Abstract
We have developed an informatics system, GeneMed, for the National Cancer Institute (NCI) molecular profiling-based assignment of cancer therapy (MPACT) clinical trial (NCT01827384) being conducted in the National Institutes of Health (NIH) Clinical Center. This trial is one of the first to use a randomized design to examine whether assigning treatment based on genomic tumor screening can improve the rate and duration of response in patients with advanced solid tumors. An analytically validated next-generation sequencing (NGS) assay is applied to DNA from patients’ tumors to identify mutations in a panel of genes that are thought likely to affect the utility of targeted therapies available for use in the clinical trial. The patients are randomized to a treatment selected to target a somatic mutation in the tumor or with a control treatment. The GeneMed system streamlines the workflow of the clinical trial and serves as a communications hub among the sequencing lab, the treatment selection team, and clinical personnel. It automates the annotation of the genomic variants identified by sequencing, predicts the functional impact of mutations, identifies the actionable mutations, and facilitates quality control by the molecular characterization lab in the review of variants. The GeneMed system collects baseline information about the patients from the clinic team to determine eligibility for the panel of drugs available. The system performs randomized treatment assignments under the oversight of a supervising treatment selection team and generates a patient report containing detected genomic alterations. NCI is planning to expand the MPACT trial to multiple cancer centers soon. In summary, the GeneMed system has been proven to be an efficient and successful informatics hub for coordinating the reliable application of NGS to precision medicine studies.
Keywords: GeneMed, MPACT, next-generation sequencing, precision medicine, informatics system, clinical trial
Introduction
With the dramatic increase in sequencing speed, rapid reduction of cost, and improvement in accuracy over the past decade, next-generation sequencing (NGS) technologies have emerged as powerful tools for the clinical diagnosis and treatment of patients with cancer and other diseases.1 Successes in the development of drugs targeting genomic alterations in tumors have provided momentum for the development of precision medicine approaches to cancer treatment guided by NGS technologies.2–7
The National Cancer Institute (NCI) is now conducting a clinical trial (NCT01827384) titled MPACT (molecular profiling based assignment of cancer therapy) to treat late-stage cancer patients guided by their targeted gene sequencing profile.8,9 Patients with progressive solid tumors who have exhausted standard treatments undergo biopsy and have their tumors characterized based on amplicon sequencing of over 380 unique actionable variants in 20 genes.9 Four treatment regimens are used in the MPACT trial: trametinib, everolimus, veliparib plus temozolomide, or AZD-1775 (MK-1775) plus carboplatin. The sequencing assay is designed to identify mutations that are likely to deregulate any of three pathways: RAS/RAF/MEK, PI3K, and DNA damage repair.9 Each mutation is designated as actionable or not for each of the four treatment regimens using pre-specified rules. The rules for actionability were derived from literature documentation and previous reporting in the Catalogue of Somatic Mutations in Cancer (COSMIC) database10 and evidence of functional effects for loss-of-function genes.9 The treatment for which a mutation is actionable is based on the mechanism of action of the drug or drug combination in the treatment. Patients whose tumors contain at least one actionable mutation are randomized to either the actionable study treatment prospectively identified to work on that mutation/pathway or a control arm consisting of one of the other treatments.
Clinical trials of this type are operationally complex, requiring a new tumor biopsy that is rapidly shipped to a Clinical Laboratory Improvement Amendments (CLIA)-accredited molecular characterization laboratory for application of an analytically validated DNA sequencing assay, development of a computational pipeline for detecting genomic variants, annotation of the variants to determine the actionable mutations, complex informed consent and randomization procedures, and documentation of the sequencing findings in the medical record. An informatics system is required for such clinical trials to enable the translation of clinical grade NGS data into clinically meaningful information in a short time frame. The informatics system also serves as a communication hub linking the sequencing lab, the treatment selection oversight team, and the clinic team. In addition to the upstream bioinformatics pipeline for raw data processing and sequence alignment, the system must have the ability to quickly annotate the sequencing data, predict the functional impact of mutations, and match the detected mutation to the pre-identified genomic changes in tumors found in previous studies (ie, the actionable mutations of interest). The informatics hub should translate the mutation finding from a sequencing result into a concise and easy-to-read actionable report for the treatment selection oversight team in a timely manner. This must occur before the team submits the mutation summary to the informatics system, which then assigns patients to different arms based on their detected mutations and a randomization table. The system also collects and stores patient data, including response data for prior therapies, that are required to evaluate eligibility of each treatment in the study and assist the treatment selection team and the clinic team in the development of a treatment plan. Finally, the system is required to make both sequencing data and clinical response data available to statisticians who are able to evaluate whether there is any improvement in response rate or progression-free survival (PFS) favoring assignment of therapy based on genetic sequencing and to determine which actionable mutation-treatment pairs lead to improved patient outcome.
We have developed an informatics system (GeneMed) that fulfills the tasks required by the MPACT clinical trial for guiding treatment based on sequencing information from patient’s tumors. Here we first report the GeneMed system design and functionality of each component within the system. We then discuss the philosophy of system development that made completion of a reliable working system feasible in a short time frame, the challenges in development process, limitations of the system, and our plans for future extensions of the system.
System Design and Architecture
Working groups for the MPACT trial
There are four groups of people who work on the MPACT trial. The tasks of these groups are as follows:
Lab group: The molecular characterization lab members perform the NGS assays and Sanger sequence validation and perform gene variant reviews on each actionable mutation of interest (aMOI) identified by the GeneMed system.
Clinic group: Treating physicians and research nurses in the National Institutes of Health (NIH) Clinical Center who recruit and register patients, gather patient clinical information, treat patients, and delink patient identification after the patient’s disease has progressed on treatment or when the patient is taken off the study.
Treatment selection group: Clinicians and scientists who submit mutation summary reports that trigger the GeneMed system to generate treatment options.
Biostatistics group: Statisticians who designed the randomized clinical trial, monitor accrual rates, and analyze the data.
Each group can only access data related to the assigned task of the group. For example, staff in lab group can only access sequence data but not patient clinical data, while members in clinic group can only access patient clinic information but not sequence data before the patient is delinked.
Workflow and communication among different working groups for the MPACT trial
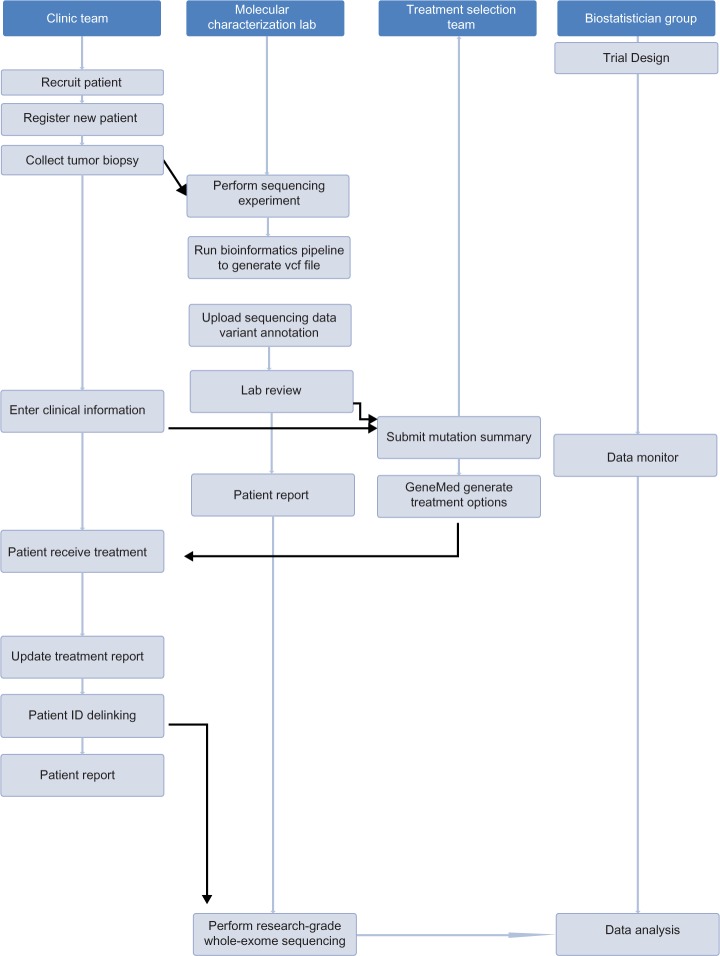
It is necessary that the four groups work collaboratively in an enterprise environment to coordinate and accomplish the tasks required by the MPACT trial. The workflow and communications among different user groups are shown in Figure 1, where all major tasks for each working group are sequentially ordered from top to bottom.
Figure 1.
Workflow for four groups who work on the MPACT trial. The order of tasks is shown as vertical from top to bottom. The tasks in the shaded area are accomplished by the GeneMed system.
When a new patient enters the trial, a member of the clinic group accesses the GeneMed system and registers the patient using the clinical trial assigned patient ID (protocol ID). Some basic demographic information for this patient is entered into the system. The patient’s tumor biopsy sample will be collected and delivered to the molecular characterization lab.
The lab receives the sample and performs the sequencing assay using the Ion Torrent PGM sequencer (Life Technologies) with the custom MPACT AmpliSeq panel. The raw data are then processed on a server in the lab using a bioinformatics pipeline for sequence alignment and variant calling. A file containing the variants in VCF (variant call format) version 4.2 format (https://github.com/samtools/hts-specs) is generated after sequence alignment and quality control. At this point, the lab member goes into the GeneMed system and uploads the VCF file. The GeneMed system stores the VCF file, automatically performs annotation of each variant, and makes functional predictions for each non-synonymous mutation. The assay for the MPACT trial is a targeted amplicon-based sequencing assay. Most of the amplicons are located entirely within exonic regions. The MPACT clinic trial only focuses on non-synonymous mutations. A variant is considered actionable if the detected genomic alteration is in the predefined aMOI list or if the alteration meets one of the predefined actionability rules. The date and user name of the submission is recorded, and an email is sent to the sequencing lab indicating a new variant file has been processed and is ready for review.
After receiving the email notice, the lab reviewer logs into the GeneMed system and begins the manual reviewing process following a documented standard operating procedure. The reviewer checks and assigns a review status of either approved or rejected to each variant identified as aMOI by the GeneMed system. The reviewer rejects aMOI calls if the variant allele frequency (VAF) is less than 5% for single nucleotide variants (SNVs) and less than 15% for short insertions or deletions (indels) or if the variant has been identified as being out of the reportable range (OORR) of the assay. If the read depth (RD) of the variant is less than 450, or, the variant is within or immediately adjacent to a homo-polymeric region, or the variant creates an insertional tandem repeat, the variant is required to be verified by Sanger sequencing to be approved.
Once the lab review is completed, the review statuses for all aMOIs of the patient are entered into GeneMed. The system will send an email to the treatment selection team to inform them that the patient’s clinical data and sequencing data are ready for their evaluation prior to submitting the mutation summary report that triggers the GeneMed system for the treatment assignment.
The treatment selection oversight team members log into GeneMed and look up the patient’s aMOI report and also the patient’s clinical data for treatment-specific ineligibilities and prior therapies. They go to the mutation summary page to fill in the summary report form. This provides physician oversight to the treatment selection process analogous to tumor boards used in settings in which decisions are made for individual cases in a non-algorithmic manner. The rules governing genomic profiling, definitions of actionable mutations, and treatment arms are all predefined in the study protocol. The rules also cover how treatments are selected in cases with multiple actionable variants. Patients whose tumors contain an actionable variant are randomized 2:1 to receive the treatment targeting the deregulated pathways.
After the mutation summary report is issued by the treatment selection team, the GeneMed system assigns the patient to either an experimental arm or a control arm, based on a pre-calculated randomization design table. An email is then sent to the clinic team to inform them that the treatment information for the patient is available on the GeneMed system. The treating physician goes into GeneMed to obtain the treatment for that patient. The patient is then treated with the designated treatment. During the trial, the clinic team updates the treatment report on the clinical information page to document the treatment response data.
The clinic team is initially blinded to the sequencing assay report. To share the sequencing results upfront would break the randomization blind and endanger the validity of the randomized comparison, since patients might preferentially drop out or be taken off the control arm prior to evaluation of the primary endpoints of disease progression and tumor response. At disease progression, the sequencing results will be shared with the patient and the treating physician.
In the MPACT trial, research-grade whole-exome sequencing will be subsequently performed for all patients once they are off-study. Since the whole-exome sequencing will not be done in a CLIA lab, the sample identification must be delinked from the patient’s clinical ID. There are three steps for the delinking process: a member of the clinic team initiates the delinking request via the GeneMed system, the study principal investigator receives the notice from the GeneMed system and issues approval after determining that the patient is ready to be delinked, and a second notice will be sent to the lab chief who will be the person to complete the delinking process. All research specimens will be relabeled with the new ID; the research lab does not keep any record that can be linked to the original patient ID. When the lab has completed the delinking process, the lab chief goes back to the GeneMed system to confirm the delinking. At that point, all records of the clinical ID will be automatically scrubbed from GeneMed and replaced with the new ID in all database tables.
During the trial, the biostatisticians can log into the GeneMed system to get the updated patient response data and sequencing data once the patient has progressed or is off-study. In addition, a separate clinical database will exist outside of GeneMed, which will remain coded by the clinical ID. The statisticians can make interim and final clinical outcome analyses, as defined in the protocol, of the effectiveness of the genomic matching strategy overall for each treatment, using this separate database.
Functional modules in GeneMed system
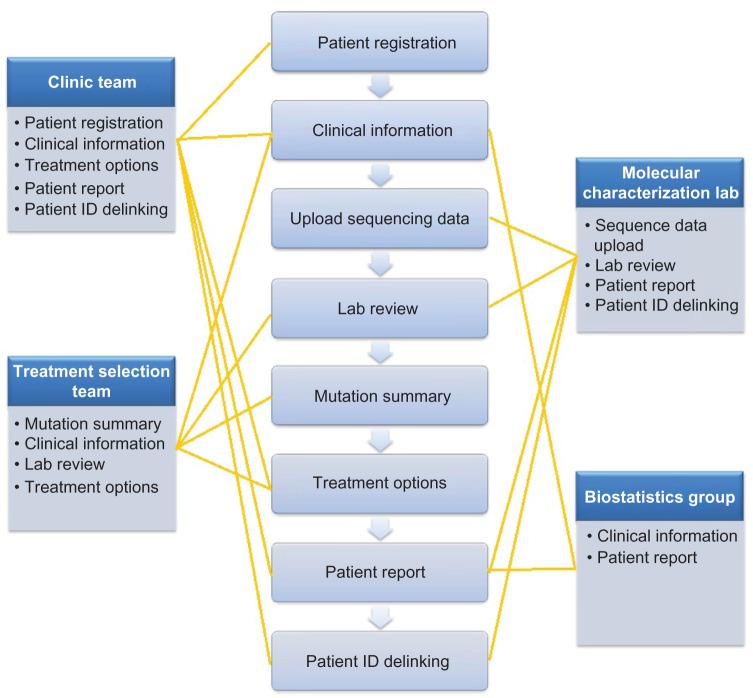
Our goal was to build a reliable, effective, and easy-to-use bioinformatics system that could provide a seamless bridge between the sequencing lab, the clinic group, and the treatment selection team in real time. We had to develop such a system with limited resources and a tight time schedule for development, multi-tier testing, and implementation. We also wanted the system to be extensible to a larger number of drugs, genes, and clinical sites. We critically reviewed the functions that were suggested for the system, prioritized those considered to be the essential tasks that the system must perform, and selected those of the highest priority that we felt could be accomplished within the time frame and limited resources. Given the assigned tasks and workflow in the MPACT clinical trial as described above, we compartmentalized the design into several functional modules, each having its own special features and functionality (Fig. 2). Each working group was given system permission to be able to access required modules according to the defined tasks (Fig. 2). At the same time, the design provides mechanisms for seamless communication among different working groups. There are eight functional modules in the GeneMed system as follows:
Patient registration module: accessible only to the Clinic Group. When a new patient enters the trial, a clinic team member registers the patient in the GeneMed system with the clinical protocol patient ID. An imaging ID is also assigned to the patient for linking the medical imaging files to the patient’s clinical and sequencing profile after delinking.
Clinical information module: edit/add/delete accessible only to the clinic group and read only to the treatment selection team and biostatistics group. The layout of the patient clinical information page is divided into four tables as shown in Figure 3. The top table is patient demographics (eg, gender, histologic diagnosis, age at diagnosis). It also contains clinical information relevant to the treatment assignment, such as known allergies to treatment components or excluded medical conditions relevant to one of the treatments. The second table is a record of the tumor biopsies performed for the MPACT trial. The first entry in the table is the phase of the trial during which the biopsy was performed, either the initial MPACT biopsy or a biopsy obtained at disease progression on the MPACT trial. Next, the user can enter the biopsy site and date of biopsy. On drug refers to the MPACT treatment the patient received prior to the biopsy. The third table lists prior systemic therapies before enrollment in the MPACT trial. The user can add the regimen, best response, regimen start month, and duration. The fourth table contains treatment information that is part of the MPACT trial. This table captures the regimen the patient receives as part of the MPACT trial. It collects the date treatment started, best response, number of cycles, date of progression or off-therapy, as well as the reason for discontinuing therapy.
Sequencing data upload module: used by the sequencing lab to upload the variant file in VCF format (VCF file produced by Torrent Suite version 4.0.2). When uploading the variant file, the system uses the combination of the patient ID and the sample ID (ie, the VCF file name) as the unique identifier. FAO is defined as flow evaluator alternate allele observations and is an integer vector with the number of elements equal to the number of different non-reference bases recorded at the position, and FRO is defined as flow evaluator reference allele observations in the VCF file generated from the Ion Torrent bioinformatics pipeline. Figure 4 shows the diagram for the workflow in the data upload and annotation module. The first step is to parse the variant with multiple changes in ALT (alternate non-reference alleles) field and split the field into lines with a single record in the ALT field.
Figure 2.
The middle panel shows eight functional modules in the GeneMed system and the left and right panels show the four working groups with assigned modules listed in each box.
Figure 3.
The web interface for collecting clinical information of patients. The clinic personnel can add, edit, and delete data in four tables on the page during the trial.
Figure 4.
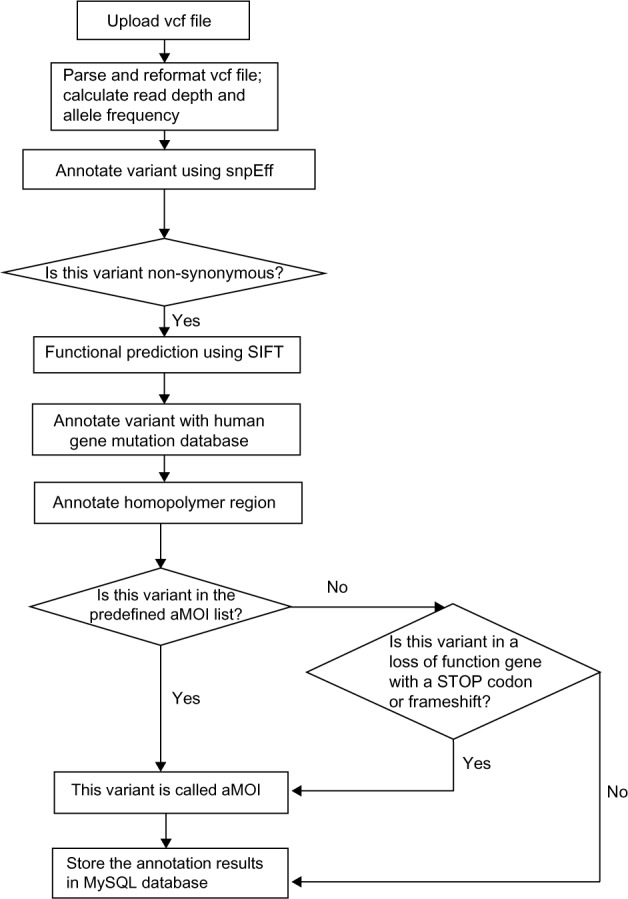
A diagram of annotation and aMOI call in the GeneMed system.
For variant i among the total of N mutations detected at a given position, we define the RD and the VAF to be calculated as follows:
The next step is to annotate the predicted effect of all variants in the VCF file. This step uses the SnpEff variant annotation and effect prediction tool.11 The output will annotate all variants as either synonymous or non-synonymous and the predicted amino acid change.
All non-synonymous variants are then further annotated with the Sift prediction algorithm.12 The Sift algorithm is incorporated into GeneMed so that the system can perform functional predictions not only for known polymorphisms but also for the de novo somatic mutations detected by our assay. It also annotates the variants if they are present in dbSNP and annotates the matching rs ID. Those non-synonymous variants are then annotated if they are found in the BIOBASE human gene mutation database (HGMD).13 In addition, all homopolymer regions within the amplicons have been annotated, and a variant is marked with the length of the homopolymer region if the reported position is within 1 bp of any homopolymer region.
The variants are finally compared to a table of aMOIs containing about 380 unique mutations that have been curated for the trial. These mutations, including SNVs and insertion/deletion variants (indels), are known to be somatically altered in solid tumors based on recent scientific and clinical literature. The observed variants are marked if they match a variant in the aMOI table with the exact COSMIC ID. If a variant results in a novel stop codon or frame shift (including loss of start codon ATG) in a gene in which a loss of function is predicted to cause the patient to be sensitive to at least one of the study treatments, then that variant will also be marked as actionable.
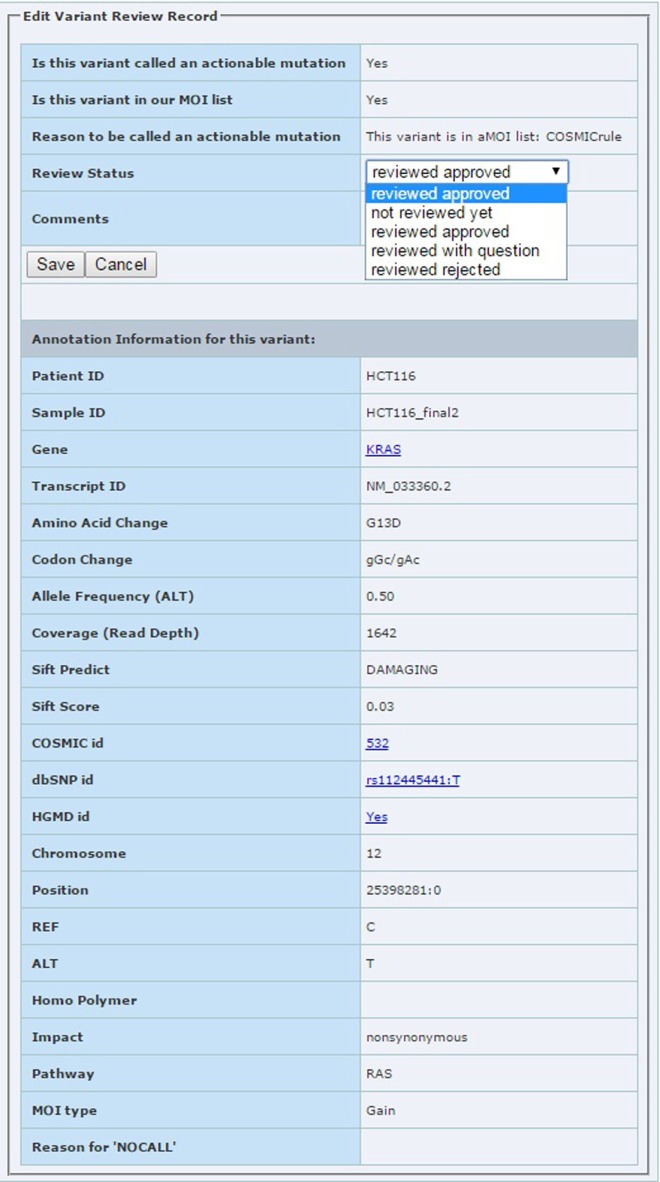
Lab review module: used by the sequencing lab to review each aMOI. The lab reviewer can assign four different review statuses to each aMOI: review approved, reviewed with question, reviewed rejected, and not reviewed yet. A screen shot of the review edit form is shown in Figure 5. When the lab review has been completed, an email will be sent to the treatment selection team to notify them that the report is ready for their review.
Mutation summary module: used by the treatment selection team. One of the team members will log into the GeneMed system to complete the summary form (Fig. 6). There are four questions that need to be addressed before GeneMed proceeds with the patient randomization. The first question is whether any actionable mutations have been detected? If yes, the team will select targetable pathways, potential targeted treatments, and appropriate control treatments. A Comment field is available for entering reasons or further explanation. Once the mutation summary is submitted by one of the treatment selection team members, GeneMed begins to randomize the patients to either the experimental arm, to receive a treatment designed to address that defect, or to the control arm, to receive a treatment from the complementary set, within the trial’s currently available treatment options. If more than one actionable mutation is identified, the patient will be assigned to a randomization stratum that has the least number of patients already observed with variants in that pathway. Patients randomized to the control arm will be assigned to the treatment that best maintains the match of treatment proportions between the two arms. The treatment assignment process has been carefully devised to enable blinding the patient and the treating physician to whether the patient has been assigned to the experimental arm or to the control arm.
Treatment option module: the clinic group uses this module to get the treatment option after the GeneMed system completes the treatment assignment. The treating physicians are blinded to which arm the patient is assigned during the trial to avoid potential biases.
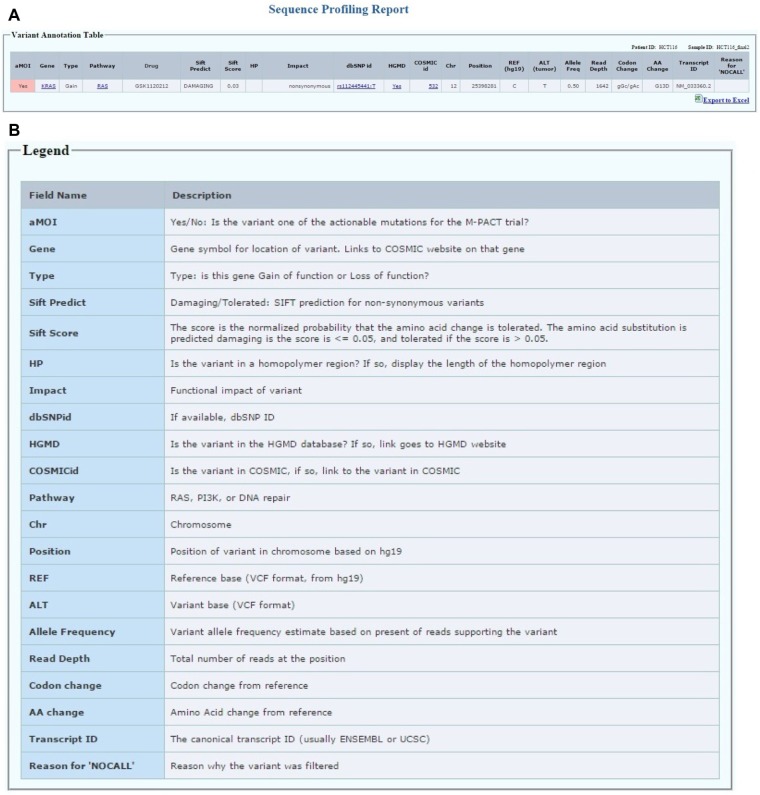
Patient sequencing report module: accessible to the clinic group and the biostatistics group only after the patient has progressed on the assigned therapy or comes off-study for other reasons (Fig. 7A). The aMOI detected by sequencing will be reported in a format shown in Figure 7A. The patient ID and sample ID are shown on the top right of the table. Each row represents a variant that has been identified as one of the actionable mutations for the MPACT trial, which has been confirmed by lab review. For each variant, we report the gene symbol for location of variant with links to the COSMIC website on that gene. The gene type indicates whether this gene has been classified as a gain of function or loss of function gene for the study. Each gene is predefined in one of the three pathways studied in the MPACT trial. The drug field lists the drug(s) assigned for the specific pathway in this trial. Impact field is the functional impact of the variant predicted by SnpEff. The Sift prediction creates two fields: the first is Sift score, which is the normalized probability that the amino acid change is tolerated; the second is Sift prediction for each non-synonymous variant and is either “damaging” if the score is ≤0.05 or “tolerated” if the score is >0.05. COSMIC id and links to COSMIC database as well as dbSNP id and links to NCBI SNP database are also listed. If the variant is in the HGMD database, a link under HGMD field is provided to access the PubMed record. If the variant is in a homopolymer region, the length of the homopolymer region is displayed under HP field. Finally, chromosome, REF, ALT, start position, allele frequency, read depth, codon change, and amino acid change for the variant are listed. The legend for each field in the patient table is shown in Figure 7B. The canonical transcript IDs are also shown in the report table to provide the isoform information. The report table can be transferred to an excel spreadsheet by clicking the export to excel button at the right bottom of the table.
Patient ID delinking module: This module uses three steps described in the workflow to replace the patient protocol ID with a randomly generated ID once the patient is off-study. The research nurse, the physician, and the lab chief are the parties involved in the delinking process.
Figure 5.
The edit page for lab review module. The reviewer can assign review status with options shown in the pull down box.
Figure 6.
The web interface for the treatment selection team to submit a mutation summary report.
Figure 7.
(A) The patient report table displays the actionable MOIs that have been reviewed and approved. (B) The legend table lists all fields in the patient report table.
Implementation of GeneMed system
The GeneMed system has been designed, developed, and managed by the Biometric Research Branch in the Division of Cancer Treatment and Diagnosis of the NCI. It is hosted and supported by the Center for Biomedical Informatics and Information Technology (CBIIT) in the NCI. There are four layers in the system development pipeline, ie, development tier, QA tier, staging tier, and production tier, attached by its own database server. The GeneMed system is operating under the Redhat Linux OS version 5.8. MySQL (v5.1.48) is used as the backend database. There are 19 MySQL tables in total for the GeneMed system. Details of the database tables are listed in Supplementary Table 1. The web client interface pages are written in html, CSS, and JavaScript. The backend programs are written using Perl. Java (version 1.6.0_18) is installed as pre-requirement to run the annotation software SnpEff (version 3.4i, released Jan 16, 2014, downloaded from http://sourceforge.net/projects/snpeff/files/). Sift version 5.0.2 is used as the functional prediction tool (downloaded from http://sift.bii.a-star.edu.sg/). In order to authenticate users and control access to the GeneMed system, SiteMinder and SSL are installed to enable the secure delivery of essential information and applications via secure single sign-on. For data quality control purposes, several applications have been developed or set up to monitor the database information transaction and user access and error history on a daily basis.
Discussion
There have been several efforts to build NGS annotation and reporting systems for clinical applications. For example, the Clinical Genomicist Workstation (CGW) was developed by Washington University Genomic and Pathology Services to support sequencing data tracking and reporting for pathologists.14 SeqReporter was developed by the University of Pittsburgh Medical Center to automate NGS results interpretation and workflow reporting in a clinical molecular lab.15 The KDI system was developed and reported recently for the SHIVA clinical trial at Institut Curie.16 Compared to other systems, the GeneMed system integrates the lab review feature and can automatically perform patient randomization based on mutation information for targeted genes. In addition, GeneMed can perform live functional prediction on both known polymorphisms and de novo somatic mutations detected by the assay. Each step or team requires a short amount of time when the required documents or information are ready. The CPU time for data upload and annotation is less than two minutes. The lab team requires 5–10 minutes to review the variant report, and the clinic team requires 10–15 minutes to enter the clinical information. After the treatment selection team reaches an agreement (usually less than half a day), one of the members submits the mutation summary in GeneMed. The treatment assignment is done by the system simultaneously after the mutation summary is submitted. GeneMed is a unique web-based informatics system that connects NGS technology to precision medicine applications in real time and addresses the complex requirements imposed by a sequencing-guided randomized clinical trial.
Current and future development of GeneMed is directed at making the system more easily generalized to other clinical trial applications. Changes include uploading VCF files having different formats, adding additional drugs during the trial, creating a user-friendly rule generator for aMOI calls, and automating the lab review process by using predefined criteria.
Currently, the GeneMed system only supports variant files generated from Life Technologies’ Torrent Suite versions 3.2 and 4.0. With rapid updates of the sequencing software, formats of the VCF files are expected to undergo significant changes. Enhancements will make it compatible with VCF files containing version changes for the same variant caller software as well as VCF files generated by systems from other popular bioinformatics pipelines. Sequencing-based clinical trials are dynamic discovery trials; adding a new drug may happen during the trial as more drugs become available. Adding a new drug potentially means that more genes and pathways will be involved in the data annotation and treatment assignment. The GeneMed system is being made more flexible so that software changes are not necessary when a new drug is added; the users can easily update the backend tables of genes, pathways, and aMOIs. The treatment assignment will also be automatically adjusted without changing the backend code. There are many rule-based clinical trials for targeted therapy being launched similar to MPACT. Different trials mean different rules for defining aMOIs and treatment assignment. Even during a trial, a rule could be modified. A user-friendly rule generator is needed so that the end user can create, modify, or delete rules for defining aMOIs using the client-side user interface. Adaptive rules for adding or dropping treatments can be built in easily at the user end.
The GeneMed system has been running successfully at the NIH Clinical Center and the NCI Division of Cancer Treatment and Diagnosis’ tumor characterization laboratory since January 2014. As of October 15, 2014, 37 patients have been enrolled on the MPACT trial. Among them, tumors from 17 patients contain at least one actionable mutation. NCI is planning to expand the MPACT trial to multiple cancer centers by the end of 2014. An optional correlative imaging study to correlate CT/MRI image phenotypic features with genomic data is under development via linking the imaging database to GeneMed system. We believe the GeneMed system could be used as a prototype of an informatics hub for coordinating NGS data in other personalized genome-based medicine clinical trials.
Supplementary Material
Supplementary Table 1. Details of 19 database tables in GeneMed.
Acknowledgments
We thank Eric Williams, Wenling Bao, Jacob Mensah, Yeon Choi, Mikol Ware, and other staff in the web team, the database team, and the security team from the CBIIT of the NCI for application deployment and information infrastructure support. We thank Dr. Boris Freidlin for providing the randomization tables for patient assignment and Ramya Antony for her input in the clinical data table design. We thank Henry Rivera and Eric Silver for technical support and assistance. We thank staff from the Center for Information Technology of the NIH for support of SiteMinder. We thank Dr. Pauline Ng’s group at the Genome Institute of Singapore for technical consultation in our implementation of SIFT.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: YZ, ECP, RMS. Made critical revisions and approved final version: JHD, CJL, BAC, SK, LVR, APC, PMW, MCL, DJS, AP. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–52. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 2.Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–6. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogelstein B, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doroshow JH. Selecting systemic cancer therapy one patient at a time: is there a role for molecular profiling of individual patients with advanced solid tumors? J Clin Oncol. 2010;28(33):4869–71. doi: 10.1200/JCO.2010.31.1472. [DOI] [PubMed] [Google Scholar]
- 5.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov. 2013;12(5):358–69. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 6.Simon R, Polley E. Clinical trials for precision oncology using next-generation sequencing. J Pers Med. 2013;10:485–95. doi: 10.2217/pme.13.36. [DOI] [PubMed] [Google Scholar]
- 7.Tran B, Brown AM, Bedard PL, et al. Feasibility of real time next generation sequencing of cancer genes linked to drug response: results from a clinical trial. Int J Cancer. 2013;132(7):1547–55. doi: 10.1002/ijc.27817. [DOI] [PubMed] [Google Scholar]
- 8.Kummar S, Williams PM, Lih CJ, et al. Application of molecular profiling in clinical trials for advanced metastatic cancers. J Natl Cancer Inst. 2015;107(4):djv003. doi: 10.1093/jnci/djv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lih CJ, Sims DJ, Harrington RD, et al. Analytical Validation and Application of a Targeted Next Generation Sequencing Mutation Detection Assay for use in Treatment Assignment in a Cancer Clinical Trial ( NCT01827384) submitted. doi: 10.1016/j.jmoldx.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso−3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN. The human gene mutation database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi0113s39. Chapter 1:Unit1.13. [DOI] [PubMed] [Google Scholar]
- 14.Sharma MK, Phillips J, Agarwal S, et al. Clinical genomicist workstation. AMIA Joint Summits Transl Sci Proc. 2013;2013:156–7. [PubMed] [Google Scholar]
- 15.Roy S, Burso MB, Wald A, Nikiforov YE, Nikiforova MN. SeqReporter: automating next-generation sequencing result interpretation and reporting workflow in a clinical Laboratory. J Mol Diagn. 2014;16:11–22. doi: 10.1016/j.jmoldx.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Servant N, Roméjon J, Gestraud P, et al. Bioinformatics for precision medicine in oncology: principles and application to the SHIVA clinical trial. Front Genet. 2014;5:152. doi: 10.3389/fgene.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Details of 19 database tables in GeneMed.