Abstract
Genome wide association studies (GWAS) have identified several low-penetrance susceptibility alleles in chronic lymphocytic leukemia (CLL). Nevertheless, these studies scarcely study regions that are implicated in non-coding molecules such as microRNAs (miRNAs). Abnormalities in miRNAs, as altered expression patterns and mutations, have been described in CLL, suggesting their implication in the development of the disease. Genetic variations in miRNAs can affect levels of miRNA expression if present in pre-miRNAs and in miRNA biogenesis genes or alter miRNA function if present in both target mRNA and miRNA sequences. Therefore, the present study aimed to evaluate whether polymorphisms in pre-miRNAs, and/or miRNA processing genes contribute to predisposition for CLL. A total of 91 SNPs in 107 CLL patients and 350 cancer-free controls were successfully analyzed using TaqMan Open Array technology. We found nine statistically significant associations with CLL risk after FDR correction, seven in miRNA processing genes (rs3805500 and rs6877842 in DROSHA, rs1057035 in DICER1, rs17676986 in SND1, rs9611280 in TNRC6B, rs784567 in TRBP and rs11866002 in CNOT1) and two in pre-miRNAs (rs11614913 in miR196a2 and rs2114358 in miR1206). These findings suggest that polymorphisms in genes involved in miRNAs biogenesis pathway as well as in pre-miRNAs contribute to the risk of CLL. Large-scale studies are needed to validate the current findings.
Introduction
Chronic lymphocytic leukemia (CLL) is the most frequent leukemia of adults in Western countries. It is characterized by the accumulation of monomorphic small round B lymphocytes in peripheral blood, bone marrow, spleen and/or lymph nodes [1]. CLL has been reported to have the highest genetic predisposition of all hematologic neoplasias [2], with an ~8.5-fold increased relative risk in first-degree relatives [3]. However, previous genetic linkage studies failed to provide evidence that rare, high-penetrance genes contribute significantly to the familial risk [4], suggesting that additional loci of modest effects could be involved. Actually, recent genome-wide association studies (GWAS) have identified several low-penetrance susceptibility alleles associated with CLL [5–9], associations that have been thoroughly validated in multiple independent series [10–13]. Nevertheless, these studies have focused their effort on identifying genes involved in the risk of CLL and scarcely study regions that are implicated in non-coding molecules, such as microRNAs (miRNAs).
MiRNAs are a group of small, functional, non-coding RNAs of approximately 20 nucleotides that regulate gene expression at the post-transcriptional level by binding to the 3´ untranslated region (UTR) of a target gene [14]. This can lead to inhibition of translation or enhanced degradation of their target mRNAs. Through this mechanism, miRNAs can regulate a large number of genes. Consequently, variations in miRNA function can affect many targets with functional consequences.
To date several reports have described altered expression of miRNAs in CLL, suggesting their implication in the development of the disease [15–17]. MiRNA expression levels can be altered by genetic polymorphisms in pre-miRNAs and/or also in miRNA biogenesis genes. Additionally, genetic polymorphisms in miRNAs or in mRNA targets altering the mRNA-miRNA binding may be also relevant, affecting the miRNA function. Indeed, mutations in miRNA transcripts have been seen to be common in CLL [17, 18], suggesting that CLL predisposition involves genetic alterations in miRNAs. Supporting this idea, a recent study has found that certain miR-SNPs show different frequency in CLL patients compared with a control population [19].
In addition, several studies have hitherto described polymorphisms in miRNAs associated with the susceptibility to different malignancies. For example, rs7372209 in miR26a-1 has been associated with the risk of throat and esophageal cancer and rs11614913 in pre-miR196a2 with breast, lung and gastric cancers, etc [14]. However, despite accumulating evidence that inherited genetic variation in miRNA genes and miRNA processing genes can contribute to the predisposition for cancer, the role of these in the risk of CLL has not been extensively studied. Therefore, the aim of this study was to evaluate whether polymorphisms in pre-miRNAs, and/or miRNA processing genes, contribute to the risk of CLL.
Methods
Study population
The study population included a total of 523 Spanish individuals, 132 CLL patients previously reported [20] and 391 cancer-free controls. Both, cases and controls, were selected from the same residing area of the north of Spain and had West European ancestry. A brief description of the clinical characteristics of CLL population is described in S1 Table.
Ethics Statement
CLL samples were obtained from the C. 0001174 collection and control samples from C.0001171 collection, registered in the Institute of Health Carlos III. All samples were collected before 2007, when Biomedical Research Act 14/2007 (http://www.boe.es/boe/dias/2007/07/04/pdfs/A28826-28848.pdf) was released in Spain. Medicine and Odontology Faculty of UPV/EHU ethics committee board approval was obtained.
Genes and polymorphisms selection
A total of 118 SNPs were selected for the study. Of them, 72 SNPs were located in 21 genes involved in the miRNAs biogenesis and processing (S2 Table) and 46 SNPs were situated in 44 miRNAs (S3 Table). The SNPs in miRNA processing genes were selected after literature review and by using Patrocles database (http://www.patrocles.org). In each gene, using tagSNPs, we covered almost all the SNPs with potentially functional effects using bioinformatics tools (F-SNP, Fast-SNP, polymirTS [21, 22] Patrocles [23], Hapmap, Haploview). We considered functional effects those causing amino acid changes, alternative splicing, those located in the promoter region in putative transcription factor binding sites, or disrupting/creating miRNAs targets. We also selected SNPs from the literature. Only SNPs with a reported minor allele frequency greater than 5% (MAF≥0.05) in European/Caucasoid populations were selected.
For the selection of SNPs in miRNAs, we had two factors in consideration. A) miRNAs can regulate a wide range of genes that are not completely defined. Therefore, any miRNA could be implicated in the regulation of genes affecting CLL risk. B) The number of miRNAs with polymorphisms described in the databases at the moment of selection with a MAF > 0.01 was affordable. Consequently, we selected all the SNPs in pre-miRNAs with a MAF>0.01 in European/Caucasoid populations, using Patrocles and Ensembl databases (http://www.ensembl.org/) (Welcome Trust Genome Campus, Cambridge, UK) and literature review. The preliminary list of SNPs was filtered using suitability for the Taqman OpenArray platform as criteria.
Genotyping
Genomic DNA was extracted from peripheral blood from controls and from granulocytes (normal cells) from CLL patients as previously described [20]. SNP genotyping was performed at the General Research Services (SGIker) of the University of the Basque Country using TaqMan Open Array technology (Applied Biosystems), following the manufacturer’s protocol. Data were analyzed with Taqman Genotyper software for genotype clustering and calling. As a genotyping control, 20 duplicate samples were placed across the plates.
Statistical analysis
Haploview 4.2 was used to search for any deviation of Hardy-Weinberg equilibrium in controls, determine haplotype block structure, calculate the genotyping success rate for each SNP and compare haplotype frequencies and clinical parameters between cases and controls. Significance of the best result was corrected for multiple testing by 1000 permutations. Genotype frequencies in the cases and controls were compared using a χ2 test. The effect sizes of the associations were estimated by the OR’s from univariate logistic regression. The most significant test among the different genetic models was used to determine the statistical significance of each SNP. The results were adjusted for multiple comparisons by the False Discovery Rate (FDR) [24]. In all cases the significance level was set at 5%. Analyses were performed by using R v2.11 software.
miRNAs secondary structures prediction
The RNAfold web tool (http://rna.tbi.univie.ac.at) [25] was used to predict the most stable secondary structures of the miRNAs showing significant SNPs. The analyzed sequences included 50 bp upstream and 50 bp downstream of the pre-miRNAs.
Results
Genotyping results
Successful genotyping was obtained in 457 DNA samples (87.4%); 107 of 132 CLL patients (81.1%) and 350 of 391 controls (89.5%). Of 118 SNPs, 91 (77.12%) were genotyped satisfactorily, 57 located in miRNAs processing genes (62.6%) and 34 in pre-miRNAs (37.4%). 27 SNPs were removed from the association study due to genotyping failures, deviations from HWE or because they were not polymorphic in our population (S4 Table). The average of success genotyping rates for all 91 SNPs was 96.2%.
Genotype association study
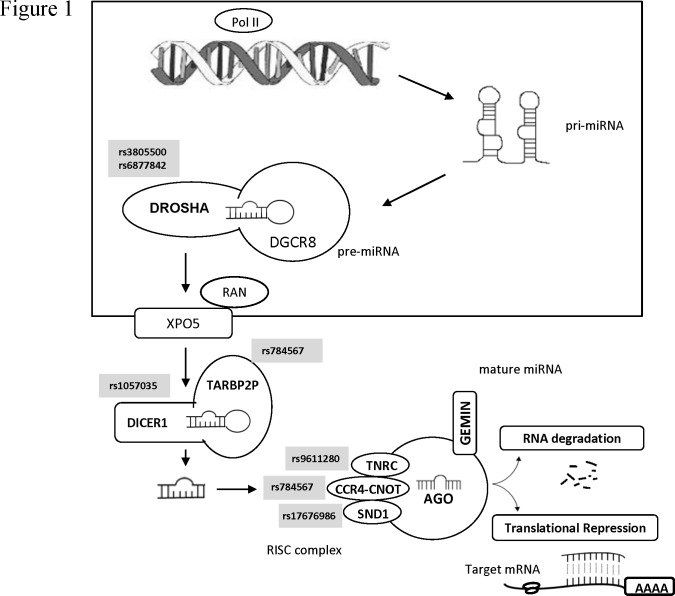
A total of 28 SNPs showed statistically significant association with CLL risk (S5 Table). After FDR correction, 9 SNPs remained significant. Among them, seven were located in six processing genes (Table 1, Fig. 1) and two in pre-miRNAs (Table 2). Two of the significant SNPs, rs3805500 and rs6877842, were located in DROSHA gene. Rs3805500 showed the most significant finding of the analyses under the log-additive genetic model (AA vs AG vs GG). The G allele showed a 0.54-fold decreased risk of CLL (95% CI: 0.38–0.77; P = 0.032). Other 5 SNPs in processing genes, rs1057035 in DICER1, rs17676986 in SND1, rs9611280 in TNRC6B, rs784567 in TRBP and rs11866002 in CNOT1, showed statistically significant results (P<0.05).
Table 1. Association of significant miRNA Processing Gene SNPs and Chronic Lymphocytic Leukemia risk.
| Gene | SNP | Best fitting Model | Genotype | Controls n (%) | Cases n (%) | OR (95% CI) | P value | Adjusted P value* |
|---|---|---|---|---|---|---|---|---|
| DROSHA | rs3805500 | Additive | AA | 125 (37.0) | 55 (55.0) | Reference | ||
| AG | 158 (46.7) | 38 (38.0) | 0.54 (0.38–0.77) | 0.0004 | 0.032 | |||
| GG | 55 (16.3) | 7 (7.0) | ||||||
| rs6877842 | Recessive | GG + CG | 330 (96.8) | 90 (88.2) | Reference | |||
| (231+99) | (64+26) | 0.002 | 0.038 | |||||
| CC | 11 (3.2) | 12 (11.8) | 4.00 (1.71–9.37) | |||||
| DICER1 | rs1057035 | Recessive | TT + CT | 308 (89.0) | 78 (75.0) | Reference | ||
| (156+152) | (46+32) | 0.0007 | 0.032 | |||||
| CC | 38 (11.0) | 26 (25.0) | 2.7 (1.55–4.72) | |||||
| SND1 | rs17676986 | Recessive | CC + CT | 341 (98.8) | 97 (92.4) | Reference | ||
| (246+95) | (69+28) | 0.001 | 0.038 | |||||
| TT | 4 (1.2) | 8 (7.6) | 7.03 (2.07–23.84) | |||||
| TNRC6B | rs9611280 | Recessive | GG + AG | 343 (98.6) | 97 (92.4) | Reference | ||
| (273+70) | (80+17) | 0.003 | 0.043 | |||||
| AA | 5 (1.4) | 8 (7.6) | 5.66 (1.81–17.69) | |||||
| TRBP | rs784567 | Dominant | AA | 80 (23.7) | 38 (39.2) | Reference | ||
| AG + GG | 258 (76.3) | 59 (60.8) | 0.48 (0.30–0.78) | 0.003 | 0.043 | |||
| (181+77) | (40+19) | |||||||
| CNOT1 | rs11866002 | Codominant | CC | 134 (38.7) | 54 (52.4) | Reference | ||
| CT | 174 (50.3) | 33 (32.0) | 0.47 (0.29–0.77) | 0.004 | 0.049 | |||
| TT | 38 (11.0) | 16 (15.5) | 1.04 (0.54–2.03) |
Abbreviations: OR, odds ratio; CI, confidence interval; S, significative; NS, no significative
* Adjusted for multiple comparisons using the False Discovery Rate (FDR) at 5% level
Fig 1. Overview of the significant SNPs in the miRNA network.
Table 2. Association of significant miRNA-related SNPs and Chronic Lymphocytic Leukemia risk.
| miRNA | SNP | SNP position | Best fitting Model | Genotype | Controls n (%) | Cases n (%) | OR (95% CI) | P value | Adjusted P value* |
|---|---|---|---|---|---|---|---|---|---|
| miR196a2 | rs11614913 | Premature | Recessive | CC + CT | 296 (85.8) | 75 (72.1) | Reference | ||
| (137+159) | (35+40) | 0.002 | 0.038 | ||||||
| TT | 49 (14.2) | 29 (27.9) | 2.34 (1.38–3.95) | ||||||
| miR1206 | rs2114358 | Premature | Dominant | AA | 110 (31.6) | 48 (47.1) | Reference | ||
| AG + GG | 238 (68.4) | 54 (52.9) | 0.52 (0.33–0.82) | 0.005 | 0.049 | ||||
| (181+57) | (36+18) |
Abbreviations: OR, odds ratio; CI, confidence interval; S, significative; NS, no significative
* Adjusted for multiple comparisons using the False Discovery Rate (FDR) at 5% level
We also observed that two SNPs in pre-miRNAs were significantly associated with CLL risk, rs11614913 and rs2114358 in the miR196a2 and miR1206, respectively. The genotype TT rs11614913 was associated with a 2.3-fold increased risk under the recessive model (95% CI: 1.38–3.95; P = 0.002) while the genotypes AA and AG rs2114358 showed 0.52-fold protective risk to CLL under the dominant model (95% CI: 0.33–0.82; P = 0.005).
When the correlation between SNP alleles and clinical parameters were analyzed (Rai stage, morphology, levels of serum markers such as β2-microglobulin or lactate dehydrogenase, presence of genomic aberrations, marrow infiltration, presence of adenopathy, lymphocyte doubling time, residual minimum disease, and treatment) no significant association was found in CLL patients (S6 Table).
Haplotype Association study
We conducted haplotype analyses for the miRNA-related genes with at least two statistically significant SNPs in this study before FDR correction, including DROSHA, DICER1, DGCR8 and TNRC6B. Two haplotype blocks, one in DROSHA and another one in DGCR8, showed statistically significant differences in their frequency in cases versus controls (χ2 test; adjusted P-value<0.05 after permutation testing) (Table 3). The significant haplotype of DROSHA gene, formed by rs3805500-rs7735863-rs6884823 (AGG), was associated with increased risk (OR = 1.63; 95% CI: 1.17–2.27), while the haplotype of DGCR8 composed by rs9606248-rs1640299 (AT) was found to confer a significant reduced risk (OR = 0.67; 95% CI: 0.49–0.91) of developing CLL.
Table 3. Significant Haplotypes in miRNA processing genes and Chronic Lymphocytic Leukemia Risk.
| Gene | SNPs | Haplotype | Freq. Controls | Freq. Cases | Chi-Square | Adjusted P-value a | OR b (CI 95%) |
|---|---|---|---|---|---|---|---|
| DROSHA | rs3805500-rs7735863-rs6884823 | AGG | 0.60 | 0.71 | 9.23 | 0.004 | 1.63 (1.17–2.27) |
| DGCR8 | rs9606248-rs1640299 | AT | 0.49 | 0.39 | 7.0 | 0.03 | 0.67 (0.49–0.91) |
aAdjusted P-value after permutation test
bOR values are referred to the rest of haplotype
miRNAs secondary structures
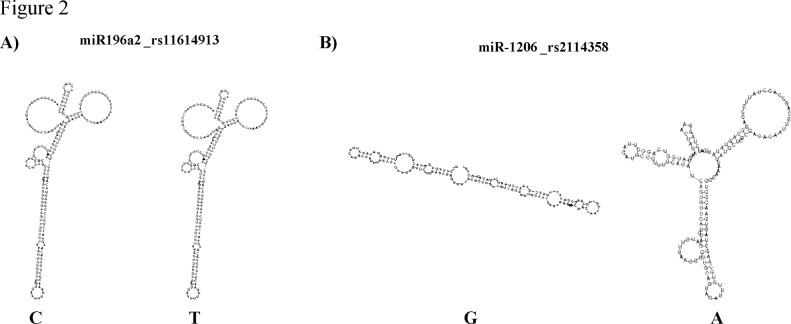
In order to check if SNPs in miR196a2 and miR1206 affected their secondary structure, we compared the minimum free energy for optimal secondary structures of both variants for each miRNA. The SNP rs2114358 in pre-miR1206 had a dramatic effect on its secondary structure, whereas the SNP rs11614913 in pre-miR196a2 had none (Fig. 2).
Fig 2. Secondary structures of the miRNAs showing significant SNPs.
(A) miR196a2 and (B) miR1206, predicted by the RNAfold tool.
Discussion
In the current study, we identified genetic variants in miRNA processing genes and pre-miRNAs that were associated with the risk of CLL. We examined a total of 91 SNPs, finding nine significantly associated after FDR correction, seven in miRNA processing genes and two in pre-miRNAs.
Interestingly, among the significant SNPs in miRNA processing genes, two were located in DROSHA; rs3805500 in the intronic region and rs6877842 upstream. Moreover, a haplotype including the SNPs rs3805500-rs7735863-rs6884823 (AGG) was also found significant in this gene. DROSHA encodes a type III Rnase enzyme, essential for the maturation of pri-miRNAs into pre-miRNAs [26]. Aberrant expression of DROSHA at mRNA or protein level was shown to be associated with patient survival and tumor progression in several types of cancer, including pleomorphic adenomas [27], esophageal cancer [28], cervical cancer [29] and ovarian cancer [30]. Moreover, the SNP rs3805500 which was the most significant finding in our study, was found by other authors to be associated with head and neck cancer recurrence [31]. In addition, a recent study in lung cancer has demonstrated that the polymorphism rs640831 in high LD with rs3805500 (r2>0.8) was associated with reduced DROSHA mRNA expression and with expression changes in 56 miRNAs out of 199 analyzed [32], some of which could also regulate genes involved in the development of CLL. Therefore, in the same way, the two polymorphisms detected in the present study could alter the expression of miRNAs involved in the regulation of genes involved in the risk of CLL.
Additionally, five extra miRNA biogenesis pathway SNPs were also found significant. One of them, rs1057035, was located within the 3’ UTR of DICER1, an important region for RNA stability. Moreover, it was located at the putative miRNA binding site of miR574-3p, which could affect the binding of this miRNA to DICER. DICER plays an important role in the cleavage of pre-miRNAs into their mature form. A recent study showed that rs1057035 C allele was associated with a lower expression of DICER and a decreased risk of oral cancer, which seemed to be due to a higher inhibition of miR574-3p on DICER mRNA [33]. In our study, in contrast, the variant genotype CC was associated with a 2.7-fold increased risk of CLL. These differences between studies suggest that DICER may have different roles in the tumorigenesis process depending on the cancer type. Supporting this idea, some studies have associated reduced expression of DICER with the development of lung cancer [34], while others have seen overexpression of DICER in oral cancer [35]. Therefore, it seems plausible that rs1057035 is affecting the binding of miR574-3p which in turn could be altering the regulation of DICER and the risk of CLL.
Another significant SNP, rs784567, was located in the 5' near gene region of TRBP, an integral component of a DICER-containing complex important to the cytoplasmic processing of pre-miRNAs into mature miRNAs. Previous studies also found rs784567 to be borderline significant in bladder cancer [36] and oral premalignant lesions [37]. Moreover, depletion of TRBP was found to result in significantly impaired miRNA biogenesis [38]. Consequently, genetic variation affecting the expression of this gene could lead to global changes in the miRNAome and subsequent deregulation of genes involved in CLL origin.
On the other hand, rs9611280 was situated in TNRC6B, gene that mediates miRNA-guided mRNA cleavage [39]. According to in silico analyses (F-SNP), this missense SNP was not predicted to abort protein function, but instead affected the regulation at splicing level. This change could perturb the maturation of the mRNA, changing the sequence of the protein, and therefore contribute to carcinogenesis in CLL.
Finally, the last two significant SNPs identified were rs17676986 at SND1 and rs1186002 at CNOT1, genes involved in RNA-induced silencing complex (RISC). Genetic variants in both genes have been previously associated with autism [40] and ventricular arrhythmias [41] respectively, but this is the first time that are associated with cancer.
In addition to SNPs from miRNA processing genes, there were also two SNPs in miR196a2 and miR1206 significantly associated with CLL risk after FDR correction. miR196a2 rs11614913 variant homozygote TT showed an increased risk of CLL. Different rs11614913 genotype variants were previously reported to be associated with cancer risk; for example, the variant CC was associated with lung [42], hepatocellular [43], glioma [44], and gastric [45] cancer, while the variants TT or TT + CT were associated with breast [46], prostate [47], and head and neck cancer [48]. Despite these differences among studies, probably due to the different populations used in the analyses [49], it seems probable that rs11614913 plays an important role in cancer development. rs11614913 is located in the 3p mature miRNA region, so it could affect maturation of 5p and 3p miRNAs, as well as the expression and function of these miRNAs. Actually, Hoffman et al already demonstrated that rs11614913 C allele led to less efficient processing of the miRNA precursor to its mature form as well as diminished capacity to regulate target genes in breast cancer [46]. Interestingly, potential targets of miR196a2 include genes with relevance in CLL, such as CDKN1B or HMOX1. Therefore, alterations in the sequence of miR196a2 which may affect miRNA function, could contribute to CLL risk due to the deregulation of its targets.
Finally, miR1206 rs2114358 AG-GG genotypes showed a reduced risk of CLL. Remarkably, miR1206 is located at 8q24.2 a frequent site of chromosomal breaks in cancer and a signature of genomic instability [50]. Moreover, miR1206 is of particular interest as it is part of long non-coding RNA (lncRNA) transcript of the PVT1 gene, locus of both tumorigenic translocations and retroviral insertions [51], which has been also shown to play a role in cell proliferation and apoptosis [52]. Overexpression of PVT1 transcript has been documented in a variety of tumor tissues [53] and miR1206 expression has been found in increased levels in B cell tumors [54]. The allelic effect of miR1206 rs2114358 has been further analyzed in different human cell lines but without concluding results, suggesting that the effect of this SNP was cell line and cell type specific [55]. We found that miR1206 rs2114358 AG-GG genotypes showed a reduced risk of CLL, so it is likely to expect that AA genotype was associated with an increased risk. Moreover, we found by in silico analysis (RNA fold), that A allele altered the miRNA secondary structure, suggesting that this variant alter the maturation of miR1206.
A recent study found that rs56103835 in pre-miR323b was associated with CLL when compared with European samples from the 1000 genome project [19]. We also analyzed this SNP in our population, finding no significant association (S5 Table), neither when CLL population was compared with the European samples from the 1000 genome project (0.143 versus 0.187), nor when our control population was used (0.143 versus 0.183).
In conclusion, the findings of the present study indicated that SNPs located at DROSHA, DICER1, SND1, TNRC6B, TRBP and CNOT1 genes, involved in miRNAs biogenesis pathway, as well as in miR196a2 and miR1206 may contribute to the risk of CLL. To our knowledge, this is the first study analyzing specifically miRNA-related SNPs in CLL, which opens a promising approach to search for new susceptibility markers in CLL. New large-scale studies including functional analyses will help to validate our findings.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Basque Government (IT661-13), UPV/EHU (UFI11/35), Saiotek (S-PE13UN079) and RTICC (RD12/0036/0060, RD12/0036/0036). AGC was supported by a predoctoral grant from the Basque Government through the grant “Programa de Formación de Personal Investigador no doctor del Departamento de Educación, Política Lingüística y Cultura del Gobierno Vasco”. ELL was supported by a postdoctoral grant from the Basque Government through the grant “Programa Posdoctoral, de Perfeccionamiento de Personal Investigador Doctor del Departamento de Educación, Política Lingüística y Cultura del Gobierno Vasco”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon; 2008. [Google Scholar]
- 2. Goldin LR, Slager SL, Caporaso NE. Familial chronic lymphocytic leukemia Curr Opin Hematol. 2010;17: 350–5. 10.1097/MOH.0b013e328338cd99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldin LR, Björkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica. 2009;94:647–53. 10.3324/haematol.2008.003632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slager SL, Skibola CF, Di Bernardo MC, Conde L, Broderick P, McDonnell SK, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120:843–6. 10.1182/blood-2012-03-413591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–10. 10.1038/ng.219 [DOI] [PubMed] [Google Scholar]
- 6. Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–5. 10.1038/ng.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010;42:661–4. 10.1038/ng.626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117:1911–6. 10.1182/blood-2010-09-308205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45:868–76. 10.1038/ng.2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–6. 10.1038/ng.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Slager SL, Goldin LR, Strom SS, Lanasa MC, Spector LG, Rassenti L, et al. Genetic susceptibility variants for chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev. 2010;19:1098–102. 10.1158/1055-9965.EPI-09-1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crowther-Swanepoel D, Broderick P, Ma Y, Robertson L, Pittman AM, Price A, et al. Fine-scale mapping of the 6p25.3 chronic lymphocytic leukaemia susceptibility locus. Hum Mol Genet. 2010;19:1840–5. 10.1093/hmg/ddq044 [DOI] [PubMed] [Google Scholar]
- 13. Crowther-Swanepoel D, Mansouri M, Enjuanes A, Vega A, Smedby KE, Ruiz-Ponte C, et al. Verification that common variation at 2q37.1, 6p25.3, 11q24.1, 15q23, and 19q13.32 influences chronic lymphocytic leukaemia risk. Br J Haematol. 2010;150:473–9. 10.1111/j.1365-2141.2010.08270.x [DOI] [PubMed] [Google Scholar]
- 14. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. 10.1038/nrc2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–51. [DOI] [PubMed] [Google Scholar]
- 16. Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. [DOI] [PubMed] [Google Scholar]
- 18. Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–15. 10.1093/carcin/bgp209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kminkova J, Mraz M, Zaprazna K, Navrkalova V, Tichy B, Plevova K, et al. Identification of novel sequence variations in microRNAs in chronic lymphocytic leukemia. Carcinogenesis. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martín-Guerrero I, Enjuanes A, Richter J, Ammerpohl O, Colomer D, Ardanaz M, et al. A putative "hepitype" in the ATM gene associated with chronic lymphocytic leukemia risk. Genes Chromosomes Cancer. 2011;50:887–95. 10.1002/gcc.20912 [DOI] [PubMed] [Google Scholar]
- 21. Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35(Database issue):D51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ziebarth JD, Bhattacharya A, Chen A, Cui Y. PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res. 2012;40(Database issue):D216–21. 10.1093/nar/gkr1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic Acids Res. 2010;38(Database issue):D640–51. 10.1093/nar/gkp926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological). 1995;57:289–300. [Google Scholar]
- 25. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70–4. 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phua SL, Sivakamasundari V, Shao Y, Cai X, Zhang LF, Lufkin T, et al. Nuclear accumulation of an uncapped RNA produced by Drosha cleavage of a transcript encoding miR-10b and HOXD4. PLoS One. 2011;6:e25689 10.1371/journal.pone.0025689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–8. [DOI] [PubMed] [Google Scholar]
- 29. Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–77. [DOI] [PubMed] [Google Scholar]
- 30. Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–50. 10.1056/NEJMoa0803785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Yang H, Lee JJ, Kim E, Lippman SM, Khuri FR, et al. MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis. 2010;31:2118–23. 10.1093/carcin/bgq177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rotunno M, Zhao Y, Bergen AW, Koshiol J, Burdette L, Rubagotti M, et al. Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br J Cancer. 2010;103:1870–4. 10.1038/sj.bjc.6605976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma H, Yuan H, Yuan Z, Yu C, Wang R, Jiang Y, et al. Genetic variations in key microRNA processing genes and risk of head and neck cancer: a case-control study in Chinese population. PLoS One. 2012;7:e47544 10.1371/journal.pone.0047544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jakymiw A, Patel RS, Deming N, Bhattacharyya I, Shah P, Lamont RJ, et al. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49:549–59. 10.1002/gcc.20765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–7. 10.1158/0008-5472.CAN-07-5991 [DOI] [PubMed] [Google Scholar]
- 37. Clague J, Lippman SM, Yang H, Hildebrandt MA, Ye Y, Lee JJ, et al. Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol Carcinog. 2010;49:183–9. 10.1002/mc.20588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15(23):2149–55. [DOI] [PubMed] [Google Scholar]
- 40. Holt R, Barnby G, Maestrini E, Bacchelli E, Brocklebank D, Sousa I, et al. Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Hum Genet. 2010;18:1013–9. 10.1038/ejhg.2010.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pfeufer A, Sanna S, Arking DE, Müller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–14. 10.1038/ng.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7. 10.1158/1055-9965.EPI-08-0814 [DOI] [PubMed] [Google Scholar]
- 43. Akkız H, Bayram S, Bekar A, Akgöllü E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat. 2011;18:e399–407. 10.1111/j.1365-2893.2010.01414.x [DOI] [PubMed] [Google Scholar]
- 44. Dou T, Wu Q, Chen X, Ribas J, Ni X, Tang C, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136:1853–9. 10.1007/s00432-010-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peng S, Kuang Z, Sheng C, Zhang Y, Xu H, Cheng Q. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55:2288–93. 10.1007/s10620-009-1007-x [DOI] [PubMed] [Google Scholar]
- 46. Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. 10.1158/0008-5472.CAN-09-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD. Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep. 2011;38:1609–15. 10.1007/s11033-010-0270-4 [DOI] [PubMed] [Google Scholar]
- 48. Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, McClean MD, et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;16:3713–20. 10.1158/1078-0432.CCR-10-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang F, Ma YL, Zhang P, Yang JJ, Chen HQ, Liu ZH, et al. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:269–75. 10.1007/s11033-011-0735-0 [DOI] [PubMed] [Google Scholar]
- 50. Brisbin AG, Asmann YW, Song H, Tsai YY, Aakre JA, Yang P, et al. Meta-analysis of 8q24 for seven cancers reveals a locus between NOV and ENPP2 associated with cancer development. BMC Med Genet. 2011;12:156 10.1186/1471-2350-12-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4 10.1186/1742-4690-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–55. [DOI] [PubMed] [Google Scholar]
- 53. Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. 2007;213:511–8. [DOI] [PubMed] [Google Scholar]
- 54. Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–21. 10.1158/1541-7786.MCR-07-0105 [DOI] [PubMed] [Google Scholar]
- 55. Kim HK, Prokunina-Olsson L, Chanock SJ. Common genetic variants in miR-1206 (8q24.2) and miR-612 (11q13.3) affect biogenesis of mature miRNA forms. PLoS One. 2012;7:e47454 10.1371/journal.pone.0047454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.