyzed with Microsoft Excel, coding Eq.(1) for non-linear fitting.
Homology modeling
The 3D structure of mCocH was modeled based on our previously refined 3D structure[16, 38] of hCocH as a template. The hCocH structure was refined through MD simulations and hybrid quantum mechanics/molecualr mechanics (QM/MM) calculations16,37 starting from the X-ray crystal structure (PDB entry code: 1P0P)[39] available for hBChE. With the refined hCocH structure as a template, a 3D structure of mCocH was constructed and refined using the Protein Modeling module of Discovery Studio (Version 2.5.5, Accelrys, San diego, CA). The amino-acid sequence of mBChE was directly extracted from the PubMed website (NCBI access No. AAH99977), with the sequence changes necessary to generate the sequence of mCocH. The sequence alignment was generated by using ClusterW with the Blosum scoring function.[40, 41] The best alignment was selected according to both the alignment score and the reciprocal positions of the conserved residues between human and mouse proteins, particularly the residues forming the catalytic triad (S198-H438-E325) and the oxyonion hole (G116-G117-A/S199). The sequence identity between mCocH and hCocH reached 80%. The coordinates of the conserved regions were directly transformed from the template structure, whereas the non-equivalent residues were mutated from the template to the corresponding ones of mCocH. The side chains of those non-conserved residues were relaxed during the process of homology modeling in order to remove the possible steric overlap or hindrance with the neighboring conserved residues. The initial structure of mCocH was subject to energy minization by using the Sander module of the Amber program[42] with a conjugate gradient energy-minimization method and a non-bonded cutoff of 10 Å. First, the structure of mCocH was solvated in an orthorhombic box of TIP3P water molecules[43] with a minimum solute-wall distance of 10 Å. Standard protonation states at physiological environment (pH ~7.4) were used for all ionizable residues of the proteins, and the proton positions were set properly on the Nδ1 atom of histidine residues. Additional Cl− ions were added to the solvent as counter ions to neutralize the system. The final system size was about 94 Å × 91 Å × 87 Å, composed of 62,489 atoms, including 18,555 water molecules. The first 2,000 steps of the energy minimization were carried out for the backbone while the side chains were fixed, and then the next 60,000 steps for the side chains and water molecules. Finally, the system (mCocH) was energy-minimized for 6,000 steps for all atoms, and a convergence criterion of 0.001 kcal mol−1 Å−1 was achieved.
Molecular dynamics (MD) simulation
Using the homology model of mCocH, we further examined how mCocH binds with (−)-cocaine. First, (−)-cocaine was docked into the binding site, giving a binding mode similar to that for the corresponding hCocH binding with (−)-cocaine through the superposition. The atomic charges for (−)-cocaine were the restrained electrostatic potential (RESP) charges determined and used in our previous studies on hBChE and hCocH interacting with (−)-cocaine.[14, 16] MD simulations were carried out on the mCocH-(−)-cocaine binding complex by using the Sander module of the Amber program. Each system was slowly heated to 300 K by the weak-coupling method[44] and then equilibrated for 50 ps. During the MD simulations, a 10 Å non-bonded interaction cutoff was used and the non-bonded list was updated every 1,000 steps. The particle-mesh Ewald (PME) method[45] was applied to treat long-range electrostatic interactions. The lengths of covalent bonds involving hydrogen atoms were fixed with the SHAKE algorithm,[46] enabling the use of a 2-fs time step to numerically integrate the equations of motion. Finally, the production MD was kept running for 4.0 ns with a periodic boundary condition in the NTP (constant temperature and pressure) ensemble at T = 300 K with Berendsen temperature coupling and at P = 1 atm with anisotropic molecule-based scaling.[47]
RESULTS AND DISCUSSION
Catalytic parameters kcat and KM
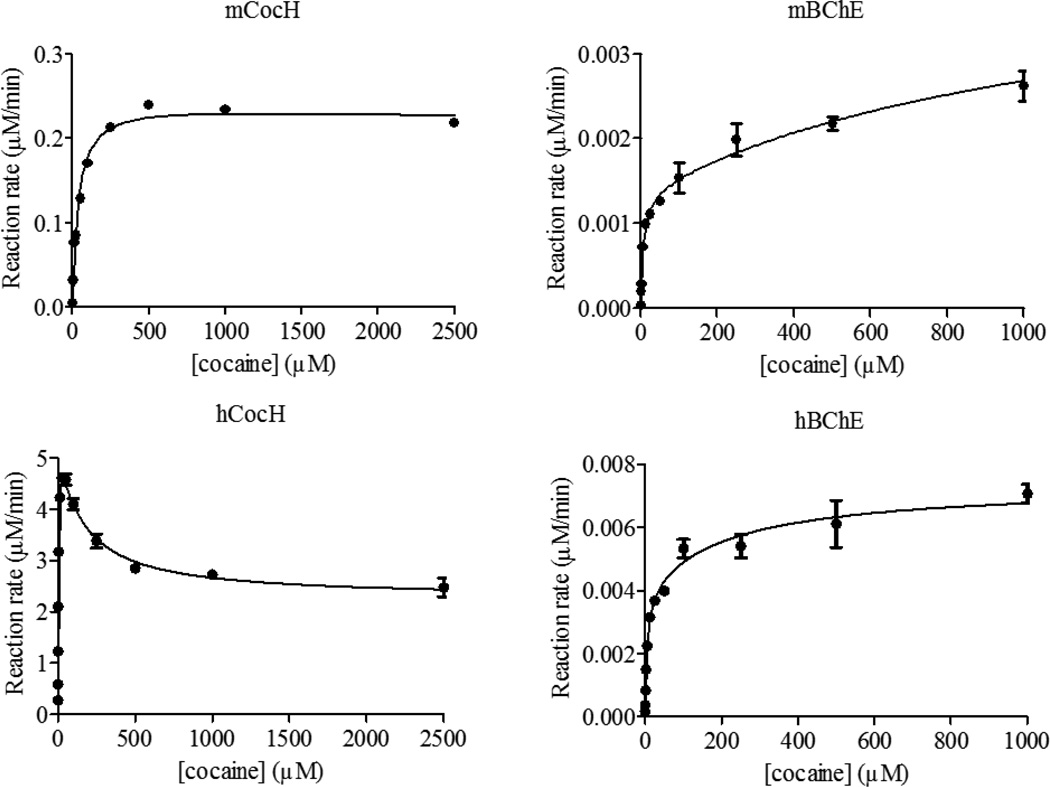
The kinetic data are depicted in Figures 1 to 3, and the obtained kinetic parameters are summarized in Table 1. As seen in Table 1, compared to hBChE, mBChE has a smaller kcat value (1.4 min−1 vs 4.1 min−1) and a smaller KM value (1.6 µM vs 4.5 µM) against (−)-cocaine. Overall, the catalytic efficiency of mBChE against (−)-cocaine (kcat/KM = 8.8 × 105 min−1 M−1) is comparable to that of hBChE (kcat/KM = 9.1 × 105 min−1 M−1). Concerning the effects of the mutations, hCocH has a ~2000-fold improved catalytic efficiency (kcat/KM) against (−)-cocaine compared to hBChE. From that standpoint, one might expect that mCocH would also have considerably greater catalytic efficiency against (−)-cocaine than mBChE. In fact, as seen in Table 1, the catalytic rate constant kcat of mCocH against (−)-cocaine is ~180-fold larger than that of mBChE against (−)-cocaine, but the KM of mCocH against (−)-cocaine is also larger than that of mBChE against (−)-cocaine (~22-fold). So, the improvement in kcat is compromised by the significant increase of KM, resulting in only ~8-fold improved catalytic efficiency over mBChE (kcat/KM = 7.1 × 106 min−1 M−1). As a result, compared to hCocH, mCocH has ~250-fold lower catalytic efficiency against (−)-cocaine.
Figure 1.
Kinetic data obtained in vitro for (−)-cocaine hydrolysis catalyzed by mCocH, mBChE, hCocH, and hBChE. The reaction rate is represented in µM min−1 per nM enzyme.
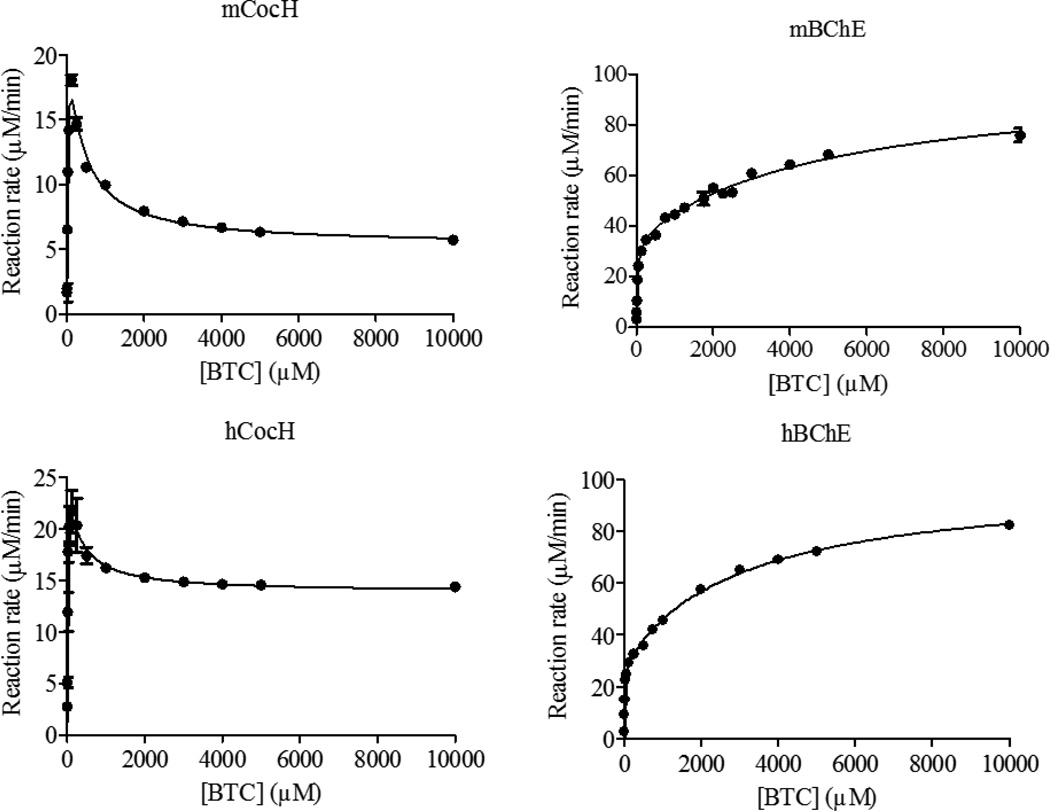
Figure 3.
Kinetic data obtained in vitro for BTC hydrolysis catalyzed by mCocH, mBChE, hCocH, and hBChE. The reaction rate is represented in µM min−1 per nM enzyme.
Table 1.
Kinetic parameters determined for (−)-cocaine, ACh, and BTC hydrolyses catalyzed by mBChE, mCocH, hBChE, and hCocH.
| Enzyme | Substrate |
kcat (min−1) |
KM (µM) |
kcat/KM (min−1 M−1) |
RCEa |
Kss (µM) |
b |
|---|---|---|---|---|---|---|---|
| mBChE | (−)-cocaine | 1.4 | 1.6 | 8.8 × 105 | 1 | 1300 | 3.13 |
| mCocH | (−)-cocaine | 250 | 35 | 7.1 × 106 | 8.2 | 1500 | 0.88 |
| hBChEb | (−)-cocaine | 4.1 | 4.5 | 9.1 × 105 | 1 | 216 | 1.80 |
| hCocHc | (−)-cocaine | 5700 | 3.1 | 1.8 × 109 | 2020 | 137 | 0.40 |
| mBChE | ACh | 38400 | 400 | 9.6× 107 | 1 | 15200 | 1.79 |
| mCocH | ACh | 19000 | 210 | 9.0 × 107 | 0.94 | 25800 | 0.30 |
| hBChEd | ACh | 61200 | 148 | 4.1 × 108 | 1 | 31600 | 2.58 |
| hCocHe | ACh | 11900 | 37 | 3.2 × 108 | 0.78 | 30000 | 0.68 |
| mBChE | BTC | 35600 | 72 | 4.9 × 108 | 1 | 5000 | 2.77 |
| mCocH | BTC | 28200 | 99 | 2.8 × 108 | 0.58 | 254 | 0.19 |
| hBChEf | BTC | 29500 | 17 | 1.7 × 109 | 1 | 3010 | 3.36 |
| hCocHe | BTC | 28000 | 13 | 2.2 × 109 | 1.24 | 243 | 0.49 |
RCE refers to the relative catalytic efficiency (kcat/KM), i.e. the ratio of the kcat/KM value of a mutant (mCocH or hCocH) to that of the corresponding wild-type enzyme (mBChE or hBChE) against the same substrate.
The kcat and KM for hBChE against (−)-cocaine were reported in ref.([8]).
The kcat and KM for hCocH against (−)-cocaine were reported in ref.([16]).
The kcat and KM for hBChE against ACh were reported in ref.([50]).
The kcat and KM for hCocH against ACh and BTC were reported in ref.([51]).
The kcat and KM for hBChE against BTC were reported in ref.([20]).
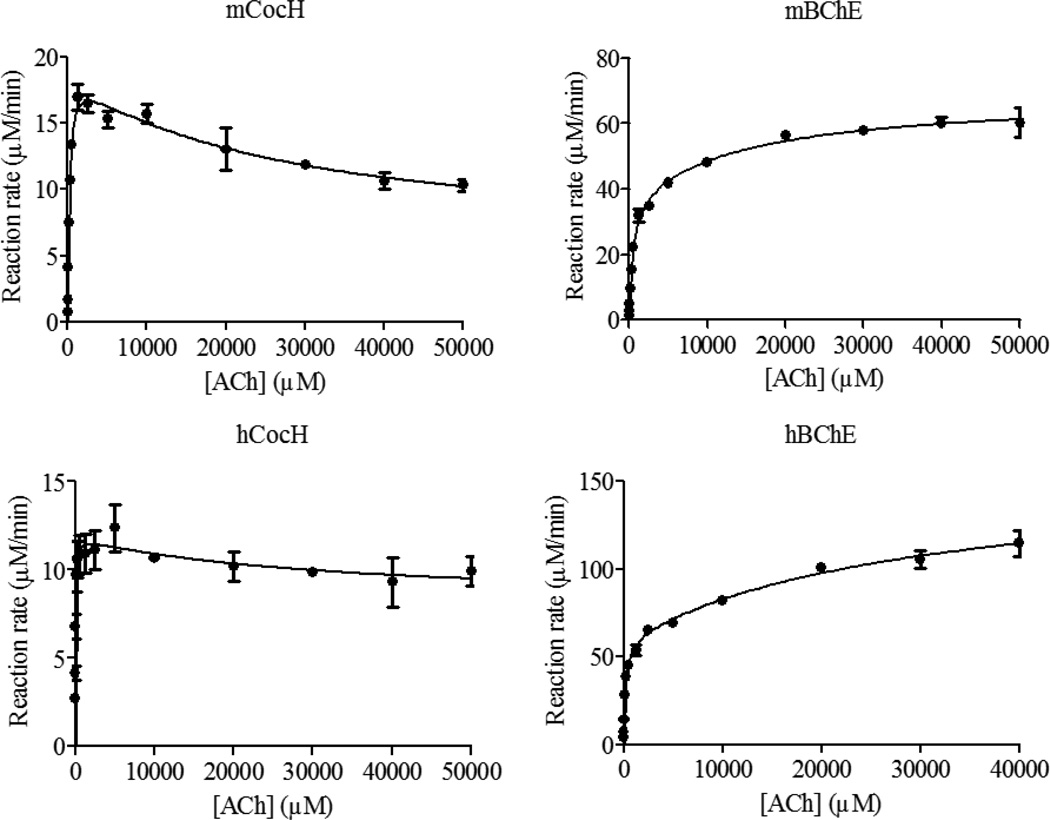
According to the kinetic parameters in Table 1, against substrate ACh, mBChE has a slightly smaller kcat value (38400 min−1 vs 61200 min−1) and a slightly larger KM value (400 µM vs 148 µM) compared to hBChE. Therefore, the catalytic efficiency (kcat/KM) of mBChE against ACh is ~4-fold lower than that of hBChE. Concerning the mutational effects on hydrolysis of ACh, both mCocH and hCocH exhibit catalytic efficiencies only slightly lower than those of the wild-type enzymes (mBChE and hBChE). In other words, the mutations caused no substantial effect.
Against substrate BTC, mBChE has a slightly larger kcat value than hBChE (35600 min−1 vs 29500 min−1) but a significantly larger KM value (72 µM vs 17 µM). Overall, the catalytic efficiency (kcat/KM) of mBChE against BTC is ~3-fold lower than that of hBChE. Concerning the mutational effects on enzyme activity against BTC, the catalytic efficiency of mCocH is only slightly lower than that of mBChE, whereas the catalytic efficiency of hCocH is only slightly higher than that of hBChE. Overall changes in the catalytic efficiency against BTC are probably not physiologically significant in either mutated enzyme.
Substrate activation/inhibition
BChE has a peripheral anionic binding site around D70, similar to acetylcholinesterase (AChE).[48] For this reason, when substrate is abundant, an additional molecule can bind during the catalytic reaction process. The binding affinity of this “side-reaction” is reflected by Kss in Eq.(1) and Table 1. Binding an additional substrate molecule at the peripheral anionic binding site may either increase catalytic activity (substrate activation, reflected by b > 1) or decrease catalytic activity (substrate inhibition, b < 1). It has long been established that BChE exhibits substrate activation with ACh, whereas AChE exhibits substrate inhibition.[48]
The data in Figures 1 to 3 reveal that substrate activation is a shared feature of both wild-type enzymes (mBChE and hBChE) with each of our three tested substrates: (b = 1.79 to 3.36). This means that the additional substrate molecule at the peripheral anionic binding site can stabilize the transition state (TS) for the rate-determining step more favorably than the corresponding reactant or intermediate associated with the TS. The result is to decrease the activation free energy and facilitate the reaction. In contrast, both mutant enzymes exhibited substrate inhibition with all three substrates (b = 0.19 to 0.88). This behavior implies that an additional substrate molecule at the peripheral anionic binding site stabilizes the TS for the rate-determining step less favorably than the corresponding reactant or intermediate associated with the TS. The mutations have converted substrate activation into substrate inhibition, with increased activation energy and slower reaction. This remarkable change may involve a shift in the rate-determining step of the enzymatic reaction. It seems reasonable that an additional substrate molecule binding to the peripheral anionic site may decrease the activation free energy for certain steps while increasing activation free energy for other reaction steps. For example, it has been known that the rate-determining step of hCocH-catalyzed hydrolysis of (−)-cocaine occurs at the acylation stage of the chemical process,[38] whereas the rate-determining step of hBChE-catalyzed (−)-cocaine hydrolysis is formation of the pre-reactive enzyme-substrate complex.[9, 49] Further experiments would be required to determine the precise steps involved.
Insights from molecular modeling
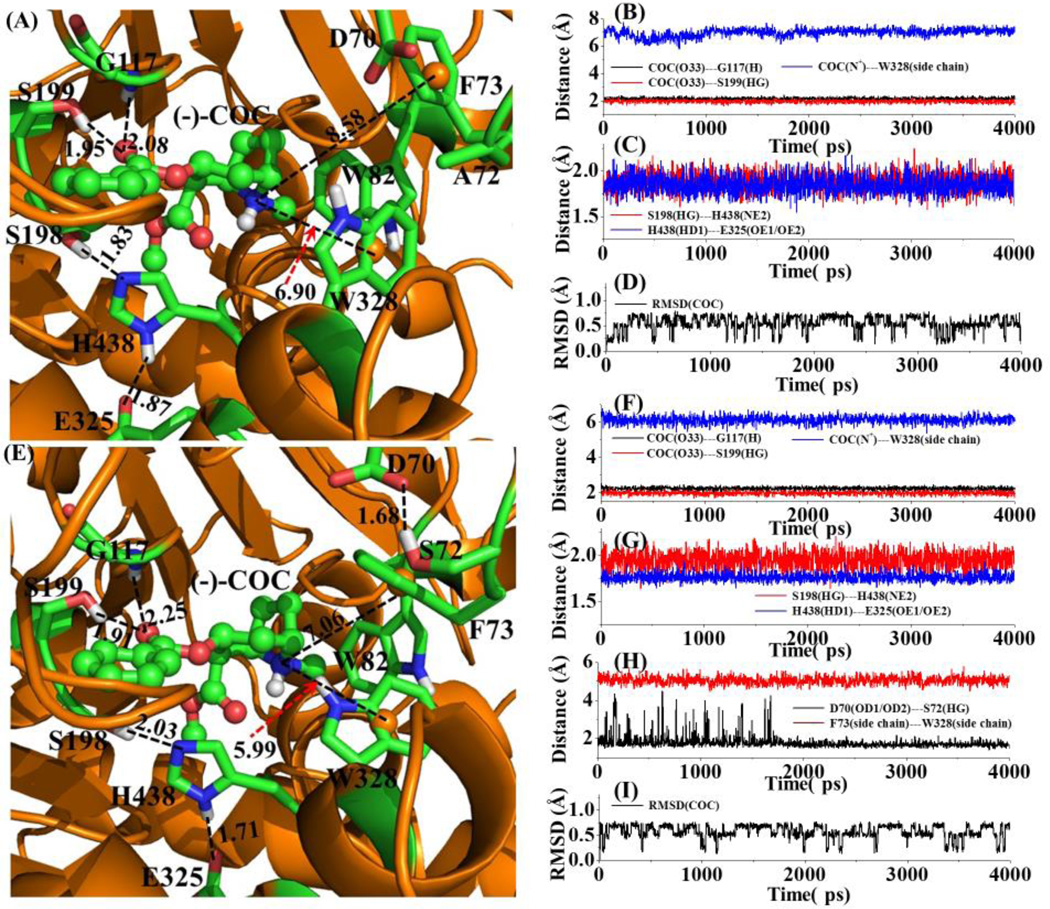
To understand why mCocH has much lower catalytic efficiency against (−)-cocaine compared to hCocH, we modeled the 3D structure of mCocH binding with (−)-cocaine for comparison with the corresponding hCocH-(−)-cocaine binding. Depicted in Figure 4 are the aligned sequences of mCocH and hCocH, showing that the overall sequence identity between these two enzymes is as high as 80%. As shown in Figure 4, mCocH and hCocH share the same residues for the catalytic triad that reacts with (−)-cocaine (S198, H438, and E325), and the same oxyanion hole residues (G116, G117, and S199) that form hydrogen bonds with the carbonyl oxygen atom on the benzoyl group of (−)-cocaine. Another common feature of cocaine binding with hCocH and mCocH is that the cationic head of (−)-cocaine has a similar cation-π interaction with the side chain of W82.
Figure 4.
Sequence alignment between mCocH and hCocH. Stars refer to identical residues, whereas filled period and double filled period refer to the conservative substitutions.
The main difference between hCocH and mCocH is that the cationic head of (−)-cocaine interacts more favorably with the protein environment including side chains of F73 and W328 in hCocH, compared to the corresponding interactions in mCocH. This appears due to a difference in the detailed shape of the binding pockets in the two enzymes. For example, residue #72 is alanine in mCocH and serine in hCocH. In hCocH, the hydroxyl group of S72 side chain forms a strong hydrogen bond with an oxygen atom in the carboxylate moiety of the D70 side chain (Figure 5C and D). This hydrogen bond apparently influences the orientation of the aromatic ring in F73 such that the cationic head of (−)-cocaine aligns nearly parallel to the vector normal to the plane of the aromatic ring of F73 side chain. As a result, the MD-simulated average distance between the positively charged N atom of (−)-cocaine and the center of the hCocH F73 side chain aromatic ring was 7.06 Å, and the MD-simulated average distance between the positively charged N atom of (−)-cocaine and the center of aromatic ring of W328 side chain was 5.99 Å.
Figure 5.
The MD-simulated structures of mCocH and hCocH binding with (−)-cocaine: (A) mCocH-(−)-cocaine binding structure; (B) and (C) plots of key distances in the mCocH-(−)-cocaine complex versus the simulation time; (D) plot of root-mean-squares deviation (RMSD) of the atomic positions of (−)-cocaine in the mCocH-(−)-cocaine complex versus the simulation time; (E) hCocH-(−)-cocaine binding structure; (F) to (H) plots of key distances in the hCocH-(−)-cocaine complex versus the simulation time; (I) plot of the RMSD of the atomic positions of (−)-cocaine in the hCocH-(−)-cocaine complex versus the simulation time. COC refers to (−)-cocaine. COC(O33)---G117(H) represents the distance between the carbonyl oxygen on the benzoyl group of (−)-cocaine and the backbone hydrogen atom of G117; COC(O33)---S199(HG) the distance between the carbonyl oxygen on the benzoyl group of (−)-cocaine and the hydroxyl hydrogen of S199 side chain; COC(N+)---W328(side chain) the distance between the positively charged nitrogen of (−)-cocaine and the center of aromatic side chain of W328; S198(HG)---H438(NE2) the distance between the hydroxyl hydrogen of S199 side chain and the nitrogen atom (NE2) of H438 side chain; H438(HD1)---E325(OE1/OE2) the distance between the hydrogen atom (HD1) on the nitrogen atom of H438 side chain; COC(C32)---S198(OG) the distance between the carbonyl carbon on the benzoyl group of (−)-cocaine and the hydroxyl oxygen of S198 side chain; D70(OD1/OD2)---S72(HG) the shortest distance between the oxygen atoms of D70 side chain and the hydroxyl hydrogen of S72 side chain; and F73(side chain)---W328(side chain) the distance between the positively charged N atom of (−)-cocaine and the center of aromatic ring of W328 side chain. All distances and RMSD are given in Å.
In mCocH, with no hydrogen bond between the side chains of A72 and D70, the side chain of altered residue #72 is farther away from D70 than it is in hCocH. This causes the orientation of the aromatic ring of the F73 side chain in mCocH to differ substantially from that in hCocH. Due to this alteration, the hydrogen atoms on the aromatic ring of F73 side chain in the initial model of mCocH-(−)-cocaine binding structure seemed too close to the (−)-cocaine atoms. After further simulation, the F73 side chain pushed away from the (−)-cocaine atoms and the MD-simulated average distance between the positively charged N atom of (−)-cocaine and the center of aromatic ring of F73 side chain grew to 8.58 Å (1.52 Å longer than that in hCocH). The difference in residue #72 also indirectly affected the interaction of (−)-cocaine with W328. The MD-simulated average distance between the positively charged N atom of (−)-cocaine and the center of the W328 side-chain aromatic ring was 6.90 Å in mCocH (0.91 Å longer than that in hCocH). Due to the less favorable interactions of the cationic head of (−)-cocaine with F73 and W328 side chains in mCocH, the overall binding of (−)-cocaine with mCocH can be expected to be weaker, which is consistent with the experimental observation that, compared to hCocH, mCocH has a significantly larger KM value and a significantly smaller kcat value against (−)-cocaine.
CONCLUSION
Kinetic analysis reveals that the catalytic efficiencies (kcat/KM) of mBChE against ACh, BTC, and (−)-cocaine resemble those of hBChE. After comparable substitutions at five homologous sites in the catalytic gorge, the corresponding mutant forms mCocH and hCocH both retain similar activities against ACh and BTC and both show enhanced hydrolysis of (−)-cocaine. However, the magnitude of enhancement differs radically between the two enzymes: ~8-fold with mCocH and ~2000-fold with hCocH, leaving the mouse protein ~250-fold less efficient with (−)-cocaine than its human counterpart. A second surprise was that ACh, BTC, and (−)-cocaine all showed substrate activation in wild-type mouse and human BChE, but uniformly caused substrate inhibition in both of the mutated enzymes. That result implies that the rate-determining step of the reactions in mCocH and hCocH may differ from that in mBChE and hBChE. These unexpected outcomes posed an interesting challenge to rational, structure and mechanism based enzyme mutation. However, homology modeling and molecular dynamics simulations shed light on the underlying causes. In other words, the observed behavior was consistent with the enzyme-(−)-cocaine binding structures obtained from molecular modeling.
Figure 2.
Kinetic data obtained in vitro for ACh hydrolysis catalyzed by mCocH, mBChE, hCocH, and hBChE. The reaction rate is represented in µM min−1 per nM enzyme.
ACKNOWLEDGMENTS
We are grateful to Dr. Palmer Taylor (Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, CA) for providing the cDNA encoding mBChE. The authors also acknowledge the Computer Center at the University of Kentucky for supercomputing time on a Dell X-series Cluster with 384 nodes or 4,768 processors.
FUNDING
This work was supported by the NIH grants R01 DA035552, R01 DA032910, R01 DA013930, R01 DA025100, and DP1 DA031340.
Footnotes
AUTHOR CONTRIBUTION
Xiabin Chen, Liyi Geng, Liu Xue, Shurong Hou, and Xirong Zheng carried out the in vivo experimental tests. Xiaoqin Huang performed the computational modeling. Stephen Brimijoin, Fang Zheng, and Chang-Guo Zhan re-analyzed the data. Xiabin Chen, Stephen Brimijoin, and Fang Zheng contributed to the manuscript preparation. Chang-Guo Zhan finalized the paper.
References
- 1.UNODC. World Drug Report 2010. 2010 (United Nations Publication, Sales No. E.10.XI.13) ed.)^eds.) [Google Scholar]
- 2.Meijler MM, Kaufmann GF, Qi L, Mee JM, Coyle AR, Moss JA, Wirsching P, Matsushita M, Janda KD. Fluorescent Cocaine Probes: A Tool for the Selection and Engineering of Therapeutic Antibodies. Journal of the American Chemical Society. 2005;127:2477–2484. doi: 10.1021/ja043935e. [DOI] [PubMed] [Google Scholar]
- 3.Carrera MRA, Kaufmann GF, Mee JM, Meijler MM, Koob GF, Janda KD. Treating cocaine addiction with viruses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10416–10421. doi: 10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landry DW, Zhao K, Yang GX, Glickman M, Georgiadis TM. Antibody-catalyzed degradation of cocaine. Science. 1993;259:1899–1901. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 5.Zhan C-G, Deng SX, Skiba JG, Hayes BA, Tschampel SM, Shields GC, Landry DW. First-principle studies of intermolecular and intramolecular catalysis of protonated cocaine. Journal of Computational Chemistry. 2005;26:980–986. doi: 10.1002/jcc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamendulis LM, Brzezinski MR, Pindel EV, Bosron WF, Dean RA. Metabolism of cocaine and heroin is catalyzed by the same human liver carboxylesterases. Journal of Pharmacology and Experimental Therapeutics. 1996;279:713–717. [PubMed] [Google Scholar]
- 7.Karila L, Gorelick D, Weinstein A, Noble F, Benyamina A, Coscas S, Blecha L, Lowenstein W, Martinot JL, Reynaud M, Lépine JP. New treatments for cocaine dependence: a focused review. The International Journal of Neuropsychopharmacology. 2008;11:425–438. doi: 10.1017/S1461145707008097. [DOI] [PubMed] [Google Scholar]
- 8.Sun H, Pang Y-P, Lockridge O, Brimijoin S. Re-engineering Butyrylcholinesterase as a Cocaine Hydrolase. Molecular Pharmacology. 2002;62:220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 9.Hamza A, Cho H, Tai H-H, Zhan C-G. Molecular Dynamics Simulation of Cocaine Binding with Human Butyrylcholinesterase and Its Mutants. The Journal of Physical Chemistry B. 2005;109:4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatley SJ. Activities of the enantiomers of cocaine and some related compounds as substrates and inhibitors of plasma butyrylcholinesterase. Biochemical Pharmacology. 1991;41:1249–1254. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 11.Darvesh S, Hopkins DA, Geula C. Neurobiology of butyrylcholinesterase. Nat Rev Neurosci. 2003;4:131–138. doi: 10.1038/nrn1035. [DOI] [PubMed] [Google Scholar]
- 12.Giacobini E. Butyrylcholinesterase: Its Function and Inhibitors. London: Martin Dunitz, an imprint of the Taylor and Francis Group plc; 2003. [Google Scholar]
- 13.Gao Y, Atanasova E, Sui N, Pancook JD, Watkins JD, Brimijoin S. Gene Transfer of Cocaine Hydrolase Suppresses Cardiovascular Responses to Cocaine in Rats. Mol. Pharmacol. 2005;67:204–211. doi: 10.1124/mol.104.006924. [DOI] [PubMed] [Google Scholar]
- 14.Pan Y, Gao D, Yang W, Cho H, Yang G, Tai H-H, Zhan C-G. Computational redesign of human butyrylcholinesterase for anticocaine medication. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16656–16661. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y, Gao D, Yang W, Cho H, Zhan C-G. Free Energy Perturbation (FEP) Simulation on the Transition States of Cocaine Hydrolysis Catalyzed by Human Butyrylcholinesterase and Its Mutants. Journal of the American Chemical Society. 2007;129:13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng F, Yang W, Ko M-C, Liu J, Cho H, Gao D, Tong M, Tai H-H, Woods JH, Zhan C-G. Most Efficient Cocaine Hydrolase Designed by Virtual Screening of Transition States. Journal of the American Chemical Society. 2008;130:12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue L, Ko M-C, Tong M, Yang W, Hou S, Fang L, Liu J, Zheng F, Woods JH, Tai H-H, Zhan C-G. Design, preparation, and characterization of high-activity mutants of human butyrylcholinesterase specific for detoxification of cocaine. Mol. Pharmacol. 2011;79:290–297. doi: 10.1124/mol.110.068494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng F, Xue L, Hou S, Liu J, Zhan M, Yang W, Zhan C-G. A highly efficient cocaine-detoxifying enzyme obtained by computational design. Nature Commun. 2014;5:3457. doi: 10.1038/ncomms4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Xue L, Fang L, Chen X, Zhan C-G. Characterization of a high-activity mutant of human butyrylcholinesterase against (−)-cocaine. Chem. Biol. Interact. 2010;187:148–152. doi: 10.1016/j.cbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, LaFleur D, Shah R, Zhao Q, Singh M, Brimijoin S. An albumin-butyrylcholinesterase for cocaine toxicity and addiction: Catalytic and pharmacokinetic properties. Chemico-Biological Interactions. 2008;175:83–87. doi: 10.1016/j.cbi.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anker JJ, Brimijoin S, Gao Y, Geng L, Zlebnik NE, Parks RJ, Carroll ME. Cocaine hydrolase encoded in viral vector blocks the reinstatement of cocaine seeking in rats for 6 months. Biol. Psychiatry. 2012;71:700–705. doi: 10.1016/j.biopsych.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brimijoin S, Orson F, Kosten TR, Kinsey B, Shen XY, White SJ, Gao Y. Anti-cocaine antibody and butyrylcholinesterase-derived cocaine hydrolase exert cooperative effects on cocaine pharmacokinetics and cocaine-induced locomotor activity in mice. Chem. Biol. Interact. 2013;203:212–216. doi: 10.1016/j.cbi.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Geng L, Orson F, Kinsey B, Kosten TR, Shen X, Brimijoin S. Effects of anti-cocaine vaccine and viral gene transfer of cocaine hydrolase in mice on cocaine toxicity including motor strength and liver damage. Chem. Biol. Interact. 2013;203:208–211. doi: 10.1016/j.cbi.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brimijoin S, Gao Y, Anker JJ, Gliddon LA, Lafleur D, Shah R, Zhao Q, Singh M, Carroll ME. A cocaine hydrolase engineered from human butyrylcholinesterase selectively blocks cocaine toxicity and reinstatement of drug seeking in rats. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2008;33:2715–2725. doi: 10.1038/sj.npp.1301666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng L, Gao Y, Chen X, Hou S, Zhan C-G, Radic Z, Parks R, Russell SJ, Pham L, Brimijoin S. Gene transfer of mutant mouse cholinesterase provides high lifetime expression and reduced cocaine responses with no evident toxicity. PLoS One. 2013;8:e67446. doi: 10.1371/journal.pone.0067446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zlebnik NE, Brimijoin S, Gao Y, Saykao AT, Parks RJ, Carroll ME. Long-term reduction of cocaine self-administration in rats treated with adenoviral vector-delivered cocaine hydrolase: evidence for enzymatic activity. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2014;39:1538–1546. doi: 10.1038/npp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue L, Hou S, Tong M, Fang L, Chen X, Jin Z, Tai H-H, Zheng F, Zhan C-G. Preparation and in vivo characterization of a cocaine hydrolase engineered from human butyrylcholinesterase for metabolizing cocaine. Biochem. J. 2013;453:447–454. doi: 10.1042/BJ20130549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhan M, Hou S, Zhan C-G, Zheng F. Kinetic characterization of high-activity mutants of human butyrylcholinesterase for cocaine metabolite norcocaine. Biochem. J. 2014;457:197–206. doi: 10.1042/BJ20131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou S, Zhan M, Zheng X, Zhan C-G, Zheng F. Kinetic characterization of human butyrylcholinesterase mutants for hydrolysis of cocaethylene. Biochem. J. 2014;460:447–457. doi: 10.1042/BJ20140360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena A, Sun W, Luo C, Doctor BP. Human serum butyrylcholinesterase: in vitro and in vivo stability, pharmacokinetics, and safety in mice. Chem Biol Interact. 2005;157–158:199–203. doi: 10.1016/j.cbi.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 31.Saxena A, Sun W, Fedorko JM, Koplovitz I, Doctor BP. Prophylaxis with human serum butyrylcholinesterase protects guinea pigs exposed to multiple lethal doses of soman or VX. Biochem Pharmacol. 2011;81:164–169. doi: 10.1016/j.bcp.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Weber A, Butterweck H, Mais-Paul U, Teschner W, Lei L, Muchitsch EM, Kolarich D, Altmann F, Ehrlich HJ, Schwarz HP. Biochemical, molecular and preclinical characterization of a double-virus-reduced human butyrylcholinesterase preparation designed for clinical use. Vox Sang. 2011;100:285–297. doi: 10.1111/j.1423-0410.2010.01415.x. [DOI] [PubMed] [Google Scholar]
- 33.Sun W, Luo C, Naik RS, Doctor BP, Saxena A. Pharmacokinetics and immunologic consequences of repeated administrations of purified heterologous and homologous butyrylcholinesterase in mice. Life Sci. 2009;85:657–661. doi: 10.1016/j.lfs.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Arpagaus M, Chatonnet A, Masson P, Newton M, Vaughan TA, Bartels CF, Nogueira CP, La Du BN, Lockridge O. Use of the polymerase chain reaction for homology probing of butyrylcholinesterase from several vertebrates. J Biol Chem. 1991;266:6966–6974. [PubMed] [Google Scholar]
- 35.Braman J, Papworth C, Greener A. Site-Directed Mutagenesis Using Double-Stranded Plasmid DNA Templates. The Nucleic Acid Protocols Handbook. 2000:835–844. doi: 10.1385/0-89603-332-5:31. [DOI] [PubMed] [Google Scholar]
- 36.Masson P, Xie W, Froment MT, Levitsky V, Fortier PL, Albaret C, Lockridge O. Interaction between the peripheral site residues of human butyrylcholinesterase, D70 and Y332, in binding and hydrolysis of substrates. Biochimica et biophysica acta. 1999;1433:281–293. doi: 10.1016/s0167-4838(99)00115-6. [DOI] [PubMed] [Google Scholar]
- 37.Boeck AT, Schopfer LM, Lockridge O. DNA sequence of butyrylcholinesterase from the rat: expression of the protein and characterization of the properties of rat butyrylcholinesterase. Biochem Pharmacol. 2002;63:2101–2110. doi: 10.1016/s0006-2952(02)01029-8. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Zhan C-G. Reaction pathway and free energy profile for cocaine hydrolase-catalyzed hydrolysis of (−)-cocaine. J. Chem. Theory Comput. 2012;8:1426–1435. doi: 10.1021/ct200810d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon FJ. Crystal Structure of Human Butyrylcholinesterase and of Its Complexes with Substrate and Products. J. Biol. Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 40.Henikoff S, Henikoff JG. Amino-acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER11. San Francisco: University of California; 2010. [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 44.Morishita T. Fluctuation formulas in molecular dynamics simulations with the weak coupling heat bath. J. Chem. Phys. 2000;113:2976–2982. [Google Scholar]
- 45.Toukmaji A, Sagui C, Board J, Darden TA. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J. Chem. Phys. 2000;113:10913–10927. [Google Scholar]
- 46.Ryckaert J, Ciccotti PG, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 47.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 48.Chiou S-Y, Wu Y-G, Lin Y-F, Lin L-Y, Lin G. Substrate Activation of Butyrylcholinesterase and Substrate Inhibition of Acetylcholinesterase by 3,3-Dimethylbutyl-N-n-butylcarbamate and 2-Trimethylsilyl-ethyl-N-n-butylcarbamate. J. Biochem. Mol. Toxicol. 2007;21:24–31. doi: 10.1002/jbt.20158. [DOI] [PubMed] [Google Scholar]
- 49.Sun H, El Yazal J, Lockridge O, Schopfer LM, Brimijoin S, Pang YP. Predicted Michaelis-Menten complexes of cocaine-butyrylcholinesterase. Engineering effective butyrylcholinesterase mutants for cocaine detoxication. J. Biol. Chem. 2001;276:9330–9336. doi: 10.1074/jbc.M006676200. [DOI] [PubMed] [Google Scholar]
- 50.Brimijoin S, Gao Y, Anker JJ, Gliddon LA, LaFleur D, Shah R, Zhao Q, Singh M, Carroll ME. A Cocaine Hydrolase Engineered from Human Butyrylcholinesterase Selectively Blocks Cocaine Toxicity and Reinstatement of Drug Seeking in Rats. Neuropsychopharmacology. 2008;33:2715–2725. doi: 10.1038/sj.npp.1301666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou S, Xue L, Yang W, Fang L, Zheng F, Zhan C-G. Substrate selectivity of high-activity mutants of human butyrylcholinesterase. Org. Biomol. Chem. 2013;11:7477–7485. doi: 10.1039/c3ob41713a. [DOI] [PMC free article] [PubMed] [Google Scholar]