Abstract
Cutaneous T-cell lymphoma (CTCL) is a potentially devastating malignancy. The pathogenesis of this cancer remains poorly elucidated. Previous studies focused on analysis of expression and function of known oncogenes and tumor suppressor genes. However, emerging reports highlight that it is also important to analyze the expression of genes that are ectopically expressed in CTCL (e.g., embryonic stem cell genes (ESC), cancer testis (CT) genes, etc.). Currently, it is not known whether ESC genes are expressed in CTCL. In the current work, we analyze by RT-PCR the expression of 26 ESC genes, many of which are known to regulate pluripotency and promote cancer stem cell-like phenotype, in a historic cohort of 60 patients from Boston and in a panel of 11 patient-derived CTCL cell lines and compare such expression to benign inflammatory dermatoses that often clinically mimic CTCL. Our findings document that many critical ESC genes including NANOG, SOX2, OCT4 (POU5F1) and their upstream and downstream signaling members are expressed in CTCL. Similarly, polycomb repressive complex 2 (PRC2) genes (i.e., EZH2, EED, and SUZ12) are also expressed in CTCL lesional skin. Furthermore, select ESC genes (OCT4, EED, TCF3, THAP11, CHD7, TIP60, TRIM28) are preferentially expressed in CTCL samples when compared to benign skin biopsies. Our work suggests that ESC genes are ectopically expressed together with CT genes, thymocyte development genes and B cell-specific genes and may be working in concert to promote tumorigenesis. Specifically, while ESC genes may be promoting cancer stem cell-like phenotype, CT genes may be contributing to aneuploidy and genomic instability by producing aberrant chromosomal translocations. Further analysis of ESC expression and function in this cancer will greatly enhance our fundamental understanding of CTCL and will help us identify novel therapeutic targets.
Keywords: cancer testis genes, cutaneous T cell lymphoma (CTCL), embryonic stem cell genes, mycosis fungoides (MF), polycomb repressive complex 2 (PRC2), sézary syndrome (SS), thymocyte development genes
Abbreviations: ALCL, Anaplastic Large Cell Lymphoma; BLK, B-lymphoid kinase; CSC, Cancer Stem Cell; C-ALCL, Cutaneous Anaplastic Large Cell Lymphoma; CTCL, Cutaneous T-Cell Lymphoma; DMC1, Disrupted Meiotic cDNA 1; ESC, Embryonic Stem Cell; EVA1, Epithelial C-like antigen 1; MF, Mycosis Fungoides; PBMC, Peripheral Blood Mononucleated Cells; PLS3, Plastin-3; PRC1, Polycomb Repressive Complex 1; PRC2, Polycomb Repressive Complex 2; SS, Sézary Syndrome; SYCP1, Synaptonemal Complex Protein 1; TOX, Thymocyte selection–associated high mobility group box; ZFX, Zinc finger protein X-linked
Introduction
CTCL is a rare, but potentially devastating malignancy. It represents a heterogeneous group of non-Hodgkin's lymphomas that are characterized by localization of malignant T lymphocytes to the skin.1 The annual incidence of CTCL was recently reported to be ∼10 cases per million individuals per year in the United States. Mycosis fungoides (MF), Anaplastic large cell lymphoma (ALCL), and Sézary syndrome (SS) represent some of the most common forms of CTCL and together account for >80% of all CTCL cases.1
The molecular pathogenesis of CTCL remains only partially understood. Recent reports elucidated the nature of cancer initiating cells for MF and SS.2 Multiple studies attempted to clarify the genetic multistep carcinogenesis of CTCL.3-6 In addition, several studies identified recurrent chromosomal aberrations in MF/SS.7-12 While much research is focused on investigation of known oncogenes and tumor suppressor genes in CTCL,6,13-15 recent reports suggest that it is also important to look for genes that could be erroneously expressed in this cancer.16 Ectopic expression of oncodevelopmental genes (e.g., α-Fetoprotein, H19 and Sonic Hedgehog signaling genes), ESC genes (e.g., OCT4 (POU5F1), SOX2, NANOG, EED, SUZ12, etc.), and CT genes have been reported in various solid and lymphoproliferative malignancies, where they are believed to play a central role in tumorigenesis and cancer progression. The expression status of these genes in CTCL remains unknown.
Recent reports demonstrated that CT genes are ectopically expressed in this malignancy. We and others have suggested that CT antigens during carcinogenesis may play an important role in maintaining cell survival (i.e., inhibition of apoptosis),17-19 promoting resistance to various forms of chemo- and radio-therapy20,21 and contributing to oncogenesis by targeting p53 and p21 tumor suppressor genes.22,23 Also, since SYCP1 (Synaptonemal Complex Protein 1), SYCP3, DMC1 (Disrupted Meiotic cDNA 1), and REC8 CT antigens under normal conditions regulate generation of double strand DNA breaks during crossing over in meiosis, it was suggested that these genes may promote aneuploidy and genomic instability in cancers by producing aberrant chromosomal translocations.24
Most importantly, in recent years a concept of cancer stem cells (CSC) has emerged, where these cells share many characteristics with normal stem cells, specifically, they possess an enhanced self-renewal capacity, resistance to apoptosis, ability to maintain undifferentiated state, overcome cellular senescence and give rise to all cell types found in a particular tumor. Like normal stem cells, CSCs divide infrequently and, therefore, are often spared by therapies that target rapidly dividing populations of cells. These CSCs are proposed to persist in tumors as a distinct population and cause relapse and metastasis by giving rise to new tumors. Hence, understanding the expression of these embryonic genes in CTCL may help us better understand the pathogenesis of this cancer and identify novel diagnostic markers and therapeutic targets.
In the current work, we investigate the expression of a panel of ESC genes in CTCL lesional skin and in 11 patient-derived CTCL cell lines and summarize expression patterns for CT antigens, thymocyte development, B cell-specific, and other ectopically expressed genes in this cancer.
Results
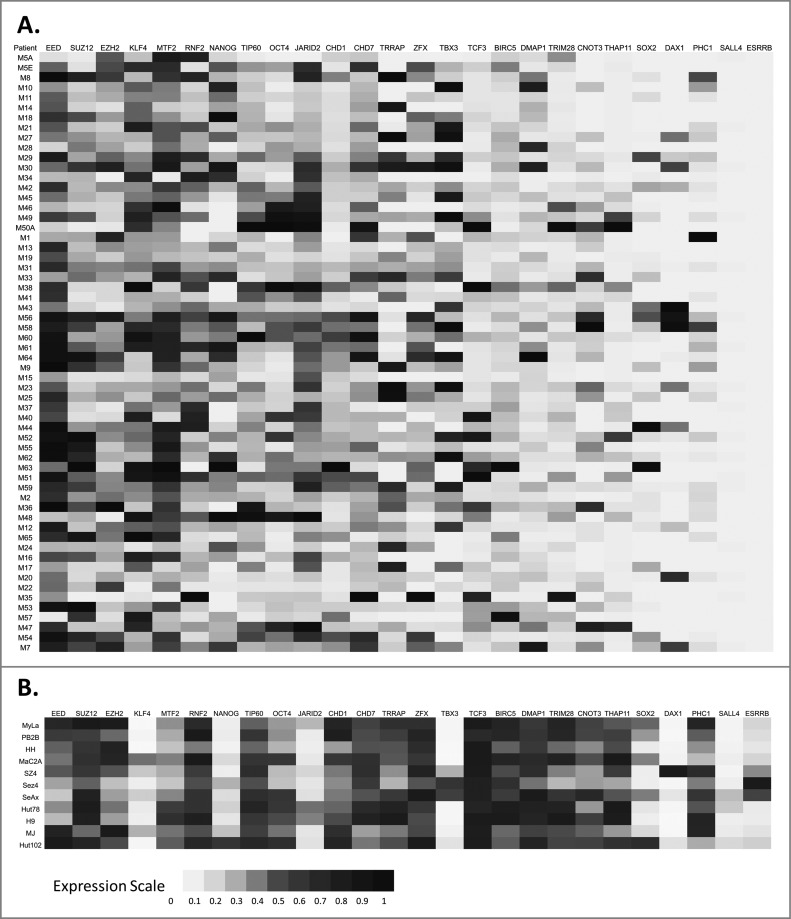
Characterization and function of CSCs has recently become the focus of cancer research. While expression of many ESC and putative CSC markers has been extensively studied in other cancers, very little experimental data are available for CTCL. To address this, we tested the expression of 26 ESC markers in CTCL lesional skin samples via RT-PCR. These results demonstrate that, while few genes are not expressed or infrequently expressed in CTCL (e.g., SALL4, ESRRB and DAX1/NR0B1), the majority of ESC markers are heterogeneously expressed in CTCL (Fig. 1A and Table S1). Furthermore, a number of genes that are critical for maintenance of pluripotency in ESCs (e.g., EED, SUZ12, EZH2, MTF2, OCT4, CHD7, and TIP60) are almost universally expressed in CTCL lesional skin samples. Normal function of these genes and their potential roles in carcinogenesis are summarized in Table S1.
Figure 1.
Expression of embryonic stem cell (ESC) genes in CTCL. (A) Lesional skin biopsies were obtained from CTCL patients and gene expression for a panel of ESC genes was analyzed via RT-PCR. (B) Similarly, the expression of ESC genes was also tested by RT-PCR in 11 patient-derived CTCL cell lines cultured under standard conditions.
Any given CTCL lesional skin biopsy contains numerous cell types including keratinocytes, fibroblasts, merkel cells, melanocytes as well as infiltrating malignant, and reactive CD4+ T cells. To confirm that the above observed ectopic expression of ESC genes takes place in malignant T cells we tested the expression of these 26 ESC genes in a panel of 11 patient-derived immortalized CTCL cell lines (Fig. 1B). Our findings confirm that many of these genes are also ectopically expressed in patient-derived CTCL cell lines.
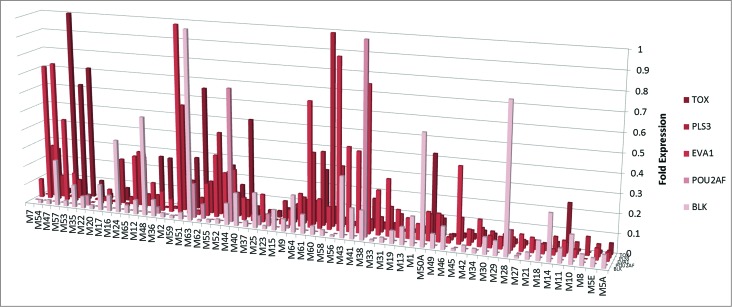
We also analyzed the expression of genes that are usually not expressed in mature CD4+ T cells. TOX (Thymocyte selection-associated high mobility group box) and EVA1 (Epithelial C-like antigen 1) are usually expressed in thymocyte development, but are subsequently downregulated in mature T cells,25,26 while BLK (B-lymphoid kinase) and POU2AF are usually specific to B cells and should not be expressed in T cells.27,28 PLS3 (Plastin 3) is an actin binding protein, which is also normally not expressed in T cells.6,29 Our RT-PCR analysis confirms that BLK, POU2AF, TOX, EVA1, and PLS3 are expressed in CTCL lesional skin biopsies (Fig. 2).
Figure 2.
Expression of selected developmental genes in CTCL. RT-PCR expression of thymocyte development genes (EVA1 and TOX), B cell-specific genes (BLK and POU2AF) and PLS3 in lesional skin biopsies from CTCL patients.
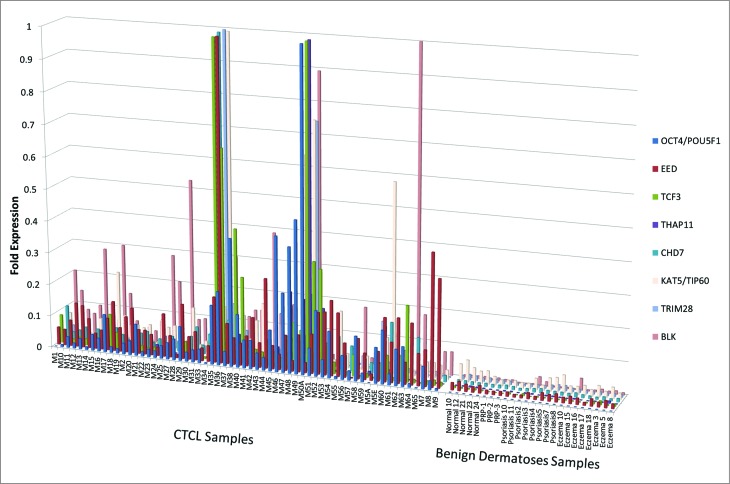
We then compared the expression of these genes between CTCL lesional skin samples, benign dermatoses that often clinically masquerade as CTCL (e.g., chronic eczema, psoriasis, and pityriasis rubra pilaris) and normal skin samples from healthy volunteers. Our findings indicate that select genes (OCT4, EED, TCF3, THAP11, CHD7, TIP60, TRIM28, and BLK) are preferentially expressed in CTCL samples when compared to benign skin biopsies (Fig. 3).
Figure 3.
Comparison by RT-PCR of gene expression for ESC and developmental genes between normal skin, skin affected by benign inflammatory dermatoses (i.e., psoriasis, chronic eczema and pityriasis rubra pilaris or PRP) vs. lesional skin from CTCL patients. OCT4, EED, TCF3, THAP11, CHD7, TIP60, TRIM28, and BLK genes are preferentially expressed on mRNA level in CTCL samples when compared to benign skin conditions.
Discussion
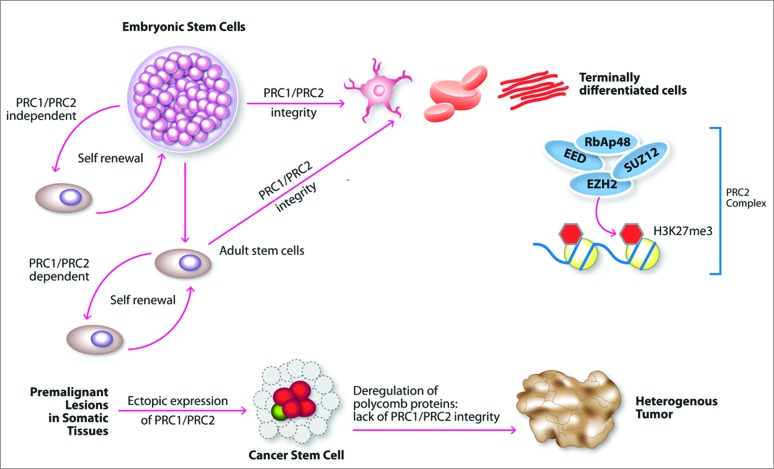
Our results for the first time document the expression of ESC genes in CTCL lesional skin biopsies and patient-derived cell lines. Furthermore, we demondtrate that OCT4, EED, TCF3, THAP11, CHD7, TIP60, and TRIM28 ESC genes are preferentially expressed in CTCL, but not in benign inflammatory dermatoses. From the extensive list of ESC genes (Table S1) few classes especially stand out. PRC2 genes (EZH2, EED, and SUZ12) were previously shown to promote pluripotency in normal stem cells, enhance self-renewal capacity, maintain de-differentiation, and resist apoptosis in normal ESCs.30-32 A schematic diagram of the role of these genes in carcinogenesis is presented in Fig. 4. Polycomb group proteins are key regulators of chromatin structure, cell identity, and development.30 Indeed, it was suggested that reprogramming of somatic cells toward pluripotency would involve extensive chromatin reorganization and changes in gene expression. These genes were documented to be ectopically expressed in multiple malignancies, where they establish/promote a cancer stem cell-like state.30-32 Such ectopic expression of PRC2 components often correlates with disease severity.30,32 As highlighted in in our study, EZH2, EED and SUZ12 are strongly expressed in CTCL samples, while EED is preferentially expressed in CTCL, but not in benign dermatoses that often clinically mimic this disease. Similarly, our study indicates that JARID2, protein that often associates with the PRC2 complex,33 as well as PHC1 member of the PRC1-like complex,34,35 are also ectopically expressed in CTCL lesional skin. JARID2 is heterogeneously expressed in cell lines derived from SS patients and in MF patient biopsies. All three genes were shown to be important in maintaining ESC identity ( Table S1).
Figure 4.
Putative function of PRC2 complex genes in carcinogenesis. Integrity of PRC1 and PRC2 signaling is critical for embryonic and adult stem cells. Ectopic expression of PRC1 and PRC2 in cancer may reprogram somatic cells toward cancer stem cell phenotype. Deregulation of these genes in somatic adult tissues may play a critical role in tumorigenesis.
NANOG, SOX2, and OCT4 are another group of key transcription factors involved in the maintenance of pluripotency of ESCs.36-38 These genes are essential for maintaining the self-renewing undifferentiated state of the inner cell mass of the blastocyst.39 OCT4, SOX2, and NANOG are ectopically expressed in many malignancies, where they are often associated with aggressive disease and poor cancer survival.40-46 Similarly to EZH2, EED, and SUZ12, these genes work in concert to induce pluripotent ESC-like state and promote cancer stem cell phenotype.47,48 Based on our RT-PCR expression results these genes are heterogeneously expressed in CTCL lesional skin, while OCT4 is preferentially expressed in CTCL lesional skin, but not in benign skin conditions. Furthermore, consistent with the above findings, upstream and downstream members of NANOG signaling are also expressed in CTCL. ZFX (Zinc finger protein X-linked), a transcription factor, is known to transactivate the promoters of NANOG and SOX2,49,50 while DAX1 (also known as NR0B1) and MTF2 pluripotency markers are induced by NANOG.39 All of the aforementioned genes are heterogeneously expressed in CTCL lesional skin. Overexpression of ZFX contributes to the ‘stemness’ and pluripotent behavior of cancers.49,50 Our study further demonstrates that CTCL lesional skin and patient-derived cell lines express other pluripotency ESC markers including CNOT3, KLF4, TBX3, and TRRAP. Detailed description of these genes and their biological activities is provided in Table S1. In contrast, few ESC genes (e.g., SALL4 and ESRRB) were not expressed in CTCL lesional biopsies.
Looking at the other classes of genes, we and others have previously documented that several CT genes were ectopically expressed in CTCL.16 Normal function of these genes is to regulate meiosis, synaptonemal complex assembly, and generation of double strand DNA breaks during crossing over.16 Ectopic expression of these genes in cancer may promote genomic instability and be an important driving force behind aneuploidy and generation of balanced and unbalanced chromosomal translocations. Hence, while ESC genes may be reprogramming cancer stem cell-like phenotype, CT genes may be promoting genomic instability enabling these cells to express important oncogenes and disrupt critical tumor suppressor genes.
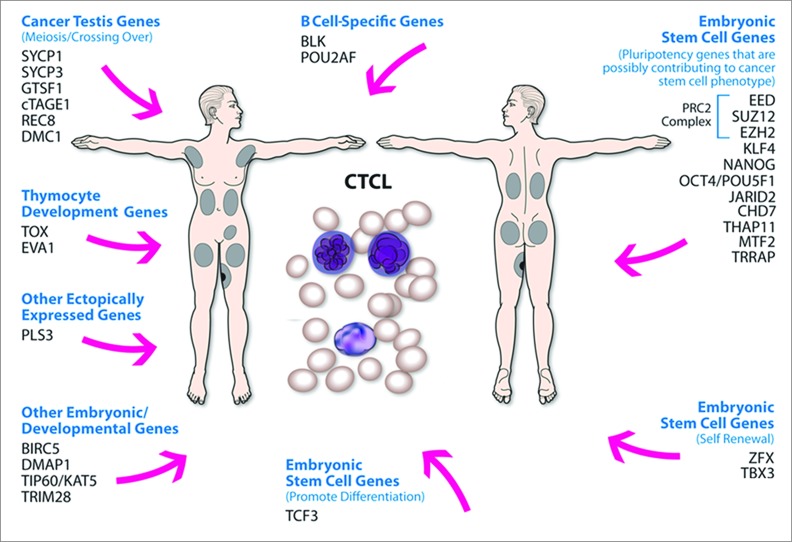
Finally, recent studies highlighted that malignant T cells in CTCL are able to express few B cell-specific genes one of which is the Src kinase BLK.27 Importantly, BLK is constitutively active in malignant T cells and appears to be a bona fide oncogene which drives malignant T cell proliferation in vitro and tumor formation in vivo.27,51 In addition, recent translational experimental work revealed that TOX expression, which is usually silenced in mature T cells, can be used as a robust prognostic and diagnostic marker for MF and SS,52,53 while PLS3 actin binding protein, which is usually not expressed in T cells is consistently expressed in CTCL.6,29,54,55 A growing body of literature documents that ectopic expression of these genes in CTCL is not a mere indication of deregulated cellular processes, but an important mechanism of tumorigenesis and cancer progression. Our study confirms that select thymocyte development genes (e.g., EVA1 and TOX), B cell-specific genes (POU2AF and BLK) and PLS3 are expressed in CTCL lesional skin and in patient-derived cell lines. A summary of different classes of ectopically expressed genes in this cancer is presented in Fig. 5.
Figure 5.
Summary of different classes of aberrantly expressed genes in CTCL. Aberrant expression of embryonic stem cells genes, cancer testis genes, B cell-specific genes and thymocyte development genes may play a role in tumorigenesis/cancer progression in CTCL. Specifically, while embryonic stem cell genes (e.g., members of the PRC2 complex) may be reprogramming cancer stem cell phenotype, cancer testis genes may be contributing to aneuploidy and genomic instability by producing aberrant chromosomal translocations.
In conclusion, our work highlights the importance of ectopic expression of ESC genes, CT antigens, B cell-specific, and thymocyte development genes in CTCL. Further analysis of how ECS genes reprogram malignant T cells and promote cancer stem cell phenotype will greatly enhance our fundamental understanding of this cancer and will help us identify novel therapeutic targets.
Materials and Methods
Patients and samples
All patients were enrolled in the IRB-approved study protocol with informed consent in accordance with the Declaration of Helsinki.3,4 CTCL patients were recruited from the Cutaneous Lymphoma Clinic at the Dana Farber Cancer Institute (DFCI)/Brigham and Women's Hospital (BWH). All tissue samples were obtained and processed as previously described.4 Briefly, punch biopsies from involved skin were collected from 60 CTCL patients between January 26, 2003 and June 1, 2005. The diagnosis and clinical staging were established according to the diagnostic criteria of CTCL.56 The obtained 6 mm biopsies were immediately snap-frozen in liquid nitrogen. Tissue was powdered in liquid nitrogen using Cryo-Press (Microtec Co, Chiba, Japan), and total RNA was extracted using Trizol reagent (Invitrogen, Catalog # 15596-026) and converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad, Catalog # 170-8896) according to the manufacturer's instructions. The historic cohort of patients from Boston (n = 60) that was initially reported in 2007,4 was at the heart of extensive research that led to multiple publications in the field.3,4,13,14,16,53,57,58 The biopsy samples analyzed in this report are the same samples that were analyzed in previous studies.3,4,13,14,16,57,58 Similarly, volunteers with normal healthy skin (N = 5) and benign inflammatory dermatoses (N = 19) were recruited from the outpatient dermatology clinic of the University of British Columbia (Vancouver, Canada) with informed consent.18,20 Full-thickness lesional skin punch biopsies were obtained under local anesthesia as previously described.14,53,57
Cell culture
HH, H9, Hut78, MJ and Hut102 patient-derived CTCL cell lines were previously described59,60 and were purchased from the American Tissue Culture Collection (ATCC). H9 is a clonal derivative of Hut78 cell line.61 MyLa, PB2B, Mac2A, SZ4, SeAx, Sez4 were a generous gift from professors K. Kaltoft and N. Ødum (Copenhagen, Denmark) and were previously described elsewhere.62-66 MJ, Hut78 cells were serially passaged in IMDM media (Invitrogen, Catalog # 12440-079) containing 10% fetal bovine serum (FBS) (Invitrogen, Catalog # 26140-079). HH, H9, Hut102, MyLa, Mac2A and SZ4 cells were grown in RPMI media (Invitrogen, Catalog # 11875-093) containing 10% FBS. Finally, Sez4 and SeAx cells were grown in RPMI media containing 10% FBS, 5 ng/mL of recombinant human IL-2 (Catalog # 202-IL-010 from R&D Systems) and IL-4 (Catalog # 204-IL-010 from R&D Systems). All cells were grown in 5% CO2, 95% air humidified incubator at 37°C. Total RNA was extracted using Trizol reagent (Invitrogen) and converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions.
Quantitative real-time reverse transcription-PCR gene expression analysis
Gene expression was tested via RT-PCR in CTCL patients’ lesional skin and in patient-derived cell lines as previously described.3,13,14 Briefly, RT-PCR was performed utilizing the obtained cDNA from patients and cell lines using iScript RT-PCR mix (Bio-Rad, Catalog #170-8893) on Bio-Rad iCycler (Bio-Rad, Catalog # 185-5201). Primer pair sequences for tested genes and control housekeeping genes are listed in Table S2. The expression was standardized using genorm method67 utilizing ACTB, B2M, SDHA, YWHAZ, and HMBS housekeeping genes.
Supplementary Material
Acknowledgments
We thank Mr. Gregory Cormack for his technical assistance in performing molecular experiments.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Canadian Dermatology Foundation research grants to Dr. Sasseville, Dr. Litvinov and Dr. Zhou, the Fonds de la recherche en santé du Québec (FRSQ) research grant to Dr. Sasseville, Canadian Institutes of Health Research to Dr. Zhou, and the National Institutes of Health SPORE program (P50 CA093683) to Dr. Kupper.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz-Perez JL, Duncan LM, Grange F, et al. WHO-EORTC classification for cutaneous lymphomas. Blood 2005; 105:3768-85; PMID:; http://dx.doi.org/ 10.1182/blood-2004-09-3502 [DOI] [PubMed] [Google Scholar]
- 2. Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood 2010; 116:767-71; PMID:; http://dx.doi.org/ 10.1182/blood-2009-11-251926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Litvinov IV, Jones DA, Sasseville D, Kupper TS. Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma. Clin Cancer Res: Off J Am Assoc Cancer Res 2010; 16:2106-14; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shin J, Monti S, Aires DJ, Duvic M, Golub T, Jones DA, Kupper TS. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood 2007; 110:3015-27; PMID:; http://dx.doi.org/ 10.1182/blood-2006-12-061507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Kester MS, Borg MK, Zoutman WH, Out-Luiting JJ, Jansen PM, Dreef EJ, Vermeer MH, van Doorn R, Willemze R, Tensen CP. A meta-analysis of gene expression data identifies a molecular signature characteristic for tumor-stage mycosis fungoides. J Invest Dermatol 2012; 132:2050-9; PMID:; http://dx.doi.org/ 10.1038/jid.2012.117 [DOI] [PubMed] [Google Scholar]
- 6. Dulmage BO, Geskin LJ. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br J Dermatol 2013; 169:1188-97; PMID:; http://dx.doi.org/ 10.1111/bjd.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barba G, Matteucci C, Girolomoni G, Brandimarte L, Varasano E, Martelli MF, Mecucci C. Comparative genomic hybridization identifies 17q11.2 approximately q12 duplication as an early event in cutaneous T-cell lymphomas. Cancer Genet Cytogen 2008; 184:48-51; PMID:; http://dx.doi.org/ 10.1016/j.cancergencyto.2008.03.007 [DOI] [PubMed] [Google Scholar]
- 8. Caprini E, Cristofoletti C, Arcelli D, Fadda P, Citterich MH, Sampogna F, Magrelli A, Censi F, Torreri P, Frontani M, et al. Identification of key regions and genes important in the pathogenesis of sezary syndrome by combining genomic and expression microarrays. Cancer Res 2009; 69:8438-46; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2367 [DOI] [PubMed] [Google Scholar]
- 9. Laharanne E, Oumouhou N, Bonnet F, Carlotti M, Gentil C, Chevret E, Jouary T, Longy M, Vergier B, Beylot-Barry M, et al. Genome-wide analysis of cutaneous T-cell lymphomas identifies three clinically relevant classes. J Invest Dermatol 2010; 130:1707-18; PMID:; http://dx.doi.org/ 10.1038/jid.2010.8 [DOI] [PubMed] [Google Scholar]
- 10. Mao X, McElwaine S. Functional copy number changes in Sezary syndrome: toward an integrated molecular cytogenetic map III. Cancer Genet Cytogen 2008; 185:86-94; PMID:; http://dx.doi.org/ 10.1016/j.cancergencyto.2008.05.006 [DOI] [PubMed] [Google Scholar]
- 11. van Doorn R, van Kester MS, Dijkman R, Vermeer MH, Mulder AA, Szuhai K, Knijnenburg J, Boer JM, Willemze R, Tensen CP. Oncogenomic analysis of mycosis fungoides reveals major differences with Sezary syndrome. Blood 2009; 113:127-36; PMID:; http://dx.doi.org/ 10.1182/blood-2008-04-153031 [DOI] [PubMed] [Google Scholar]
- 12. Vermeer MH, van Doorn R, Dijkman R, Mao X, Whittaker S, van Voorst Vader PC, Gerritsen MJ, Geerts ML, Gellrich S, Söderberg O, et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res 2008; 68:2689-98; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6398 [DOI] [PubMed] [Google Scholar]
- 13. Litvinov IV, Kupper TS, Sasseville D. The role of AHI1 and CDKN1C in cutaneous T-cell lymphoma progression. Exp Dermatol 2012; 21:964-6; PMID:; http://dx.doi.org/ 10.1111/exd.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Litvinov IV, Zhou Y, Kupper TS, Sasseville D. Loss of BCL7A expression correlates with poor disease prognosis in patients with early-stage cutaneous T-cell lymphoma. Leukemia Lymphoma 2013; 54:653-4; PMID:; http://dx.doi.org/ 10.3109/10428194.2012.717695 [DOI] [PubMed] [Google Scholar]
- 15. Litvinov IV, Pehr K, Sasseville D. Connecting the dots in cutaneous T cell lymphoma (CTCL): STAT5 regulates malignant T cell proliferation via miR-155. Cell Cycle 2013; 12:2172-3; PMID:; http://dx.doi.org/ 10.4161/cc.25550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Litvinov IV, Cordeiro B, Huang Y, Zargham H, Pehr K, Dore MA, Litvinov IV, Cordeiro B, Huang Y, Zargham H, et al. Ectopic expression of cancer-testis antigens in cutaneous T-cell lymphoma patients. Clin Cancer Res: Off J Am Assoc Cancer Res 2014; 20:3799-808; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-14-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kular RK, Yehiely F, Kotlo KU, Cilensek ZM, Bedi R, Deiss LP. GAGE, an antiapoptotic protein binds and modulates the expression of nucleophosminB23 and interferon regulatory factor 1. J Interferon Cytokine Res: Off J Int Soc Interferon Cytokine Res 2009; 29:645-55; http://dx.doi.org/ 10.1089/jir.2008.0099 [DOI] [PubMed] [Google Scholar]
- 18. Nylund C, Rappu P, Pakula E, Heino A, Laato L, Elo LL, Vihinen P, Pyrhönen S, Owen GR, Larjava H, et al. Melanoma-associated cancer-testis antigen 16 (CT16) regulates the expression of apoptotic and antiapoptotic genes and promotes cell survival. PloS One 2012; 7:e45382; PMID:; http://dx.doi.org/ 10.1371/journal.pone.0045382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng Y, He Y, Yang F, Mooney SM, Getzenberg RH, Orban J, Kulkarni P. The cancertestis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J Biol Chem 2011; 286:13985-94; PMID:; http://dx.doi.org/ 10.1074/jbc.M110.210765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cilensek ZM, Yehiely F, Kular RK, Deiss LP. A member of the GAGE family of tumor antigens is an anti-apoptotic gene that confers resistance to FasCD95APO-1, Interferon-gamma, taxol and gamma-irradiation. Cancer Biol Ther 2002; 1:380-7; PMID:; http://dx.doi.org/ 10.4161/cbt.1.4.11 [DOI] [PubMed] [Google Scholar]
- 21. Duan Z, Duan Y, Lamendola DE, Yusuf RZ, Naeem R, Penson RT, Seiden MV. Overexpression of MAGEGAGE genes in paclitaxeldoxorubicin-resistant human cancer cell lines. Clin Cancer Res: Off J Am Assoc Cancer Res 2003; 9:2778-85; PMID: [PubMed] [Google Scholar]
- 22. Caballero OL, Chen YT. Cancertestis (CT) antigens: potential targets for immunotherapy. Cancer Sci 2009; 100:2014-21; PMID:; http://dx.doi.org/ 10.1111/j.1349-7006.2009.01303.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu X, Asa SL, Ezzat S. Fibroblast growth factor 2 and estrogen control the balance of histone 3 modifications targeting MAGE-A3 in pituitary neoplasia. Clin Cancer Res: Off J Am Assoc Cancer Res 2008; 14:1984-96; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-2003 [DOI] [PubMed] [Google Scholar]
- 24. Kalejs M, Ivanov A, Plakhins G, Cragg MS, Emzinsh D, Illidge TM, Erenpreisa J. Upregulation of meiosis-specific genes in lymphoma cell lines following genotoxic insult and induction of mitotic catastrophe. BMC Cancer 2006; 6:6; PMID:; http://dx.doi.org/ 10.1186/1471-2407-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gunther C, Zimmermann N, Berndt N, Grosser M, Stein A, Koch A, Meurer M. Up-regulation of the chemokine CCL18 by macrophages is a potential immunomodulatory pathway in cutaneous T-cell lymphoma. Am J Pathol 2011; 179:1434-42; PMID:; http://dx.doi.org/ 10.1016/j.ajpath.2011.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y, Litvinov IV, Wang Y, Su M-W, Tu P, Jiang X, Kupper TS, Dutz JP, Sasseville D, Zhou Y. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget 2014 June 30; 5:4418-25; PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krejsgaard T, Vetter-Kauczok CS, Woetmann A, Kneitz H, Eriksen KW, Lovato P, Zhang Q, Wasik MA, Geisler C, Ralfkiaer E, et al. Ectopic expression of B-lymphoid kinase in cutaneous T-cell lymphoma. Blood 2009; 113:5896-904; PMID:; http://dx.doi.org/ 10.1182/blood-2008-09-181024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 1995; 80:497-506; PMID:; http://dx.doi.org/ 10.1016/0092-8674(95)90500-6 [DOI] [PubMed] [Google Scholar]
- 29. Booken N, Gratchev A, Utikal J, Weiss C, Yu X, Qadoumi M, Schmuth M, Sepp N, Nashan D, Rass K, et al. Sezary syndrome is a unique cutaneous T-cell lymphoma as identified by an expanded gene signature including diagnostic marker molecules CDO1 and DNM3. Leukemia 2008; 22:393-9; PMID:; http://dx.doi.org/ 10.1038/sj.leu.2405044 [DOI] [PubMed] [Google Scholar]
- 30. Liu C, Shi X, Wang L, Wu Y, Jin F, Bai C, Song Y. SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol: J Int Soc Oncodev Biol Med 2014; 35:6073-82; PMID:; http://dx.doi.org/ 10.1007/s13277-014-1804-5 [DOI] [PubMed] [Google Scholar]
- 31. Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia 2014; 28:44-9; PMID:; http://dx.doi.org/ 10.1038/leu.2013.288 [DOI] [PubMed] [Google Scholar]
- 32. Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis 2011; 2:e204; PMID:; http://dx.doi.org/ 10.1038/cddis.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Landeira D, Fisher AG. Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol 2011; 21:74-80; PMID:; http://dx.doi.org/ 10.1016/j.tcb.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dukers DF, van Galen JC, Giroth C, Jansen P, Sewalt RG, Otte AP, Kluin-Nelemans HC, Meijer CJ, Raaphorst FM. Unique polycomb gene expression pattern in Hodgkin's lymphoma and Hodgkin's lymphoma-derived cell lines. Am J Pathol 2004; 164:873-81; PMID:; http://dx.doi.org/ 10.1016/S0002-9440(10)63175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 2013; 495:236-40; PMID:; http://dx.doi.org/ 10.1038/nature11918 [DOI] [PubMed] [Google Scholar]
- 36. Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005; 121:465-77; PMID:; http://dx.doi.org/ 10.1016/j.cell.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 37. Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007; 1:55-70; PMID:; http://dx.doi.org/ 10.1016/j.stem.2007.05.014 [DOI] [PubMed] [Google Scholar]
- 38. Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007; 448:318-24; PMID:; http://dx.doi.org/ 10.1038/nature05944 [DOI] [PubMed] [Google Scholar]
- 39. Choi SC, Choi JH, Park CY, Ahn CM, Hong SJ, Lim DS. Nanog regulates molecules involved in stemness and cell cycle-signaling pathway for maintenance of pluripotency of P19 embryonal carcinoma stem cells. J Cell Physiol 2012; 227:3678-92; PMID:; http://dx.doi.org/ 10.1002/jcp.24076 [DOI] [PubMed] [Google Scholar]
- 40. Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing L, Zhang Y, Ling EA, Gao J, Hao A. Expression profile of embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human gliomas. Histopathology 2011; 59:763-75; PMID:; http://dx.doi.org/ 10.1111/j.1365-2559.2011.03993.x [DOI] [PubMed] [Google Scholar]
- 41. Huang P, Qiu J, Li B, Hong J, Lu C, Wang L, Wang J, Hu Y, Jia W, Yuan Y. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin Biochem 2011; 44:582-9; PMID:; http://dx.doi.org/ 10.1016/j.clinbiochem.2011.02.012 [DOI] [PubMed] [Google Scholar]
- 42. Han X, Fang X, Lou X, Hua D, Ding W, Foltz G, Hood L, Yuan Y, Lin B. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PloS One 2012; 7:e41335; PMID:; http://dx.doi.org/ 10.1371/journal.pone.0041335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matsuoka J, Yashiro M, Sakurai K, Kubo N, Tanaka H, Muguruma K, Sawada T, Ohira M, Hirakawa K. Role of the stemness factors sox2, oct34, and nanog in gastric carcinoma. JJ Surg Res 2012; 174:130-5; PMID:; http://dx.doi.org/ 10.1016/j.jss.2010.11.903 [DOI] [PubMed] [Google Scholar]
- 44. Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, Hardisson D, Reis-Filho JS, Palacios J. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol : Off J U S Can Acad Pathol, Inc 2007; 20:474-81; PMID:; http://dx.doi.org/ 10.1038/modpathol.3800760 [DOI] [PubMed] [Google Scholar]
- 45. Velcheti V, Schalper K, Yao X, Cheng H, Kocoglu M, Dhodapkar K, Deng Y, Gettinger S, Rimm DL. High SOX2 levels predict better outcome in non-small cell lung carcinomas. PloS One 2013; 8:e61427; PMID:; http://dx.doi.org/ 10.1371/journal.pone.0061427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun Q, Li J, Wang G, Xie Y. Role of the embryonic protein SOX2 in cholangiocarcinoma. Cell Biochem Biophys 2014; 70:1311-6; PMID:; http://dx.doi.org/ 10.1007/s12013-014-0056-8 [DOI] [PubMed] [Google Scholar]
- 47. Lee MR, Ju HJ, Kim BS, Ko YH, Kim WS, Kim SJ. Isolation of side population cells in B-cell non-Hodgkin's lymphomas. Acta Haematologica 2013; 129:10-7; PMID:; http://dx.doi.org/ 10.1159/000341284 [DOI] [PubMed] [Google Scholar]
- 48. Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, Ivanov A, Jascenko E, Scherthan H, Cragg M, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res 2010; 316:2099-112; PMID:; http://dx.doi.org/ 10.1016/j.yexcr.2010.04.030 [DOI] [PubMed] [Google Scholar]
- 49. Lai KP, Chen J, He M, Ching AK, Lau C, Lai PB, To KF, Wong N. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer J Int du Cancer 2014; 135:1790-9; PMID:; http://dx.doi.org/ 10.1002/ijc.28819 [DOI] [PubMed] [Google Scholar]
- 50. Weisberg SP, Smith-Raska MR, Esquilin JM, Zhang J, Arenzana TL, Lau CM, Churchill M, Pan H, Klinakis A, Dixon JE, et al. ZFX controls propagation and prevents differentiation of acute T-lymphoblastic and myeloid leukemia. Cell Rep 2014; 6:528-40; PMID:; http://dx.doi.org/ 10.1016/j.celrep.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Petersen DL, Krejsgaard T, Berthelsen J, Fredholm S, Willerslev-Olsen A, Sibbesen NA, Bonefeld CM, Andersen MH, Francavilla C, Olsen JV, et al. B-lymphoid tyrosine kinase (Blk) is an oncogene and a potential target for therapy with dasatinib in cutaneous T-cell lymphoma (CTCL). Leukemia 2014; 28:2109-12; PMID:; http://dx.doi.org/ 10.1038/leu.2014.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, Martinka M, Dutz JP, Zhang X, Zheng Z, et al. Molecular markers of early-stage mycosis fungoides. J Invest Dermatol 2012; 132:1698-706; PMID:; http://dx.doi.org/ 10.1038/jid.2012.13 [DOI] [PubMed] [Google Scholar]
- 53. Huang Y, Litvinov IV, Wang Y, Su MW, Tu P, Jiang X, Kupper TS, Dutz JP, Sasseville D, Zhou Y. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget 2014; 5:4418-25; PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson VE, Vonderheid EC, Hess AD, Eischen CM, McGirt LY. Genetic markers associated with progression in early mycosis fungoides. J Eur Acad Dermatol Venereol. 2014 Nov; 28(11):1431-5; http://dx.doi.org/ 10.1111/jdv.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nebozhyn M, Loboda A, Kari L, Rook AH, Vonderheid EC, Lessin S, Berger C, Edelson R, Nichols C, Yousef M, et al. Quantitative PCR on 5 genes reliably identifies CTCL patients with 5% to 99% circulating tumor cells with 90% accuracy. Blood 2006; 107:3189-96; PMID:; http://dx.doi.org/ 10.1182/blood-2005-07-2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, Zackheim H, Duvic M, Estrach T, Lamberg S, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007; 110:1713-22; PMID:; http://dx.doi.org/ 10.1182/blood-2007-03-055749 [DOI] [PubMed] [Google Scholar]
- 57. Krejsgaard T, Litvinov IV, Wang Y, Xia L, Willerslev-Olsen A, Koralov SB, Kopp KL, Bonefeld CM, Wasik MA, Geisler C, et al. Elucidating the role of interleukin-17F in cutaneous T-cell lymphoma. Blood 2013; 122:943-50; PMID:; http://dx.doi.org/ 10.1182/blood-2013-01-480889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Willerslev-Olsen A, Litvinov IV, Fredholm SM, Petersen DL, Sibbesen NA, Gniadecki R, Zhang Q, Bonefeld CM, Wasik MA, Geisler C, et al. IL-15 and IL-17F are differentially regulated and expressed in mycosis fungoides (MF). Cell Cycle 2014; 13:1306-12; PMID:; http://dx.doi.org/ 10.4161/cc.28256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang C, Hazarika P, Ni X, Weidner DA, Duvic M. Induction of apoptosis by bexarotene in cutaneous T-cell lymphoma cells: relevance to mechanism of therapeutic action. Clin Cancer Res: Off J Am Assoc Cancer Res 2002; 8:1234-40; PMID: [PubMed] [Google Scholar]
- 60. Bunn PA, Jr, Foss FM. T-cell lymphoma cell lines (HUT102 and HUT78) established at the National Cancer Institute: history and importance to understanding the biology, clinical features, and therapy of cutaneous T-cell lymphomas (CTCL) and adult T-cell leukemia-lymphomas (ATLL). J Cell Biochem Suppl 1996; 24:12-23; PMID:; http://dx.doi.org/ 10.1002/jcb.240630503 [DOI] [PubMed] [Google Scholar]
- 61. Chen TR. Karyotypic derivation of H9 cell line expressing human immunodeficiency virus susceptibility. J Natl Cancer Inst 1992; 84:1922-6; PMID:; http://dx.doi.org/ 10.1093/jnci/84.24.1922 [DOI] [PubMed] [Google Scholar]
- 62. Abrams JT, Lessin S, Ghosh SK, Ju W, Vonderheid EC, Nowell P, Murphy G, Elfenbein B, DeFreitas E. A clonal CD4-positive T-cell line established from the blood of a patient with Sezary syndrome. J Invest Dermat 1991; 96:31-7; PMID:; http://dx.doi.org/ 10.1111/1523-1747.ep12514693 [DOI] [PubMed] [Google Scholar]
- 63. Kaltoft K, Bisballe S, Dyrberg T, Boel E, Rasmussen PB, Thestrup-Pedersen K. Establishment of two continuous T-cell strains from a single plaque of a patient with mycosis fungoides. In vitro Cell Dev Biol: J Tissue Cult Assoc 1992; 28A:161-7; http://dx.doi.org/ 10.1007/BF02631086 [DOI] [PubMed] [Google Scholar]
- 64. Kaltoft K, Bisballe S, Rasmussen HF, Thestrup-Pedersen K, Thomsen K, Sterry W. A continuous T-cell line from a patient with Sezary syndrome. Arch Dermatol Res 1987; 279:293-8; PMID:; http://dx.doi.org/ 10.1007/BF00431220 [DOI] [PubMed] [Google Scholar]
- 65. Starkebaum G, Loughran TP, Jr, Waters CA, Ruscetti FW. Establishment of an IL-2 independent, human T-cell line possessing only the p70 IL-2 receptor. Int J Cancer J Int du Cancer 1991; 49:246-53; PMID:; http://dx.doi.org/ 10.1002/ijc.2910490218 [DOI] [PubMed] [Google Scholar]
- 66. Wasik MA, Seldin DC, Butmarc JR, Gertz R, Marti R, Maslinski W, Kadin ME. Analysis of IL-2, IL-4 and their receptors in clonally-related cell lines derived from a patient with a progressive cutaneous T-cell lymphoproliferative disorder. Leukemia Lymphoma 1996; 23:125-36; PMID:; http://dx.doi.org/ 10.3109/10428199609054811 [DOI] [PubMed] [Google Scholar]
- 67. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3:RESEARCH0034; PMID:; http://dx.doi.org/ 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.