Abstract
Nakagawa & Schielzeth extended the widely used goodness-of-fit statistic R2 to apply to generalized linear mixed models (GLMMs). However, their R2GLMM method is restricted to models with the simplest random effects structure, known as random intercepts models. It is not applicable to another common random effects structure, random slopes models.
I show that R2GLMM can be extended to random slopes models using a simple formula that is straightforward to implement in statistical software. This extension substantially widens the potential application of R2GLMM.
Keywords: coefficient of determination, generalized linear mixed model, random slopes model, random regression
Introduction
The coefficient of determination, R2, is a widely used statistic for assessing the goodness-of-fit, on a scale from 0 to 1, of a linear regression model (LM). It is defined as the proportion of variance in the response variable that is explained by the explanatory variables or, equivalently, the proportional reduction in unexplained variance. Unexplained variance can be viewed as variance in model prediction error, so R2 can also be defined in terms of reduction in prediction error variance. Insofar as it is justifiable to make the leap from ‘prediction’ to ‘understanding’, R2 can be intuitively interpreted as a measure of how much better we understand a system once we have measured and modelled some of its components.
R2 has been extended to apply to generalized linear models (GLMs) (Maddala 1983) and linear mixed effects models (LMMs) (Snijders & Bosker 1994) [reviewed by (Nakagawa & Schielzeth 2013)]. Nakagawa & Schielzeth (2013) proposed a further generalization of R2 to generalized linear mixed effects models (GLMMs), a useful advance given the ubiquity of GLMMs for data analysis in ecology and evolution (Bolker et al. 2009). A function to estimate this R2GLMM statistic, r.squaredGLMM, has been included in the MuMIn package (Bartoń 2014) for the R statistical software (R Core Team 2014). However, Nakagawa and Schielzeth's R2GLMM formula is applicable to only a subset of GLMMs known as random intercepts models. Random intercepts models are used to model clustered observations, for example, where multiple observations are taken on each of a sample of individuals. Correlations between clustered observations within individuals are accounted for by allowing each subject to have a different intercept representing the deviation of that subject from the global intercept. Random intercepts are typically modelled as being sampled from a normal distribution with mean zero and a variance parameter that is estimated from the data. Although random intercepts are probably the most popular random effects models in ecology and evolution, other random effect specifications are also common, in particular random slopes models, where not only the intercept but also the slope of the regression line is allowed to vary between individuals. Random intercepts and slopes are typically modelled as normally distributed deviations from the global intercept and slope, respectively. For example, random slopes models, under the name of ‘random regression’ models, are used to investigate individual variation in response to different environments (Nussey, Wilson & Brommer 2007). The aim of this article is to show how Nakagawa and Schielzeth's R2GLMM can be further extended to encompass random slopes models.
Nakagawa and Schielzeth's R2GLMM
Nakagawa & Schielzeth (2013) defined two R2 statistics for GLMMs, marginal and conditional R2GLMM, that allow separation of the contributions of fixed and random effects to explaining variation in the responses. Marginal R2GLMM gauges the variance explained by the fixed effects as a proportion of the sum of all the variance components:
| eqn 1 |
where  is the variance attributable to the fixed
effects,
is the variance attributable to the fixed
effects,  is the variance of the lth of
u random effects,
is the variance of the lth of
u random effects,  is
the variance due to additive dispersion and
is
the variance due to additive dispersion and  is
the distribution-specific variance. The residual variance,
is
the distribution-specific variance. The residual variance,  , is
defined as
, is
defined as  for the purposes of this manuscript but see
Nakagawa & Schielzeth (2013) for an alternative
definition of dispersion. Conditional R2 additionally includes in the
numerator the variance explained by the random effects:
for the purposes of this manuscript but see
Nakagawa & Schielzeth (2013) for an alternative
definition of dispersion. Conditional R2 additionally includes in the
numerator the variance explained by the random effects:
| eqn 2 |
It is the definition of the random effect variances, the  ,
that requires generalization to allow R2GLMM (m) and
R2GLMM (c) to be extended beyond random intercepts models. In
Nakagawa and Schielzeth's formula,
,
that requires generalization to allow R2GLMM (m) and
R2GLMM (c) to be extended beyond random intercepts models. In
Nakagawa and Schielzeth's formula,  is
simply the variance of the l th random intercept. This formula is correct for
random intercept models because each observation has the same random effect variance. However, in
other random effects specifications, the random effect variance can differ between observations,
and, as pointed out by Nakagawa and Schielzeth, this causes difficulties in computing a single
random effect variance component.
is
simply the variance of the l th random intercept. This formula is correct for
random intercept models because each observation has the same random effect variance. However, in
other random effects specifications, the random effect variance can differ between observations,
and, as pointed out by Nakagawa and Schielzeth, this causes difficulties in computing a single
random effect variance component.
Extension of R2GLMM to random slopes models
Consider the simplest and most familiar random slopes GLMM, a LMM with a single random intercept and a single random slope:
| eqn 3 |
| eqn 4 |
| eqn 5 |
| eqn 6 |
where
Yij and
xij are, respectively, the response and predictor
values (covariates) for the ith observation on the jth individual.
Random deviation of the jth individual from the fixed global intercept,
β0, is represented by α0j,
while random deviation from the fixed global slope, β1, is
represented by α1j. Because intercepts and slopes are typically
correlated, three parameters are required to model the random effect, which are represented by the
covariance matrix Σ. The leading diagonal of Σ consists of
the random intercept variance,  ,
and the random slope variance,
,
and the random slope variance,  ,
while the off-diagonal element is the covariance, σα0α1, between the
random intercept and random slope. Finally, ɛij
is the residual of the ith observation on the jth individual and
,
while the off-diagonal element is the covariance, σα0α1, between the
random intercept and random slope. Finally, ɛij
is the residual of the ith observation on the jth individual and
 is the residual variance. For LMMs,
is the residual variance. For LMMs,
 , so that
, so that  .
.
The difficulty of defining  for this model arises from the dependence of the random effect variance component on
xij, which implies that
for this model arises from the dependence of the random effect variance component on
xij, which implies that  cannot be defined from Σ alone, but requires input from the
xij. An observation-specific random effect variance,
cannot be defined from Σ alone, but requires input from the
xij. An observation-specific random effect variance,
 , can be defined, given
xij, as
, can be defined, given
xij, as
| eqn 7 |
showing the dependence of  on
xij. For example, when
xij = 0 (i.e. at the intercept),
on
xij. For example, when
xij = 0 (i.e. at the intercept),
| eqn 8 |
while when xij = 1,
 |
eqn 9 |
(Snijders
& Bosker 2012). In the most extreme case where the
xij values are unique, there will be as many random
effect variances as observations. The first step to estimating the random effect variance component
is to estimate each  . The random effect portion of the model,
α0j + α1jxij,
can then be viewed as a mixture of n normal distributions with a common mean of
zero but up to n different variances, where n is the number of
observations. When the mean is constant, the variance of a mixture is simply the mean of the
individual variances (Behboodian 1970). The mean random effect
variance is therefore
. The random effect portion of the model,
α0j + α1jxij,
can then be viewed as a mixture of n normal distributions with a common mean of
zero but up to n different variances, where n is the number of
observations. When the mean is constant, the variance of a mixture is simply the mean of the
individual variances (Behboodian 1970). The mean random effect
variance is therefore
|
|
eqn 9 |
A simple and general formula for  given any value of xij can be derived as follows. For
any random effects specification, let Z be the design matrix of the random effects of a
GLMM with n rows and k columns corresponding to the
k random effects, and Σ the covariance matrix of the random
effects of dimension k. For example, in the simple random slopes model in equations
3-6, the first column of Z is a vector of ones corresponding to the random intercept,
while the second is the predictor variable, the xij.
The vector of observation-level random effect variances is the leading diagonal of the
n × n matrix ZΣZ′,
where Z′ is the transpose of Z (Laird & Ware 1982). The mean random effect variance,
given any value of xij can be derived as follows. For
any random effects specification, let Z be the design matrix of the random effects of a
GLMM with n rows and k columns corresponding to the
k random effects, and Σ the covariance matrix of the random
effects of dimension k. For example, in the simple random slopes model in equations
3-6, the first column of Z is a vector of ones corresponding to the random intercept,
while the second is the predictor variable, the xij.
The vector of observation-level random effect variances is the leading diagonal of the
n × n matrix ZΣZ′,
where Z′ is the transpose of Z (Laird & Ware 1982). The mean random effect variance,  ,
is the mean of this vector, that is,
,
is the mean of this vector, that is,
| eqn 11 |
where
the Tr denotes the trace operation, which sums the leading diagonal. An index notation version of
the matrix notation equation 11 is contained within equation 20 of Snijders & Bosker (1994). The advantage of the matrix version is computational
simplicity. Equation 11 gives the same results as Nakagawa & Schielzeth's method for
random intercepts models but can also be used for random slopes models as well as models with no
intercept. An estimate of  for use in Equations 1 and 2 can be easily
computed from the estimated covariance matrix of the lth random effect. Examples of
the application of this procedure to estimating R2GLMM from
random slopes GLMMs using R are provided as Data S1.
for use in Equations 1 and 2 can be easily
computed from the estimated covariance matrix of the lth random effect. Examples of
the application of this procedure to estimating R2GLMM from
random slopes GLMMs using R are provided as Data S1.
The Supplementary R code also illustrates a simplified method of estimating the term
β0 in equation A6 of Nakagawa
& Schielzeth (2013), which approximates 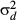 for a Poisson GLMM. Rather than refit the model after centring or dropping the covariates as
recommended, β0 can be more easily estimated by taking the mean
of
for a Poisson GLMM. Rather than refit the model after centring or dropping the covariates as
recommended, β0 can be more easily estimated by taking the mean
of  , the linear predictor, where X
is the design matrix for the fixed effects and
, the linear predictor, where X
is the design matrix for the fixed effects and  is
the vector of fixed effect estimates.
is
the vector of fixed effect estimates.
These extensions to R2GLMM have been incorporated into the r.squaredGLMM function in version 1.10.0 of the MuMIn package (Bartoń 2014).
Discussion
The extension described above allows both marginal and conditional
R2GLMM to be estimated from a random slopes model, obviating
the need to approximate R2GLMM from the corresponding random
intercepts model as recommended by Nakagawa & Schielzeth
(2013). It is clearly preferable to estimate R2GLMM
from the correct model given that there is no computational cost but is the improvement in either
marginal or conditional R2GLMM likely to be substantial?
Nakagawa & Schielzeth (2013) suggest that marginal and
condition R2GLMM will usually be very similar when
approximated from a random intercepts fit, and Snijders &
Bosker (2012) make a similar claim for their related
R21 and R22
statistics. Not surprisingly, the gain in accuracy in both
R2GLMM statistics will depend on how well the random
intercepts model approximates the random slopes model. The accuracy of the marginal
R2GLMM approximation will depend on the accuracy of the
global slope (or slopes) estimate from the random intercepts model, because the scale of the global
slope (or slopes) estimate determines  (Nakagawa & Schielzeth 2013), which in turn determines
marginal R2GLMM. For balanced data, where the numbers of
observations and the covariate distributions are balanced between groups, this approximation should
be good, so the estimates of the global slope and marginal
R2GLMM are likely to be very similar under both models.
However, unbalanced data are common in ecology, for example where sampling strategies are
constrained in space by variable access to sampling sites or in time by fluctuating resources, and
in such cases the improvement in marginal R2GLMM could be
considerable. For example, if one individual (or site, etc.) yields an unusually large number of
observations, the global slope estimate will be biased towards that individual in a random
intercepts model but not in a random slopes model. Examples of both scenarios are given in the
Supplementary R code (Data S1).
(Nakagawa & Schielzeth 2013), which in turn determines
marginal R2GLMM. For balanced data, where the numbers of
observations and the covariate distributions are balanced between groups, this approximation should
be good, so the estimates of the global slope and marginal
R2GLMM are likely to be very similar under both models.
However, unbalanced data are common in ecology, for example where sampling strategies are
constrained in space by variable access to sampling sites or in time by fluctuating resources, and
in such cases the improvement in marginal R2GLMM could be
considerable. For example, if one individual (or site, etc.) yields an unusually large number of
observations, the global slope estimate will be biased towards that individual in a random
intercepts model but not in a random slopes model. Examples of both scenarios are given in the
Supplementary R code (Data S1).
Improvement in conditional R2GLMM is easier to predict and explain. Regardless of the adequacy of the marginal R2GLMM approximation, if the random slopes model fits substantially better than the random intercepts model, it should have lower residual variance (or less overdispersion, in the context of overdispersed Poisson or binomial GLMMs) and therefore higher conditional R2GLMM.
This extension will apply to other statistics that incorporate a random effects variance component calculated from a random slopes model, including the intraclass correlation coefficient (ICC), which gauges variance between groups (e.g. individuals or sites) as a proportion of the total variance. ICC can be used to measure intraindividual repeatability, also known as consistency, and has been applied widely in ecology and evolutionary biology (Nakagawa & Schielzeth 2010). Like R2, ICC has also been generalized to random intercepts GLMMs by Nakagawa & Schielzeth (2010), but not to random slopes GLMMs. Equation 11 could also be applied to calculating repeatability (Nakagawa & Schielzeth 2010) by fixing a column of Z to a single value. For example, age dependence in phenotypic consistency could be investigated by estimating ICC conditioned on a range of ages.
In conclusion, the extension of R2GLMM to random slopes GLMMs substantially widens the range of models to which this useful measure can be applied.
Acknowledgments
I am grateful to S. Nakagawa, K. Bartoń and J. Lindström for helpful discussions, and to H. Schielzeth and two anonymous reviewers, whose comments greatly improved this manuscript. This work was supported by a BBSRC project grant (BB/K004484/1).
Data accessibility
R scripts: uploaded as online supporting information.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
R code illustrating the calculation of R2GLMM.
References
- Bartoń K. MuMIn: Multi-model inference. R package version 1.10.0. 2014. Retrieved May 14, 2014, from http://cran.r-project.org/package=MuMIn.
- Behboodian J. On a Mixture of Normal Distributions. Biometrika. 1970;57:215–217. [Google Scholar]
- Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH. White J-SS. Generalized linear mixed models: a practical guide for ecology and evolution. Trends in Ecology & Evolution. 2009;24:127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Laird NM. Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Maddala GS. Limited-Dependent and Qualitative Variables in Econometrics. Cambridge, UK: Cambridge University Press; 1983. 1st edn. [Google Scholar]
- Nakagawa S. Schielzeth H. Repeatability for Gaussian and non-Gaussian data: a practical guide for biologists. Biological Reviews of the Cambridge Philosophical Society. 2010;85:935–956. doi: 10.1111/j.1469-185X.2010.00141.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa S. Schielzeth H. A general and simple method for obtaining R^2 from generalized linear mixed-effects models. Methods in Ecology and Evolution. 2013;4:133–142. [Google Scholar]
- Nussey DH, Wilson AJ. Brommer JE. The evolutionary ecology of individual phenotypic plasticity in wild populations. Journal of Evolutionary Biology. 2007;20:831–844. doi: 10.1111/j.1420-9101.2007.01300.x. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. Retrieved April 10, 2014, from http://www.r-project.org/ [Google Scholar]
- Snijders TAB. Bosker RJ. Modeled variance in two-level models. Sociological Methods & Research. 1994;22:342–363. [Google Scholar]
- Snijders TAB. Bosker RJ. Multilevel Analysis: An Introduction to Basic and Advanced Multilevel Modeling. London: Sage; 2012. 2nd edn. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R code illustrating the calculation of R2GLMM.
Data Availability Statement
R scripts: uploaded as online supporting information.


