Abstract
The pigmentation of many Aeromonas species has been thought to be due to the production of a L-DOPA (L-3,4-dihydroxyphenylalanine) based melanin. However, in this study we found that although L-DOPA synthesis occurs in the high-melanin-yielding Aeromonas media strain WS, it plays a minor, if any, role in pigmentation. Instead, the pigmentation of A. media strain WS is due to the production of pyomelanin through HGA (homogentisate). Gene products of phhA (encodes phenylalanine hydroxylase), tyrB and aspC (both encode aromatic amino acid aminotransferase), and hppD (encodes 4-hydroxyphenylpyruvate dioxygenase) constitute a linear pathway of converting phenylalanine to HGA and disruption of any one of these genes impairs or blocks pigmentation of A. media strain WS. This HGA biosynthesis pathway is widely distributed in Aeromonas, but HGA is only detectable in the cultures of pigmented Aeromonas species. Heterologous expression of HppD from both pigmented and non-pigmented Aeromonas species in E. coli leads to the production of pyomelanin and thus pigmentation, suggesting that most Aeromonas species have the critical enzymes to produce pyomelanin through HGA. Taken together, we have identified a widely conserved biosynthesis pathway of HGA based pyomelanin in Aeromonas that may be responsible for pigmentation of many Aeromonas species.
Introduction
Melanin is a group of negatively charged hydrophobic macromolecules formed by the enzymatic oxidation and subsequent polymerization of phenolic and/or indolic compounds [1, 2]. Production of melanin occurs in almost every taxon of living organisms ranging from bacteria to human [3, 4]. Although melanin is normally not essential for the growth of organisms, it can help the organisms compete and survive under certain environmental stress conditions, such as UV radiation and toxic free radicals [5, 6]. Melanin produced by some pathogenic microbes is also believed to protect the microbes from host defenses [7, 8]. Meanwhile, microbial melanin has multiple biotechnological applications, such as photoprotectant, antioxidant, semiconductor, energy transducer, drugs carriers, and cation exchangers [9–11].
Melanin can be classified into four categories based on the intermediates of melanogenesis: eumelanin, pheomelanin, allomelanin and pyomelanin. Eumelanin is derived from the L-DOPA, pheomelanin forms from cysteinylation of L-DOPA, allomelanin results from nitrogen-free precursors, and pyomelanin is produced from HGA [12–14]. In bacteria, melanin is usually synthesized from either L-DOPA or HGA. DOPA based melanin which contains eumelanin and pheomelanin is catalyzed by tyrosinase, which converts tyrosine to L-DOPA and then to dopaquinone. The latter then undergoes auto-oxidization and polymerization to form melanin. Production of DOPA based melanin has been described to occur in a wide range of bacteria, including Bacillus, Marinomonas, Rhizobium, Streptomyces and Thermomicrobium [15–19]. HGA based melanin, or the so-called pyomelanin, is carried out by the 4-hydroxyphenylpyruvate dioxygenase (HppD) which catalyzes the reaction from 4-hydroxyphenylpyruvate to HGA. The latter then auto-oxidizes to form benzoquinoneacetic acid and then self polymerizes to produce the pigment [20]. Bacteria known to synthesize HGA-based pyomelanin are Hyphomonas, Pseudomonas, Ralstonia, Shewanella, Sinorhizobium, Vibrio and Xanthomonas [21–25].
Aeromonas are rod-shaped, gram-negative, facultative anaerobic bacteria that are widely distributed in terrestrial and aquatic environments [26]. The Aeromonas genus currently contains 31 species (http://www.bacterio.net/-allnamesac.html), some of which are able to synthesize melanin, such as A. salmonicida, A. media and A. liquefaciens [27, 28]. However, a number of Aeromonas species are believed to never undergo melanogenesis, such as A. allosaccharophila, A. encheleia and A. hydrophila [29–31]. The melanin produced in Aeromonas had been considered to be DOPA based melanin because L-DOPA but not HGA had been detected in the bacterial cultures [27, 28, 32]. Consistent with the documents, we also detected the melanin precursor L-DOPA in the culture of a high-melanin-yielding strain A. media strain WS isolated from the East Lake, Wuhan, China. A distinct tyrosinase TyrA was also identified from this bacterium [33]. However, subsequent study found that deletion of tyrA from the bacterium does not significantly affect pigment production, suggesting that TyrA is not necessary for pigmentation and DOPA-based melanin is likely not the primary melanin produced by A. media strain WS [34].
In order to unravel the molecular determinants involved in the pigment formation in A. media strain WS, we screened for A. media WS mutants that were defective in pigmentation by transposon mutagenesis. Through studies of some of the isolated mutants, we found that the major melanin constituents produced by this bacterium is HGA-based pyomelanin rather than L-DOPA based melanin as previously believed. Phenylalanine 4-monooxygenase, aromatic amino acid aminotransferase and 4-hydroxyphenylpyruvate dioxygenase are important enzymes required for HGA based melanin synthesis. Through bioinformatics analysis, we found that genes encoding for these enzymes are widely distributed in Aeromonas, including both pigmented and non-pigmented Aeromonas species. However, we found that HGA is detectable in the cultures of pigmented Aeromonas species but not in that of the non-pigmented Aeromonas hydrophila, although both have functional HppD. Consistent with the result in A. media strain WS, L-DOPA is detected in the culture of A. hydrophila, indicating that the presence of L-DOPA does not correlate with pigmentation. Based on these results, we propose that pigmentation of many Aeromonas species, if not all of them, is due to the production of pyomelanin through HGA rather than the production of L-DOPA based melanin.
Materials and Methods
Bacterial strains and media
The strains used in this study are listed in Table 1. The high-melanin-yielding A. media strain WS was isolated previously from the East Lake, Wuhan, China [35]. A. salmonicida_AB98041 and A. salmonicida KACC14791 were obtained from the China Center for Type Culture Collection (CCTCC) and Korean Agricultural Culture Collection (KACC), respectively. Both of them could produce melanin when grown at 22°C but not at 30°C or above. A. hydrophila_XS91–4–1, which does not produce melanin, was a kind gift by Prof. Aihua Li of Institute of Hydrobiology, Chinese Academy of Sciences. All Aeromonas strains were generally cultured in Luria-Bertani (LB) medium at 30°C, but the two A. salmonicida strains were cultured at 22°C for the production of melanin. The strains of Escherichia coli were grown in LB at 37°C unless otherwise stated. If necessary, this medium was supplemented with kanamycin (50 μg ml-1), ampicillin (100 μg ml-1), chloramphenicol (34 μg ml-1) or gentamicin (50 μg ml-1), respectively.
Table 1. Bacterial strains used in this study.
| Strains | Characteristic(s) | Reference or source |
|---|---|---|
| A. media strain WS | wild type, high-melanin-yielding strain, Ampr Cms | 35 |
| WS-M1/ WS-M2/ WS-M3/ WS-M4/ WS-M5/ WS-M6/ WS-M7/ WS-M8/ WS-M9/ WS-M11/ | A. media strain WS with a Tn5 transposon insertion, exhibiting reduced melanin production (about 40%-70%) but with no detectable growth defect | This study |
| WS-M12/ WS-M14 | A. media strain WS with a Tn5 transposon insertion, exhibiting reduction in both melanin production and growth capacity | This study |
| WS-M10/ WS-M13 | A. media strain WS with a Tn5 transposon insertion, exhibiting abolished melanin production | This study |
| WSΔphhA | A. media strain WS with a deletion of phhA | This study |
| WSΔtyrB | A. media strain WS with a deletion of tyrB | This study |
| WSΔaspC | A. media strain WS with a deletion of aspC | This study |
| WSΔtyrBΔaspC | A. media strain WSΔtyrB with a deletion of aspC | This study |
| WSΔhppD | A. media strain WS with a deletion of hppD | This study |
| A. salmonicida_AB98041 | Pigment-producing Aeromonas strain | CCTCC |
| A. salmonicidaKACC14791 | Pigment-producing Aeromonas strain | KACC |
| A. hydrophila_XS91-4-1 | Non-pigment-producing Aeromonas strain | Kindly donated by Prof. Aihua Li from Institute of Hydrobiology, Chinese Academy of Sciences |
| E. coli DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 ΔlacU169 (φ80lacZΔM15) | Commercially available |
| E. coli BL21 | F- ompT hsdSB(rB-mB -)gal dcm (DE3) | Commercially available |
| E. coli S17-1 (λpir) | recA thi hsdRM +, λpir phage lysogen RP4::Mu::Km Tn7 TprSmr | 38 |
Conjugation and transposon mutagenesis
The plasmid pUTKm containing the Tn5 transposon, which has been reported to work in a wide range of gram-negative bacteria and exhibited no bias for any specific DNA sequence [36, 37], was chosen for random mutagenesis in A. media strain WS. Nevertheless, since the antibiotic resistance cassettes in pUTKm are not suitable for the targeting strain, its ampicillin cassette was deleted and the kanamycin cassette was replaced by a chloramphenicol cassette. The resulting transposon delivery vector was named as pTnCm, its molecular manipulation flowchart was shown in S1 Fig.
The plasmid pTnCm was first introduced into E.coli S17-1 (λpir) [38] by artificial transformation with Ca2+ [39]. The E. coli S17-1 (pTnCm) and A. media strain WS were cultivated in 5 ml LB-ampicillin medium and LB-chloramphenicol medium overnight, respectively. Conjugation was performed between E. coli S17-1 (pTnCm) as the donor strain and A. media strain WS as the recipient strain. The donor and recipient strain cultures were mixed in a 2 mL volume in the ratio 1:1, washed twice with 1 ml of 10% glycerol, resuspended in 50 μl of 10% glycerol, and then transferred onto 0.45-μm-pore-size membranes overlaid on LB plates. After being incubated at 30°C for approximate 12 h, the membranes were washed with 1 ml LB and the collected culture mixture was serial diluted and spread on LB agar supplemented with ampicillin and chloramphenicol to select the transposon mutants. In order to identify the possible genes involved in melanogenesis, separated transposon mutants were picked, inoculated into 96-well plates and cultured at 30°C for 72 h. Every well in the 96-well plates contained 200 μl LB-ampicillin-chloramphenicol agar medium. Then, the mutants exhibiting attenuated melanin production compared to the parent strain A. media strain WS were selected and inoculated into LB to detect the general growth (see below). The strains with no detectable growth defect relative to the level of wild-type A. media strain WS were considered as potential candidate for further study. All isolated mutants were confirmed to be A. media strain WS by sequencing the 16S rRNA and other conserved genes such as motB (encoding the flagellar motor protein), the gene encoding protease YbbK or the gene encoding acetyltransferase.
Measurement of the bacterial growth ability and the melanin production
The growth capacity and the pigment production of the wild-type A. media strain WS and its mutants were all evaluated according to optical density (OD). To monitor the growth capacity of the strains, bacteria were inoculated to 25 ml of LB, incubated with shaking at 30°C at 200 rpm, and then at various periods of post-inoculation, the optical density of the cultures was determined at 600 nm (OD600) [40]. To quantify melanin production of the strains, the cultures from different cultivation periods were filtered through a Millipore filter (pore size, 0.45 μm) and the filter-sterilized culture supernatants were tested for their absorbance at 400 nm (OD400) [41].
Identification of the Tn5 transposon insertion sites
Tn5 transposon insertion sites were identified by each of the following two methods: 1) Transposon rescue by taking advantage of the chloramphenicol acetyltransferase encoding gene within the transposon’s inverted repeats [42]. The chromosomal DNA from the strain WS Tn5 mutants was extracted and digested with BamHI, PstI or XbaI, then ligated into the vector pUC18 (Fermentas). The ligation mixtures were transformed to E. coli DH5α and spread on LB plates containing ampicillin and chloramphenicol. The obtained transformants may harbor the recombinant plasmid containing the transposon with flanking insertions from strain WS. The cloned DNA fragments were determined by automated DNA sequencing (Invitrogen). 2) Thermal asymmetric interlaced PCR (TAIL-PCR) [43] by three sequential amplification steps with primers complementary to mini-Tn5 ends and arbitrary degenerate primers (S1 Table) to clone the potential DNA flanking to the transposon. PCR products were purified by Gel Extraction Kit (OMEGA), cloned into pMD19-T vector (TAKARA) and sequenced. The interrupted sequences were analyzed by comparison of the whole genome of A. media strain WS (gi: 615550237).
Identification of the melanin intermediates in culture supernatants by high-performance liquid chromatography (HPLC) analysis
Strains were grown in LB media at 30°C or 22°C, taken at different times, and were centrifuged (16, 000 ×g, 10 min) to remove bacterial cells. Supernatants (1 ml) were mixed with 100 μl of glacial acetic acid, clarified by centrifugation, and then stored at −20°C until assayed. The frozen samples were thawed, diluted threefold with 10 mM acetic acid, and then filtered through a Millipore filter (pore size, 0.45 μm).
Twenty microliters of filtrate was loaded directly onto an Agilent Eclipse Plus C18 reverse phase column (5 μm particle size; 4.6 by 250 mm) in an Agilent 1100 liquid chromatograph equipped with a photodiode array detector (G1315B). The mobile phase was 10 mM acetic acid-methanol (90:10 [vol/vol]). The flow rate was 1 ml min-1, and the eluate was monitored at 260 nm and 290 nm, as previously described [8, 32, 44]. The absorption maxima of L-DOPA and HGA are 260 nm and 290 nm, respectively. Additionally, the chromatograms of standard solutions of L-DOPA and HGA (both from Sigma) were used as references to identify the corresponding HPLC peaks.
Identification of the melanin intermediate products by mass spectrometry (MS)
The eluting peaks of cell-free culture filtrates corresponding to standard L-DOPA and HGA peaks by HPLC were collected and further analyzed by mass spectrometry (MS). MS analysis was performed using a Thermo-Finnigan LCQ advantage ion trap mass spectrometer (San Jose, CA, USA) using an ESI interface in negative-ion mode.
Construction of the targeted gene deletion mutants
The homologous recombination method was employed to knock out the genes which may affect pigment production, namely phhA, tyrB, aspC, hppD and hmgA predicted to encode phenylalanine hydroxylase, aromatic amino acid aminotransferase, aromatic amino acid aminotransferase, 4-hydroxyphenylpyruvate dioxygenase and homogentisate dioxygenase, respectively. The primers used for constructing the mutants were listed in S1 Table.
To get the WSΔphhA mutant, the fragments located upstream and downstream of phhA gene were amplified from chromosomal DNA of strain WS with the primers phhA(S)-S/phhA(S)-A and phhA(X)-S/phhA(X)-A, respectively. The fragments obtained were 242 bp (primers phhA(S)-S/phhA(S)-A) and 255 bp (phhA(X)-S/phhA(X)-A) (S1 Table), and were then subcloned into the suicide vector pDM4 [45]. The recombinant plasmid pDM-phhA was transformed into E. coli S17-1 (λpir) and the transformants were selected for chloramphenicol resistance (Cmr). Subsequently, parental mating was used to transfer the recombinant plasmid pDM-phhA into A. media strain WS (Ampr) strains. The transconjugants with the first allelic exchange were selected on LB agar plates with ampicillin and chloramphenicol. Colonies were transferred to LB agar plates for 24 h growth, and then transferred to LB agar plates containing 15% sucrose. The phhA deletion mutant WSΔphhA was then screened and identified by PCR.
With the same approach, individual gene (tyrB, aspC, hppD, hmgA1, hmgA2) deletion mutants and a double gene (tyrB, aspC) deletion mutant were also created.
Complementation of the targeted gene deletion mutants
To complement the function of the deleted genes in the mutants (WSΔphhA, WSΔtyrB, WSΔtyrBΔaspC, WSΔhppD), the DNA fragments containing the Shine–Dalgarno (SD) sequences and open reading frames (ORFs) of the target genes were amplified by PCR from chromosomal DNA of A. media strain WS. The primers were shown in S1 Table. PCR products were ligated to pBBR1MCS-5 [46]. The resulting complementary plasmids pBBR1MCS-5-phhA, pBBR1MCS-5-tyrB, pBBR1MCS-5-aspC, pBBR1MCS-5-hppD and pBBR1MCS-5-phh(A+B) which contained the Shine–Dalgarno (SD) sequence, complete phhA gene and phhB gene (encoding 4a-carbinolamine dehydratase, supplying cofactor for PhhA), were introduced into E. coli S17-1, then transferred to the deletion mutants by conjugation, respectively.
In order to complement A. media strain WS, the HmgA coding sequences and Shine–Dalgarno (SD) sequences were amplified from A. salmonicida _AB98041, A. salmonicida KACC14791 and A. hydrophila_XS91-4-1, then subcloned into pBBR1MCS-5, respectively [46]. The complementary plasmids pBBR1MCS-5-hmgA-AS, pBBR1MCS-5-hmgA-KACC and pBBR1MCS-5-hmgA-AH were transformed to E. coli S17-1, then transferred to A. media strain WS by conjugation, respectively.
Cloning, sequence analysis of the hppD genes from A. media strain WS, A. salmonicida_AB98041, A. salmonicida KACC14791, A. hydrophila_XS91-4-1 and heterologous expression of HppD in E. coli BL21 (DE3)
The hppD gene from A. media strain WS was PCR amplified from chromosomal DNA using the primers hppD(WS)-S/hppD(WS)-A (S1 Table). The PCR product was digested with NdeI and HindIII and ligated to NdeI/HindIII-digested pET26b(+) (Novagen). Then the ligations were transformed into E. coli BL21. The transformants were confirmed by PCR. The correct colonies containing the expression plasmid pET26b(+)-hppD-WS were inoculated to LB with kanamycin, grown at 37°C overnight, then transferred to fresh medium. When the optical density at 600 nm reached 0.5 to 0.8, 1 mM IPTG (isopropyl—D-thiogalactopyranoside) was added to induce the expression of hppD [44].
Based on the GenBank sequences (gi: 142852228, gi: 507222057, gi: 117562568, gi: 569548302), the primers AS(hppD)-S / AS(hppD)-A, AS(hppD)-S / AS(hppD)-A, AH(hppD)-S /A (S1 Table) were designed to clone the hppD genes from A. salmonicida_AB98041, A. salmonicida KACC14791 and A. hydrophila_XS91-4-1, respectively. The resulting fragments were cloned into pMD19-T vector (TAKARA) and sequenced. It is noted that the sequences of the hppD genes from A. salmonicida_AB98041 and A. salmonicida KACC14791 were completely identical. The PCR products were cloned to pET26b(+) to construct expression plasmids pET26b(+)-hppD-AS and pET26b(+)-hppD-AH, transformed to E. coli BL21 and then sequenced. The following procedures were the same as above.
RT-PCR analysis of the genes (phhA, phhB, tyrB, aspC, hppD) transcription in Aeromonas strains
The transcription of the genes (phhA, phhB, tyrB, aspC, hppD) involved in pyomelanin synthesis was detected with reverse transcription (RT)-PCR. Total RNA was extracted from A. salmonicida_AB98041, A. salmonicida KACC14791, A. hydrophila_XS91-4-1 and A. media strain WS, respectively, at different times using Trizol reagent (Invitrogen). The isolated RNA was reverse transcribed, utilizing PrimeScript RT reagent Kit with gDNA Eraser (TAKARA). Control experiments in which reverse transcriptase was omitted from the reaction were performed to confirm the absence of contaminating DNA in the RNA samples. The resulting cDNAs were used as PCR templates. Gene specific primers, phhA(RT)-S/phhA(RT)-A, phhB(RT)-S/phhB(RT)-A, tyrB(RT)-S/tyrB(RT)-A, aspC(RT)-S/aspC(RT)-A (S1 Table) were used to detect the transcription of phhA, phhB, tyrB, aspC in A. salmonicida_AB98041, A. salmonicida KACC14791, A. hydrophila_XS91-4-1 and A. media strain WS, respectively. Moreover, the primers hppD(AS)-S/hppD(AS)-A, hppD(AS)-S/hppD(AS)-A, hppD(AH)-S/hppD(AH)-A and hppD(WS)-S/hppD(WS)-A (S1 Table) were used to analyse hppD transcription in A. salmonicida_AB98041, A. salmonicidaKACC14791, A. hydrophila_XS91-4-1 and A.media WS, respectively. Transcription of 16S rRNA was detected using primers 16sRNA(S)/16sRNA(A) as a positive control. All the PCR products were detected by gel electrophoresis.
Results
Screening and identification of the genes involved in melanogenesis in A. media strain WS by transposon mutagenesis
Bacteria usually produce melanin via either HGA or L-DOPA as intermediate [8]. Consistent with the literature [27, 28, 32], we detected L-DOPA in the culture of A. media strain WS in our previous work [35]. However, subsequent study found that deletion of tyrA, the gene encoding the enzyme converting tyrosine to L-DOPA, does not affect pigmentation of A. media strain WS [34], suggesting that A. media strain WS produces melanin through a non-DOPA pathway.
To identify the factors involved in pigmentation of A. media strain WS, we decided to isolate mutants that were defective in pigmentation by transposon mutagenesis. When grown in LB medium, A. media strain WS colony and culture turned black because of the production of melanin [35]. Thus, A. media strain WS mutants defective in pigmentation could be easily isolated by comparing the color of the colonies or cultures to that of the wild-type WS strain. 14 mutants showing significantly less or no pigmentation were obtained from about 20, 000 transposon insertion mutants (see materials and methods for a detailed description of the screen). As listed in Table 1, while most mutants exhibited reduced melanin production, ranging from 40% to 70% relative to the level of the wild-type strain WS, two of them, namely WS-M10 and WS-M13, lost their pigmentation ability completely (S2 Fig.). All the mutated strains grew as well as wild-type strain WS except for strains WS-M12 and WS-M14, indicating that their attenuated melanin production may be caused by the growth defects (S2 Fig.). Thus, WS-M12 and WS-M14 were not kept for further analysis.
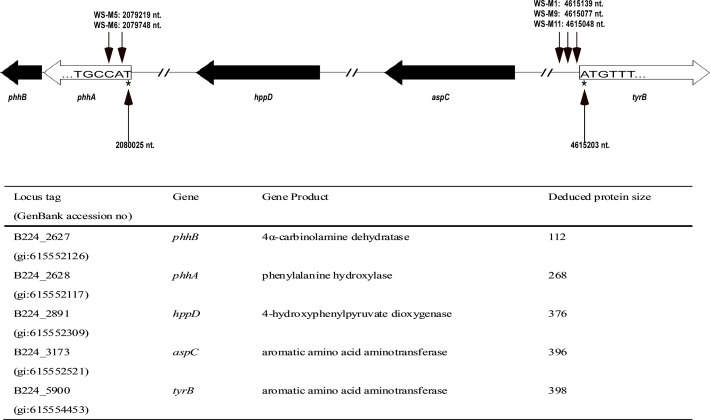
As shown in Table 2, the transposon insertion sites in 9 of the 12 WS mutated strains have been identified by transposon rescue or thermal asymmetric interlaced PCR (TAIL-PCR). However, transposon-flanking sequences for 3 mutants were not obtained, including the two non-pigmented mutants, WS-M10 and WS-M13. The most frequent insertion site of the miniTn5 transposon (hit three times independently) was upstream of the start codon of tyrB, in mutants WS-M1, WS-M9, WS-M11 (Fig. 1). tyrB was predicted to encode an aromatic amino acid aminotransferase which shows 53% amino acid sequence identity to its homolog PhhC from Pseudomonas aeruginosa PAO1. In Pseudomonas aeruginosa PAO1, PhhC has been shown to transform tyrosine to 4-hydroxyphenylpyruvate, which is an intermediate in the HGA based pyomelanin biosynthesis pathway [47]. Besides tyrB, gene phhA was also hit by the miniTn5 transposon twice independently, in WS-M5 and WS-M6 (Fig. 1). phhA was predicted to encode a Phenylalanine 4-monooxygenase which is 50% and 62% identical to the phenylalanine hydroxylase from Legionella pneumophila 130b and Pseudomonas aeruginosa PAO1, respectively [48, 49]. Phenylalanine hydroxylase has been reported to catalyze the transformation of phenylalanine to tyrosine and promote pyomelanin production by supplying additional tyrosine [49]. The phenotypes of the transposon insertion mutants of tyrB and phhA and the functions of their homologs in other bacteria suggest that A. media strain WS may synthesize pyomelanin through the HGA pathway with PhhA providing tyrosine and TyrB providing 4-hydroxyphenylpyruvate for HGA synthesis.
Table 2. Melanin-deficient A. media strain WS miniTn5 transposon mutants identified in this study a .
| Locus of insertion | No. of multiple hits | Function or product name | Phenotype | Mutants |
|---|---|---|---|---|
| B224_5900 (gi: 615554453) | 3 | Aromatic amino acid aminotransferase | Reduced | WS-M1/WS-M9/WS-M11 |
| B224_2628 (gi: 615552127) | 2 | Phenylalanine 4-monooxygenase | Reduced | WS-M5/WS-M6 |
| B224_0715 (gi: 615550742) | 1 | RNA methyltransferase | Reduced | WS-M3 |
| B224_1944 (gi: 615551635) | 1 | type III restriction enzyme, res subunit | Reduced | WS-M7 |
| B224_0883 (gi: 615550862) | 1 | Hypothetical protein | Reduced | WS-M8 |
| B224_5895 (gi: 615554450) | 1 | Hypothetical protein | Reduced | WS-M2 |
a Three mutants (WS-M4, WS-M10, WS-M13) were unable to generate the sequences of transposon insertion sites.
Fig 1. Locations of the genes in pyomealnin-synthesis pathway.
The transposon insertion sites of five mutants in the genome of A. media strain WS are marked with small black arrows. The numbers after the mutant names stand for the nucleotide (nt) site just following the transposon in the genome of A. media strain WS. Locus tags, putative protein function, and the number of amino acids of the deduced proteins are listed.
In the other 4 pigmentation mutants WS-M2, WS-M3, WS-M7 and WS-M8, the miniTn5 was found to insert into gene B224_5895 encoding a hypothetical protein, gene B224_0715 encoding a RNA methyltransferase, gene B224_1944 encoding a type III restriction enzyme and B224_0883 encoding a hypothetical protein, respectively (Table 2). However, these mutants were not included in this study because of the presence of tyrB and phhA and identification of HGA in the culture supernatant of A. media strain WS (see below).
Identification of the intermediate products involved in melanin synthesis in culture supernatants of A. media strain WS by high-performance liquid chromatography (HPLC)
Although the transposon insertion sites of the two non-pigmented mutants WS-M10 and WS-M13 could not be determined, the two mutants are great tools to determine which type of melanin is produced by A. media strain WS. As the presence of tyrB and phhA strongly suggested that A. media strain WS produces the HGA based melanin, pyomelanin, we expected to detect the presence of HGA in the culture supernatant of wild-type A. media strain WS but not in those of the two mutant strains WS-M10 and WS-M13. Methods developed to detect HGA from cultures of Aeromonas in previous studies have been unsuccessful [32, 35], so we followed a HPLC protocol developed by Keith KE, with slight modifications, which has successfully identified HGA in the culture of Burkholderia cencepacia [8]. We also analyzed cultures at different stages of pigmentation to make sure we would not miss HGA if it was only transiently available in the culture.
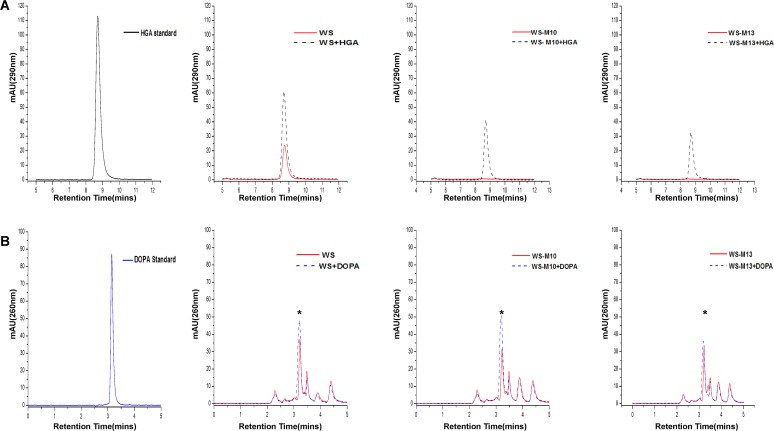
Wild-type A. media strain WS and the two mutant strains were cultured in LB liquid media with shaking at 30°C and culture samples were taken for HPLC analysis every 12 h until 72 h when melanin production reached its maximum in wild-type strain WS. Commercially available HGA was used as standard, which showed a single peak at 8.707 min (Fig. 2A). As shown in Fig. 2A and S3 Fig., a peak corresponding to HGA was identified in the 12 h, 24 h and 36 h culture supernatants of the wild-type A. media strain WS but not in those of the two mutant strains WS-M10 and WS-M13. However, it should be noted that although the peak corresponding to HGA appeared in the 12 h and 36 h culture supernatants, the values of the peak were substantially lower than that of the 24 h culture supernatant (S3 Fig.), suggesting that these two time points are either too early or too late to detect large amounts of HGA from the culture. We did not detect any HGA from the culture supernatants after the cells were grown for more than 36 h, suggesting that HGA is quickly oxidized and polymerized into pyomelanin once it is synthesized and secreted to the medium. Although these results are a little surprising given that pigment production continue to increase until 72 h, it is reasonable considering the time course of pigmentation. These results may also explain the failure to detect HGA from culture of A. media strain WS in previous studies because the time point of sampling is extremely critical for successful detection of HGA [32, 35]. Addition of HGA standard to the culture supernatant of the wild- type strain WS resulted in a peak slightly higher than the culture sample without addition of HGA standard, while addition of HGA standard to the samples of the two mutant strains WS-M10 and WS-M13 restored the peak corresponding to HGA (Fig. 2A), further confirming the correlation of pigmentation with the presence of HGA. Taken together, these results suggest that HGA is synthesized by A. media strain WS and the two non-pigmented mutants WS-M10 and WS-M13 are defective in HGA synthesis or secretion.
Fig 2. HPLC analysis.
HPLC analysis of the supernatants of wild-type A. media strain WS and non-pigmented mutants WS-M10 and WS-M13. Samples were taken after 24 h of growth at 30°C in LB. (A) Analysis of HGA. (B) Analysis of L-DOPA. HGA Standard: commercial HGA (Sigma); DOPA Standard: commercial L-DOPA (Sigma). The asterisk indicates the peak of L-DOPA.
As L-DOPA has been detected in the culture of A. media strain WS [35], we would like to know whether L-DOPA synthesis was disrupted in the non-pigmented strains WS-M10 and WS-M13. As showed in Fig. 2B, a peak corresponding to L-DOPA was detected not only in the culture supernatant of wild-type A. media strain WS, but also in those of the two mutants (Fig. 2B and data not shown), suggesting that L-DOPA-based melanogenesis pathway exists in strain WS and the L-DOPA pathway has not been disrupted in the two non-pigmented mutants. However, this result also suggests that L-DOPA based melanogenesis plays almost no role in the pigmentation of A. media strain WS.
To further confirm that HGA and L-DOPA were produced by A. media strain WS, eluted fractions corresponding to the L-DOPA and HGA peaks from the wild-type A. media strain WS supernatant were collected for MS analysis. The spectra obtained showed the presence of molecular ion peaks at 196 and 167, similar to those in the spectrum of authentic standards, L-DOPA and HGA, respectively (S4 Fig.). Thus, although both L-DOPA and HGA are synthesized in A. media strain WS, pigmentation is largely due to HGA based pyomelanin synthesis and the L-DOPA pathway plays a very minor, if any, role in pigmentation.
Identification of the genes involved in pyomelanin synthesis in the genome of A. media strain WS
Identification of HGA in the culture supernatant of A. media strain WS suggests that genes required for HGA synthesis must exist in the genome of A. media strain WS besides tyrB and phhA. Therefore, we searched the genome of A. media strain WS for genes whose homologs in other bacteria have been known to be involved in HGA synthesis. Analysis of the genome resulted in the identification of two other genes, aspC and hppD, which seem highly likely to be required for pyomelanin synthesis (Fig. 1). aspC is predicted to encode an aromatic amino acid aminotransferase that shares 50% similarity with its homolog from Pseudomonas aeruginosa PAO1 [47]. Aromatic amino acid aminotransferase has been reported to be involved in conversion of tyrosine to 4-hydroxyphenylpyruvate in Pseudomonas aeruginosa PAO1 [47]. hppD is assumed to encode a putative 4-hydroxyphenylpyruvate dioxygenase (HppD) whose amino acid sequence is 67%, 62%, 46% and 37% identical to HppD from Pseudomonas aeruginosa PAO1, Burkholderia cenocepacia C5424, Shewanella colwelliana D and Streptomyces avermitilis ATCC317272, respectively. The HppDs from these bacteria have been shown to catalyze the reaction from 4-hydroxyphenylpyruvate to HGA [8, 44, 50,51]. Identification of aspC and hppD confirmed our hypothesis that A. media strain WS contains all the genes required for HGA synthesis. Based on these findings, we hypothesized that PhhA converts phenylalanine to tyrosine, TyrB and AspC transform tyrosine into 4-hydroxyphenylpyruvate and HppD catalyzes the last reaction from 4-hydroxyphenylpyruvate to HGA, which in turn is further oxidized and polymerized to form pyomelanin. To confirm our hypothesis, we characterized these genes one by one in the following sections of this study.
PhhA promotes pyomelanin production by providing additional tyrosine in A. media strain WS
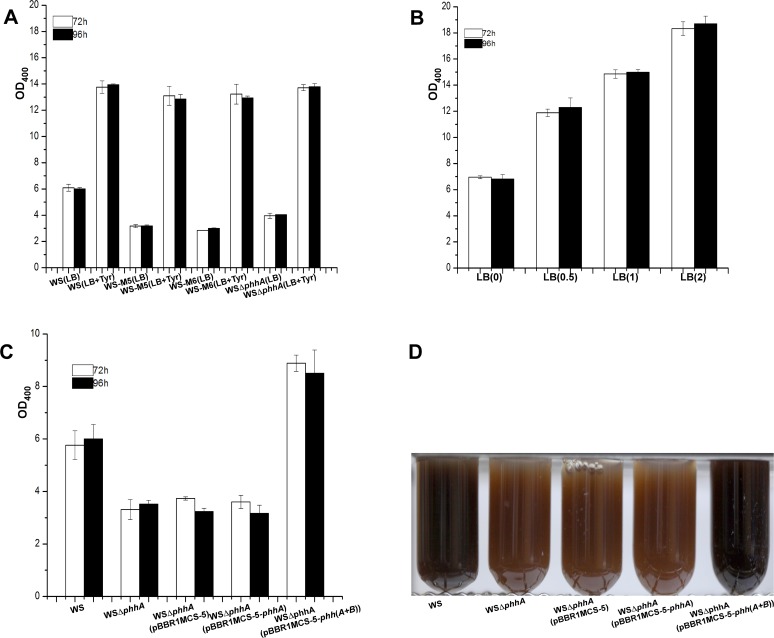
In our model, PhhA promotes pyomelanin synthesis by converting phenylalanine to tyrosine. If that is true, then addition of tyrosine to the medium of the phhA mutant strains should restore melanin production to the wild-type level. To this end, we compared melanin production of wild-type A. media strain WS and the phhA mutants (WS-M5, WS-M6, WSΔphhA) with or without tyrosine in the medium. As showed in Fig. 3A, all the phhA mutants exhibited a dramatic reduction in pigmentation when cultured without tyrosine, with strains WS-M5, WS-M6 and WSΔphhA producing only 52%, 50% and 64% of that of wild-type strain WS. However, when tyrosine was present in the medium, all the phhA mutant strains (WS-M5, WS-M6, WSΔphhA) produced as much melanin as wild-type strain WS (Fig. 3A). It is noteworthy that all the strains produced significantly more melanin in the presence of tyrosine, which is not surprising because we had showed previously that addition of tyrosine to the medium could increase melanin production by wild-type A. media strain WS and this phenomenon was confirmed again in this work [35] (Fig. 3B). These results suggest that disruption of the phhA gene results in a reduction of tyrosine which in turn leads to a reduction of melanin production.
Fig 3. The function of phhA in pigmentation in A. media strain WS.
(A) Wild-type A. media strain WS, phhA mutants WS-M5, WS-M6 and WSΔphhA were inoculated into LB or LB with 1mg ml-1 tyrosine, and then at 72 and 96 h post-inoculation, the OD400 of the cultures were determined. (B) Wild-type A. media strain WS was inoculated into LB by addition of different amounts of tyrosine (0, 0.5, 1, 2 mg ml-1), and then at 72 and 96 h post-inoculation, the OD400 of the cultures were determined. (C) A. media strain WS, WSΔphhA (pBBR1MCS-5), WSΔphhA (pBBR1MCS-5-phhA) and WSΔphhA (pBBR1MCS-5-phh(A+B)) were cultured in LB, and then at 72 h and 96 h post-inoculation, the OD400 of the cultures were determined. (D) Photographs of 72 h LB cultures of A. media strain WS, WSΔphhA, WSΔphhA (pBBR1MCS-5), WSΔphhA (pBBR1MCS-5-phhA) and WSΔphhA (pBBR1MCS-5-phh(A+B)). The data presented are the means and standard deviations from triplicate cultures.
To further confirm the role of phhA, we tried to complement the phhA deletion strain with a plasmid carrying phhA, pBBR1MCS-5-phhA. To our surprise, pigmentation of the phhA deletion mutant WSΔphhA was not restored by the introduction of pBBR1MCS-5-phhA (Fig. 3C). As phhA forms an operon with the downstream gene phhB (predicted to encode 4a-carbinolaine dehydratase that could synthesize a cofactor tetrahydrobiopterin (BH4) for PhhA [52]), we speculated that the inability of pBBR1MCS-5-phhA to complement WSΔphhA was due to a polar effect of the deletion on phhB (Fig. 1). Therefore, we cloned a fragment containing both phhA and phhB, and ligated it into pBBR1MCS-5. As shown in Fig. 3C and 3D, pigmentation was restored when plasmid pBBR1MCS-5-phh(A+B) was introduced into the phhA deletion strain, indicating that both phhA and phhB are important for optimal pyomelanin production in A. media strain WS.
Both TyrB and AspC could transform tyrosine to 4-hydroxyphenylpyruvate and contribute to pyomelanin production in A. media strain WS
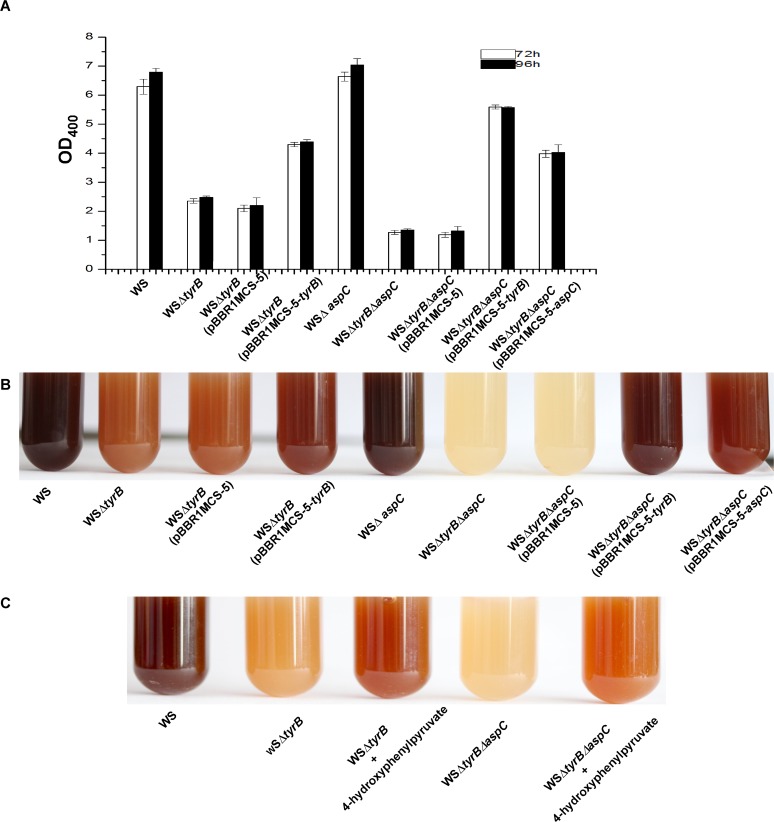
Both tyrB and aspC in strain WS are predicted to encode aromatic amino acid aminotransferase which is believed to be able to transform tyrosine into 4-hydroxyphenylpyruvate and contribute to pyomelanin production [47]. To confirm that both of these two genes are involved in pyomelanin biosynthesis in A. media strain WS, we constructed single and double deletion strains of tyrB and aspC (WSΔtyrB, WSΔaspC, WSΔtyrBΔaspC) and the effect of each deletion on pigmentation was compared. Consistent with the phenotypes of the transposon insertion mutants of tyrB, deletion of tyrB resulted in the reduction of melanin production to 37% of that of wild-type strain WS (Fig. 4A and 4B, S2 Fig.). However, deletion of aspC had no discernable effect on melanization (Fig. 4A and 4B). The tyrB and aspC double mutant WSΔtyrBΔaspC showed no sign of pigmentation after being cultured in LB for 72 h (Fig. 4B), suggesting that aspC is responsible for pigmentation in the absence of tyrB. Thus, tyrB and aspC play redundant roles in pigmentation but tyrB is more important than aspC.
Fig 4. The function of tyrB and aspC in pigmentation in A. media strain WS.
(A) Wild-type A. media strain WS, WSΔtyrB, WSΔaspC, WSΔtyrBΔaspC, WSΔtyrB (pBBR1MCS-5), WSΔtyrB (pBBR1MCS-5-tyrB), WSΔtyrBΔaspC (pBBR1MCS-5), WSΔtyrBΔaspC (pBBR1MCS-5-tyrB), WSΔtyrBΔaspC (pBBR1MCS-5-aspC) were cultured in LB, and then at 72 h and 96h post-inoculation, the OD400 of the cultures were determined. (B) Photographs of cultures from 72 h LB cultures of the strains. (C) Pigment production can be restored to WSΔtyrB and WSΔtyrBΔaspC by the addition of 5 mM 4-hydroxyphenylpyruvate to the LB.
The function of the two genes in pigmentation was further confirmed by the complementation experiments. As showed in Fig. 4A and 4B, while pigmentation of WSΔtyrB could be restored by supplying the tyrB gene in trans on the plasmid pBBR1MCS-5, pigmentation of the double mutant WSΔtyrBΔaspC could only be partly recovered by a plasmid containing tyrB (pBBR1MCS-5-tyrB) or aspC (pBBR1MCS-5-aspC) (Fig. 4A and 4B). Consistent with the above result that tyrB is more important for pigment production, complementation of the double mutant WSΔtyrBΔaspC with tyrB always showed a better recovery of pigmentation compared to the complementation with aspC (Fig. 4A and 4B). Addition of 4-hydroxyphenylpyruvate to the cultures of WSΔtyrB and WSΔtyrBΔaspC strains also partly restored pigmentation, suggesting that the pigmentation defect of the WSΔtyrB mutant and the double mutant WSΔtyrBΔaspC is due to a reduction or the absence of 4-hydroxyphenylpyruvate (Fig. 4C). Taken together, these results confirm that TyrB and AspC converts tyrosine to 4-hydroxyphenylpyruvate for pyomelanin biosynthesis in A. media strain WS.
HppD could convert 4-hydroxyphenylpyruvate to HGA and plays the key role in pyomelanin biosynthesis in A. media strain WS
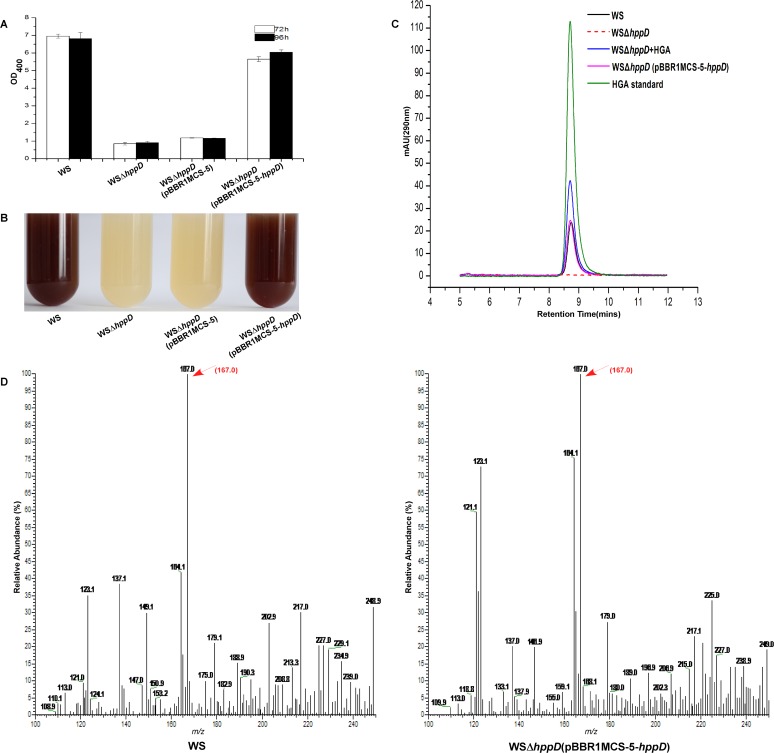
hppD encodes a putative 4-hydroxyphenylpyruvate dioxygenase (HppD) that could transform 4-hydroxyphenylpyruvate to HGA, which then could form pyomelanin by self-oxidization and polymerization [8]. To determine the function of HppD in A. media strain WS, an hppD deletion strain (WSΔhppD) was constructed. As shown in Fig. 5A and 5B, while the wild-type A. media strain WS produced a significant amount of pigment when grew in LB for 72 h, WSΔhppD lost the ability of pigmentation completely, suggesting that hppD plays a crucial role in pigmentation in A. media strain WS. As expected, pigmentation of WSΔhppD was restored when a plasmid carrying hppD was introduced into the strain (Fig. 5A and 5B).
Fig 5. The function of hppD in pigmentation in A. media strain WS.
(A) Wild-type A. media strain WS, WSΔhppD, WSΔhppD (pBBR1MCS-5) and WSΔhppD (pBBR1MCS-5-hppD) were cultured in LB, and then at 72 h and 96 h post-inoculation, the OD400 of the cultures were determined. (B) Photographs of 72 h LB cultures of the strains. (C) HPLC chromatograms of culture supernatants of wild-type A. media strain WS, WSΔhppD and WSΔhppD (pBBR1MCS-5-hppD). Samples were taken after 24 h of growth at 30°C in LB. (D) MS analysis of the sample from wild-type A. media strain WS and WSΔhppD (pBBR1MCS-5-hppD), the arrow indicates the molecular ion peaks at 167, the same to that of HGA.
As HppD is predicted to catalyze the reaction from 4-hydroxyphenylpyruvate to HGA, we should not be able to detect HGA in the culture supernatant when hppD is deleted from the chromosome. To test this, we employed HPLC to detect HGA in the 24 h culture supernatant of wild-type A. media strain WS, the deletion mutant WSΔhppD and the complementary strain WSΔhppD (pBBR1MCS-5-hppD). As shown in Fig. 5C, a peak matching the HGA standard could be detected in the culture supernatant of wild-type strain WS but not in the culture supernatant of the WSΔhppD. Introduction of the plasmid pBBR1MCS-5-hppD into WSΔhppD restored the peak corresponding to HGA in the culture supernatant (Fig. 5C). MS analysis of the elution fractions matching with the HGA peaks from wild-type A. media strain WS and WSΔhppD (pBBR1MCS-5-hppD) showed the presence of molecular ion peaks at 167, the same to the spectrum of that of standard HGA (Sigma) (Fig. 5D). Therefore, these results strongly argue that HppD convert 4-hydroxyphenylpyruvate to HGA and plays a critical role in pigmentation in A. media strain WS.
The HGA synthesis pathway is widely distributed in Aeromonas
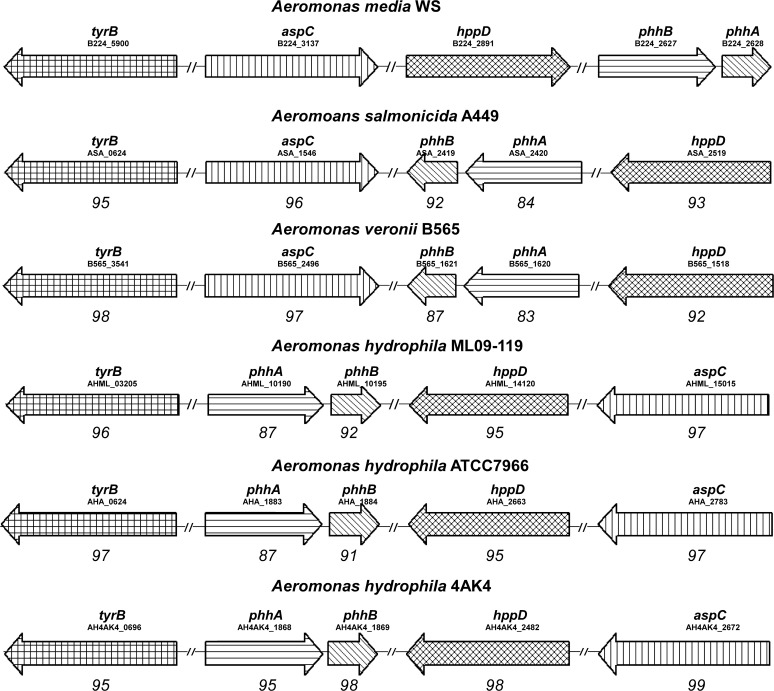
Pigmented Aeromonas species have been thought to synthesize melanin through the L-DOPA pathway because L-DOPA was detected in the cultures of these species [27, 28, 32]. However, our work in A. media strain WS showed that although L-DOPA is present in the culture, it plays almost no role in pigmentation. Instead, pigmentation of A. media strain WS is due to the production of pyomelanin through HGA, although HGA is only transiently detected. This suggests that the other pigmented Aeromonas species may also synthesize HGA based melanin rather than the predicted L-DOPA based melanin. Thus, we analyzed the genomes of many Aeromonas species for the presence of the genes involved in HGA synthesis in A. media strain WS. Up to present, whole genome sequencing has been finished in several Aeromonas species, such as A. salmonicida A449 (gi: 142849896), A. veronii B565 (gi: 328802836), A. hydrophila ML09-119 (gi: 507219248), A. hydrophila ATCC7966 (gi: 117558854) and A. hydrophila 4AK4 (gi: 569545899). As showed in Fig. 6, homologs of the genes responsible for HGA synthesis in A. media strain WS exist in all the examined Aeromonas species, including both pigmented and non-pigmented. This is surprising because A. hydrophila ML09-119, A. hydrophila ATCC7966 and A. hydrophila 4AK4 have been considered to be non-pigmented Aeromonas species. However, it is possible that A. hydrophila ML09-119, A. hydrophila ATCC7966 and A. hydrophila 4AK4 have been misclassified because we have not found the right conditions for them to produce pigment. Similar to A. media strain WS, these genes are scattered in the genomes of Aeromonas species with phhA always forming an operon with phhB (Fig. 6). In addition, alignments of each gene family showed that they display a high degree of identity, for example, hppD from A. salmonicida A449, A. veronii B565, A. hydrophila ML09-119, A. hydrophila ATCC7966 and A. hydrophila 4AK4 was 93%, 92%, 95%, 95%, 98% identical to hppD from A. media strain WS at the amino acid level, respectively. Based on these findings, we argued that the HGA biosynthesis pathway identified from A. media strain WS is conserved in the genus of Aeromonas and pigmentation of most pigmented Aeromonas species is likely due to the production of pyomelanin through HGA.
Fig 6. Gene organization of those genes encoding the pyomelanin synthesis pathway in Aeromonas strains.
Genes are represented by arrows as follows: phhA, gene encoding the phenylalanine hydroxylase; phhB, gene encoding the 4a-carbinolamine dehydratase; tyrB, gene encoding the aromatic amino acid aminotransferase; aspC, gene encoding the aromatic amino acid aminotransferase; hppD, gene encoding the 4-hydroxyphenylpyruvate dioxygenase. The numbers beneath the arrows indicate the levels of amino acid sequence identity (expressed as percentages) between the encoded gene products and the equivalent products from A. media strain WS.
Heterologous expression of hppD genes from different Aermonas species in E. coli BL21 results in pigmentation
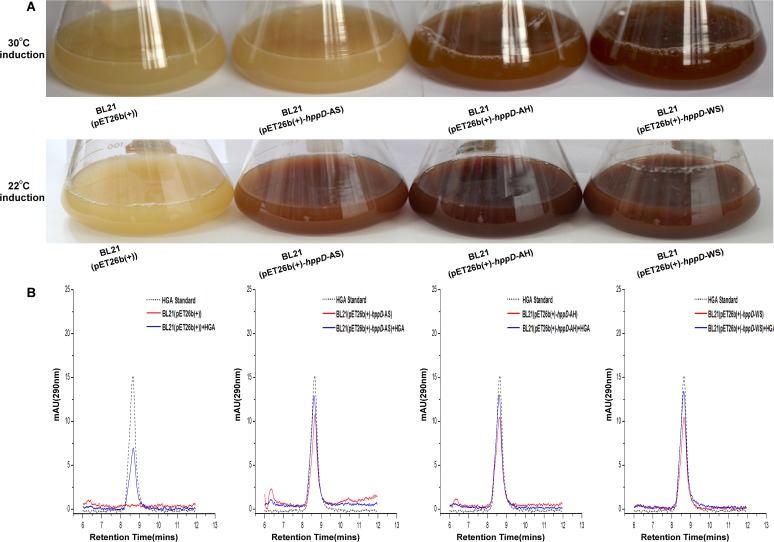
To test our hypothesis that pigmentation of Aeromonas species is dependent on pyomelanin synthesis through HGA, we examined whether HppD, the critical enzymes in HGA synthesis, from several Aeromonas species are functional. It has been showed that heterologous expression of 4-hydroxyphenylpyruvate dioxygenase (HppD) from other bacteria in E. coli could result in pigmentation of E. coli [8, 44]. We thus cloned the hppD genes from the melanin producing A. media strain WS, A. salmonicida_AB98041 and A. salmonicida KACC14791, and the non-pigmented strain A. hydrophila_XS91-4-1 into pET26b(+) and expressed them in E. coli BL21. As listed in S5 Fig., the hppD gene from A. hydrophila_XS91-4-1 was 99%, 99%, 94% identical to hppD from A. hydrophila ML09-119, A. hydrophila ATCC7966, A. hydrophila 4AK4 at the amino acid level. On the other hand, the hppD gene seems highly conserved in the species of A. salmonicida, since the DNA sequences of the gene from the two laboratory preserved strains (A. salmonicida_AB98041, A. salmonicida KACC14791) share 100% identity with that of A. salmonicida A449.
As shown in Fig. 7A, when induced with 1 mM IPTG at 30°C, BL21 (pET26b(+)-hppD-AH) and BL21 (pET26b(+)-hppD-WS) produced large amounts of pigment, indicating that both the hppD genes from the pigmented strain WS and the non-pigmented A. hydrophila are functional. On the contrary, almost no pigment formation could be observed in the culture of BL21 (pET26b(+)-hppD-AS) under the same condition (Fig. 7A), although the hppD gene contained in the recombinant plasmid was cloned from a pigmented strain. Since the strains of A. salmonicida usually produce melanin at 20–25°C [53, 54], we changed the induction temperature accordingly. As shown in Fig. 7A, all of the three recombinant E. coli bacteria turned black when they were cultivated at 22°C, suggesting that the enzyme activity of HppD from A. salmonicida is probably temperature sensitive. In agreement with these results, we were able to detect HGA from the cultures of E. coli BL21 expressing these HppDs but not in cultures of E. coli BL21 containing just the vector (Fig. 7B). The finding that expression of hppD from the non-pigmented A. hydrophila in E. coli resulted in pigmentation indicates that A. hydrophila contains functional HppD, but for unknown reasons it does not pigment under the laboratory conditions.
Fig 7. Expression of hppD genes from multiple Aeromonas sp. in E. coli BL21 results in pigmentation.
(A) Photographs of E. coli BL21 (pET26b(+)), E. coli BL21 (pET26b(+)-hppD-AS), E. coli BL21 (pET26b(+)-hppD-AH) and E. coli BL21 (pET26b(+)-hppD-WS), incubated at 30°C and 22°C, respectively. (B) HPLC analysis of the supernatants of BL21 (pET26b(+)), BL21 (pET26b(+)-hppD-AS), BL21 (pET26b(+)-hppD-AH), BL21 (pET26b(+)-hppD-WS), incubated at 22°C.
HGA but not L-DOPA production correlates with pigmentation in Aeromonas
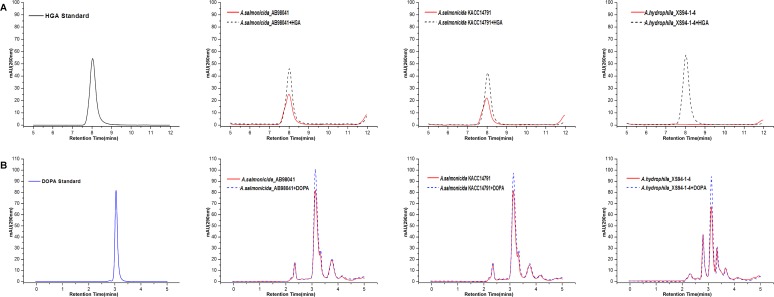
To further test our hypothesis that pigmentation of Aeromonas species is dependent on pyomelanin synthesis through HGA, we examined whether HGA production correlates with pigmentation in various Aeromonas species. We expected to detect HGA in the cultures of pigmented Aeromonas species but not in those of the non-pigmented ones. To this end, we cultured the pigmented A. salmonicida, and the non-pigmented strain A. hydrophila in LB at 22°C, 30°C, respectively, and tried to detect HGA in their culture supernatants by HPLC. As showed in Fig. 8A, HGA was detected in the 36 h culture supernatants of A. salmonicida_AB98041 and A. salmonicida KACC14791 but not in that of A. hydrophila (a pattern similar to A. media strain WS was found for the two pigmented A. salmonicida, but only the data for 36 h culture supernatant was showed for simplicity). As a control, we also tested the presence of L-DOPA in the culture supernatants of these three species. Unlike HGA, L-DOPA was detected in the culture supernatants of both A. salmonicida_AB98041 and A. salmonicida KACC14791 as well as in that of A. hydrophila (Fig. 8B). Therefore, there is a strong correlation between the production of HGA and pigmentation but no correlation between the production of L-DOPA and pigmentation in many Aeromonas species.
Fig 8. HPLC analysis.
Detection of the intermediates from cultures of A. salmonicida_AB98041, A. salmonicida KACC14791 and A. hydrophila_XS91-4-1. Samples were taken after 36 h of growth in LB. (A) Analysis of HGA. (B) Analysis of L-DOPA. HGA Standard: commercial HGA (Sigma); DOPA Standard: commercial L-DOPA (Sigma).
Identification of the transcription of the genes involved in pyomelanin formation in Aeromonas strains
The hppD genes cloned from pigment-producing (A. salmonicida_AB98041, A. salmonicida KACC14791, A. media strain WS) and non-pigment-producing Aeromonas strains (A. hydrophila_XS91-4-1) were expressed to mediate the melanin production in E.coli BL21. We want to investigate whether the differences of transcriptional level of the genes contributing to pyomelanin synthesis lead to the differences in melanin phenotype in the Aeromonas strains. RT-PCR analysis detected phhA, phhB, tyrB, aspC, hppD mRNA in A. salmonicida _AB98041, A. salmonicida KACC14791, A. hydrophila_XS94-1-4 and A. media strain WS, respectively, with expression at 12, 24, 48 and 72 h post-incubation (S6 Fig.). Taken together, these data indicated that there were no differences in the transcriptional level of phhA, phhB, tyrB, aspC, hppD in the pigment-producing and non-pigment-producing Aeromonas strains.
The function of hmgA in A. media strain WS
The gene hmgA encodes the homogentisate dioxygenase that converts HGA to maleylacetoacetate [23] (Fig. 9). By analysis of the hmgA sequences from different Aeromonas strains, we found that the hmgA is interrupted into two parts, hmgA1 and hmgA2 in A. media strain WS (S7A Fig.). To investigate whether the genes of hmgA1 and hmgA2 are functional, we constructed the deletion mutants WSΔhmgA1 and WSΔhmgA2. Pigmentation of A. media strain WS was not affected by the deletion of hmgA1 or hmgA2 (S7B Fig.). Moreover, we cloned the intact hmgA from other Aeromonas strains (A. salmonicida _AB98041, A. salmonicida KACC14791, A. hydrophila_XS91-4-1) and introduced them into A. media strain WS, respectively. By complementation with the hmgA gene, A. media strain WS exhibited reduced melanin production (S7C Fig.). Taken together, these data indicate that the hmgA was disrupted and lost the function in A. media strain WS.
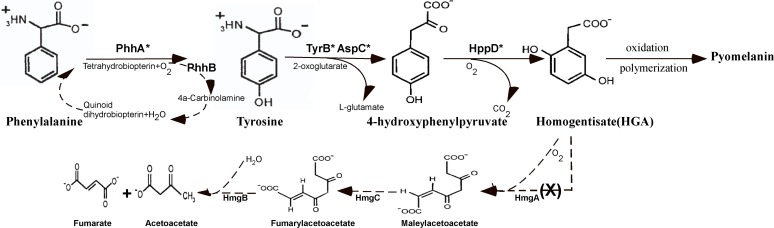
Fig 9. Pathway for pyomelanin synthesis and phenylalanine/tyrosine catabolism in A. media strain WS (modified from reference 49).
The intermediates of the pathway are indicated. PhhA, phenylalanine hydroxylase; PhhB, 4a-carbinolamine dehydratase; AspC, aromatic amino acid aminotransferase; TyrB, aromatic amino acid aminotransferase; HppD, 4-hydroxyphenylpyruvate dioxygenase; HmgA, homogentisate dioxygenase; HmgB, fumarylacetoacetate hydrolase; HmgC, maleylacetoacetate isomerase. The asterisk indicates the location of the related genes mutated in this study.
Discussion
In this study, we have determined that A. media strain WS produces pyomelanin through HGA rather than L-DOPA based melanin as previously thought. PhhA, TyrB and AspC, and HppD constitute a linear pathway of converting phenylalanine to HGA and this HGA biosynthesis pathway is widely conserved in the genus of Aeromonas, including both the pigmented and non-pigmented Aeromonas. Heterologous expression showed that hppD genes from both pigmented and non-pigmented Aeromonas species encode functional enzymes, suggesting that even the non-pigmented Aeromonas may contain necessary enzymes for pyomelanin synthesis. Moreover, we found that the presence of HGA in the culture correlates with pigmentation but there is no correlation between L-DOPA production and pigmentation. Based on these results, we propose that most of the pigmented Aeromonas, if not all of them, produce the HGA based pyomelanin. Aeromonas may also produce L-DOPA based melanin, but this kind of melanin does not seem to play a role in pigmentation. In addition, our work also suggests that there are many additional factors involved in pigmentation in A. media strain WS.
Pigmentation of Aeromonas is due to the synthesis of pyomelanin through HGA
The melanogenic Aeromonas species had been considered to synthesize DOPA based melanin because only DOPA but not HGA was detected in the culture extracts [27, 28, 32, 35]. However, using A. media strain WS as a model we found that pigmentation of many Aeromonas species is likely due to the production of pyomelanin through HGA. In support of this idea, we have found that: 1) HGA is detected in the culture of wild-type A. media strain WS in the early phase of melanogenesis but not in those of the non-pigmented mutants WS-M10 and WS-M13; 2) deletion of any one of the genes involved in HGA biosynthesis impairs or blocks pigmentation of A. media strain WS dependent on the gene deleted; 3) genes encoding for enzymes required for HGA biosynthesis are widely distributed in Aeromonas; 4) HGA is detected in the cultures of pigmented A. salmonicida strains but not in that of the non-pigmented A. hydrophila. Similar to previous reports, we were able to detect DOPA in the cultures of A. media strain WS and cultures of pigmented A. salmonicida strains [28, 32, 35]. However, there is no correlation between the presence of L-DOPA in the culture and pigmentation of Aeromonas because L-DOPA was identified at high levels in the cultures of the non-pigmented A. media strain WS mutants and the non-pigmented A. hydrophila. Therefore, these results suggest that pigmentation of many Aeromonas species is largely due to the production of pyomelanin through HGA rather than the production of L-DOPA based melanin.
One of the reasons that many pigmented Aeromonas species had been considered to synthesize melanin through L-DOPA is because HGA had never been detected in the cultures of Aeromonas species before this study [27, 28, 32]. We also failed to detect HGA from the culture of A. media strain WS in a previous study [35]. However, the identification of phhA and tyrB from the screen for mutants defective in pigmentation led us to believe that A. media strain WS produces HGA based melanin because homologs of phhA and tyrB have been found to be involved in HGA synthesis in Pseudomonas aeruginosa PAO1 and Legionella pneumophila 130b [47, 49]. Following a HPLC protocol developed by Keith KE to detect HGA in the culture of Burkholderia cencepacia [8], we successfully detected HGA in the culture of wild-type A. media strain WS but not in those of the non-pigmented mutants. Analysis of cultures at different stages of melanogenesis appears to be critical for our success in detecting HGA because HGA is only detectable in the culture of the wild-type strain WS in the early phase of pigmentation (12 to 36 h). It should be noted that although HGA is present in the culture of A. media strain WS when it is grown in LB for 12 h or 36 h, the level of HGA is very low compared to that at 24 h, a time point pigment starts to appear in the culture. This result may explain why HGA has never been identified in the cultures of Aeromonas species because only samples from late exponential-phase were taken for analysis in the previous studies [32, 35]. We also successfully identified HGA from the cultures of pigmented A. salmonicida but not in that of non-pigmented A. hydrophila. Thus, we believe that previous failures to identify HGA from the cultures of pigmented Aeromonas are due to the low presence of HGA in the cultures and the way the experiments were performed.
A widely conserved pyomelanin synthesis pathway in Aeromonas
With the characterization of phhA, tyrB, aspC and hppD, a pathway of HGA based pyomelanin production in A. media strain WS is established: 1) phenylalanine is transformed into tyrosine by phenylalanine 4-monooxygenase (PhhA) and 4a-carbinolamine dehydratase (PhhB), the latter provides the cofactor for the reaction; 2) tyrosine is then converted to 4-hydroxyphenylpyruvate by two aromatic amino acid aminotransferases (TyrB and AspC); 3) 4-hydroxyphenylpyruvate is transformed into HGA by 4-hydroxyphenylpyruvate dioxygenase (HppD); 4) the secreted HGA spontaneously polymerizes to pyomelanin (Fig. 9). In this pathway, PhhA is helpful but not necessary for melanin production because deletion of it does not eliminate pigmentation of A. media strain WS. TyrB and AspC play a redundant role in converting tyrosine into 4-hydroxyphenylpyruvateas as deletion of tyrB impairs pigmentation but deletion of both tyrB and aspC results in a complete loss of pigmentation. It is generally believed that there is only one aromatic amino acid aminotransferase in the pyomelanin synthesis pathway [8, 47, 49], but our work suggests that there could be more than one enzyme catalyzing the transformation of tyrosine to 4-hydroxyphenylpyruvate in pyomelanin producing bacteria. HppD appears to be the most critical enzyme in this pathway because its deletion results in the complete abrogation of pigmentation. This is not surprising as 4-hydroxyphenylpyruvate dioxygenase has been found to be essential for melanin production in Shewanella colwelliana and Pseudomonas sp. And it is ubiquitously present among pyomelanogenic microorganisms [51, 55, 56].
Genomic analysis indicates that the identified HGA synthesis pathway is widely present in Aeromonas, including both the pigmented and non-pigmented Aeromonas species. In agreement with this, heterologous expression of HppD from multiple Aeromonas species in E. coli results in pigmentation of E. coli, suggesting that HppDs from these bacteria are functional. Moreover, HGA is also detected in the cultures of several pigmented Aeromonas species. Thus, we propose that this HGA based pyomelanin synthesis pathway is widely conserved in Aeromonas.
Regulation of pigmentation in Aeromonas
In addition to mutants of phhA and tyrB, we have also isolated additional A. media strain WS mutants that are impaired or defective in pigmentation. Although we were unable to determine the transposon insertion sites of the two non-pigmented mutants, WS-M10 and WS-M13, our pilot analysis suggests that the pigmentation defect of these two mutants may be due to the absence of HppD because addition of a plasmid carrying hppD into these two mutants restored pigmentation (data not shown). PCR and sequencing analysis of the genomic region around hppD showed that the hppD gene and its regulatory sequences are intact (data not shown). However, the transcript of hppD is not detectable in these two mutants (data not shown), suggesting that a critical transcription factor required for hppD expression is likely disrupted by the transposon in these two mutants. Efforts to identify the transposon insertion sites of these two mutants are ongoing, and we believe that we will learn more about the transcription regulation of pigmentation in A. media strain WS once we identify the insertion sites.
Besides these two non-pigmented mutants, the other pigmentation mutants have the miniTn5 transposon inserted in genes encoding hypothetical proteins or proteins that have never been suggested to be involved in HGA synthesis, indicating that there are many factors contributing to the regulation of pigmentation in A. media strain WS. Considering the potential functions of melanin for microbes, one should not be surprised to see that many other factors might affect melanin production in addition to the enzymes involved in biosynthesis. A recent study in Pseudomonas aeruginosa identified 26 genes contributing to pyomelanogenesis, including genes encoding ABC transporter, transcriptional regulator and hypothetical proteins [57]. Some of the genes identified in P. aeruginosa overlaps with those identified in this study, such as phhA, tyrB and hppD, which are conserved enzymes required for HGA biosynthesis. However, a number of genes seem to be unique to each screen, possibly reflecting the different lifestyles of the two bacteria. It will be interesting to determine how the genes identified in our study affect melanin production in A. media strain WS in the future.
One surprising finding from our study is that even the non-pigmented A. hydrophila contains the HGA synthesis pathway. Heterologous expression of HppD from this bacterium in E. coli leads to pigmentation of E. coli, suggesting that the genes in the HGA synthesis pathway may encode functional enzymes. However, we were unable to detect HGA in the culture of A. hydrophila, suggesting that HGA is not synthesized or not secreted in this bacterium. One possible reason why A. hydrophila does not produce pigment could be that the genes in the HGA synthesis pathway are not transcribed. However, RT-PCR showed otherwise (S6 Fig.). Another possibility is that HGA is quickly broken down by HGA oxidase, HmgA, because pyomelanin production not only depends on the synthesis of HGA, but also relies on a reduced ability of the bacterium to degrade HGA. Inactivation of hmgA has been reported to be responsible for the hyperproduction of pyomelanin in many bacteria and eukaryotes [58, 59]. In fact, we found that the hyperproduction of melanin by A. media strain WS was partly due to the inactivation of hmgA. While the hmgA genes in most of the Aermonas species are intact, the hmgA gene in A. media strain WS is disrupted into two parts by a gene in opposite orientation (S7A Fig.). Deletion of the two disrupted hmgA parts, named hmgA1 and hmgA2, did not affect pigmentation, but introduction of the hmgA gene from A. salmonicida or A. hydrophila into A. media strain WS resulted in a 20% reduction of pigmentation (S7B and S7C Fig.). As A. hydrophila has functional hmgA, it is possible that HGA is produced but that it is quickly degraded by HmgA such that it does not pigment. However, this doesn’t seem to be the case, given that A. salmonicida still pigments even though it encodes functional HmgA. Moreover, HmgA activity is controlled by many factors, such as iron concentration [60, 61].
One interesting phenomenon we observed when the hppD gene from A. salmonicida is expressed in E. coli BL21 is that HppD only results in pigmentation of E. coli at 22°C but not at 30°C. Previous study has reported that the optimum growth temperature for A. salmonicida to produce melanin was 20–25°C and the bacterium lost the ability to produce pigment when it was cultured at supra-optimal temperature, i.e., 30–37°C [54]. Thus, it is possible that the enzymes required for HGA synthesis in A. hydrophila require a certain condition, for example, a specific temperature or pH, to be active. It is also possible that A. hydrophila is able to synthesize HGA, but HGA is not secreted into the medium due to a lack of appropriate transporter such that no pigmentation is observed. As A. hydrophila infects many organisms, including animals and humans, it may only synthesize melanin when it encounters the hosts given that melanin production has also been associated with virulence and pathogenicity in many pathogenic microbes [3, 4].
It is noteworthy to mention that the biological functions of melanin in many pigmented Aeromonas have never been studied, although pigmentation of Aeromonas has been known for a long time [31]. Melanin produced by many microorganisms has been suggested to protect the microbes from UV radiation, reactive oxygen species and host immune defense. Using the non-pigmented mutants of A. media strain WS constructed in this study, we should be able to determine whether pyomelanin has a role for Aeromonas under such stressed conditions. Lastly, previous studies from our lab have found that the melanin produced by A. media strain WS has many potential applications, such as photoprotection for pesticide [35], therefore, identification of the pyomelanin biosynthesis pathway provides a starting point for molecular manipulations of A. media strain WS to increase melanin production for industrial use.
Supporting Information
Arrows and boxed text indicate the construction manipulations. The chloramphenicol resistance cassette from pBeloBAC11 is represented by an open arrow. All plasmids are drawn to scale. IR, inverted repeat.
(DOC)
(A) Photographs of cultures from 72 h LB cultures of wild-type A. media strain WS and the 14 transposon mutants (WS-M1, WS-M2, WS-M3, WS-M4, WS-M5, WS-M6, WS-M7, WS-M8, WS-M9, WS-M10, WS-M11, WS-M12, WS-M13, WS-M14). (B) Wild-type A. media strain WS and the 14 transposon mutants were cultured in LB, and then at 72 h post-inoculation, the OD400 of the cultures were determined. (C) Wild-type A. media strain WS and the 14 transposon mutants were cultured in LB, and then at 72 h post-inoculation, the OD600 of the cultures were determined.
(DOC)
Detection of HGA from cultures of wild-type A. media strain WS and non-pigmented mutants WS-M10 and WS-M13. Samples were taken at 12-h intervals until 72 h at 30°C in LB. (A) Analysis of the culture from wild-type A. media strain WS. (B) Analysis of the culture from non-pigmented mutant WS-M10. (C) Analysis of the culture from non-pigmented mutant WS-M13. The asterisk indicates the peak of HGA.
(DOC)
(A) MS analysis of an authentic L-DOPA sample. (B) MS analysis of the sample from culture of wild-type A. media strain WS. (C) MS analysis of an authentic HGA sample. (D) MS analysis of the sample from culture of wild-type A. media strain WS.
(DOC)
The primary structure and conservation of HppD from A. hydrophila_XS91-4-1 compared to that from A. hydrophila ML09-119, A. hydrophila ATCC7966, A. hydrophila 4AK4. Amino acids depicted in gray are little conserved.
(DOC)
Expression of phhA, phhB, tyrB, aspC and hppD transcripts in A. salmonicida_AB98041, A. salmonicida KACC14791, A. hydrophila_XS94-1-4 and A. media strain WS, which were cultivated in LB at 30°C for 12, 24, 48 and 72 h cultivation. And then RT-PCR was done using primers that amplify the specific transcripts. That the PCR products obtained resulted from mRNA templates was confirmed by the lack of product obtained when the PCR did not incorporate reverse transcriptase (- RT). PCR products obtained from genomic DNA appear in the left-most lane, indicating that the mRNAs observed are full-length. RT-PCR analysis of 16S rRNA served as a positive control.
(DOC)
(A) The hmgA gene in A. media strain WS is interrupted into two parts, hmgA1 and hmgA2. hmgA, gene encoding the homogentisate dioxygenase. B224_2888, gene encoding a transposase. (B) Wild-type A. media strain WS, WSΔhmgA1, WSΔhmgA2 were cultured in LB, and then at 72 h and 96 h post-inoculation, the OD400 of the cultures were determined. (C) Wild-type A. media strain WS, WS (pBBR1MCS-5), WS (pBBR1MCS-5-hmgA-AS), WS (pBBR1MCS-5-hmgA-KACC) and WS (pBBR1MCS-5-hmgA-AH) were cultured in LB, and then at 72 h and 96 h post-inoculation, the OD400 of the cultures were determined.
(DOC)
(DOC)
Acknowledgments
We thank Dr. Aihua Li (Institute of Hydrobiology, Chinese Academy of Sciences) for kindly providing us the strain A. hydrophila_XS91-4-1, and Dr. Kenneth N. Timmis (GBF-National Research Centre for Biotechnology, Braunschweig, Germany) who kindly permitted us to use the plasmid pUTKm2. We also thank Dr. Shishen Du (Department of Microbiology, Molecular Genetics and Immunology, University of Kansas Medical Center) and Dr. Alex Miller (CSIRO Agriculture Flagship) for many helpful discussions and for revising the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Basic Research Program of China (973 Program) (No. 2011CB808800) and National Natural Science Foundation of China (No. 31070077). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. White LP. Melanin: a naturally occurring cation exchange material. Nature. 1958; 182: 1427–1428. [DOI] [PubMed] [Google Scholar]
- 2. Riley P. Melanin. Int J Biochem Cell Biol. 1997; 29: 1235–1239. [DOI] [PubMed] [Google Scholar]
- 3. Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 2003; 5: 203–223. [DOI] [PubMed] [Google Scholar]
- 4. Nosanchuk JD, Casadevall A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Ch. 2006; 50: 3519–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rundgren M. Tritium isotope effects in the reaction catalyzed by 4-hydroxyphenylpyruvate dioxygenase from Pseudomonassp. strain PJ 874. Biochim Biophys Acta. 1982; 704: 59–65. [DOI] [PubMed] [Google Scholar]
- 6. Bell AA, Wheeler MH. Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol. 1986; 24: 411–451. [Google Scholar]
- 7. Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans . Mol Microbiol. 2005; 57: 1381–1396. [DOI] [PubMed] [Google Scholar]
- 8. Keith KE, Killip L, He P, Moran GR, Valvano MA. Burkholderia cenocepacia C5424 produces a pigment with antioxidant properties using a homogentisate intermediate. J Bacteriol. 2007; 189: 9057–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarna T, Hyde JS, Swartz HM. Ion-exchange in melanin: an electron spin resonance study with lanthanide probes. Science. 1976; 192: 1132–1134. [DOI] [PubMed] [Google Scholar]
- 10. Krol ES, Liebler DC. Photoprotective actions of natural and synthetic melanins. Chem Res Toxicol. 1998; 11: 1434–1440. [DOI] [PubMed] [Google Scholar]
- 11. Różanowska M, Sarna T, Land EJ, Truscott TG. Free radical scavenging properties of melanin: interaction of eu-and pheo-melanin models with reducing and oxidising radicals. Free Radic Biol Med. 1999; 26: 518–525. [DOI] [PubMed] [Google Scholar]
- 12. Yabuuchi E, Ohyama A. Characterization of pyomelanin-producing strains of Pseudomonas aeruginosa . Int J Syst Bacteriol. 1972; 22: 53–64. [Google Scholar]
- 13. Swan G. Structure, chemistry, and biosynthesis of the melanins, Fortschritte der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products Springer; 1974. pp. 521–582 [DOI] [PubMed] [Google Scholar]
- 14. Wakamatsu K, Ito S. Advanced chemical methods in melanin determination. Pigment Cell Res. 2002; 15: 174–183. [DOI] [PubMed] [Google Scholar]
- 15. Leu W-M, Chen L-Y, Liaw L-L, Lee Y. Secretion of the Streptomyces tyrosinase is mediated through its trans-activator protein, MelC1. J Biol Chem. 1992; 267: 20108–20113. [PubMed] [Google Scholar]
- 16. Kong KH, Hong MP, Choi SS, Kim YT, Cho SH. Purification and characterization of a highly stable tyrosinase from Thermomicrobium roseum . Biotechnol Appl Biochem. 2000; 31: 113–118. [DOI] [PubMed] [Google Scholar]
- 17. López-Serrano D, SAnchez-Amat A, Solano F. Cloning and Molecular Characterization of a SDS-Activated Tyrosinase from Marinomonas mediterranea . Pigment Cell Res. 2002; 15: 104–111. [DOI] [PubMed] [Google Scholar]
- 18. Ruan L, He W, He J, Sun M, Yu Z. Cloning and expression of mel gene from Bacillus thuringiensis in Escherichia coli . Anton Leeuw Int J G. 2005; 87: 283–288. [DOI] [PubMed] [Google Scholar]
- 19. Piñero S, Rivera J, Romero D, Cevallos MA, Martínez A, Bolívar F, et al. Tyrosinase from Rhizobium etli is involved in nodulation efficiency and symbiosis-associated stress resistance. J Mol Microbiol Biotechnol. 2007; 13: 35–44. [DOI] [PubMed] [Google Scholar]
- 20. Rundgren M. Steady state kinetics of 4-hydroxyphenylpyruvate dioxygenase from human liver (III). J Biol Chem. 1977; 252: 5094–5099. [PubMed] [Google Scholar]
- 21. Goodwin P, Sopher C. Brown pigmentation of Xanthomonas campestris pv. phaseoli associated with homogentisic acid. Can J Microbiol. 1994; 40: 28–34. [Google Scholar]
- 22. Kotob SI, Coon SL, Quintero EJ, Weiner RM. Homogentisic acid is the primary precursor of melanin synthesis in Vibrio cholerae, a Hyphomonas strain, and Shewanella colwelliana . Appl Environ Microbiol. 1995; 61: 1620–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milcamps A, de Bruijn FJ. Identification of a novel nutrient-deprivation-induced Sinorhizobium meliloti gene (hmgA) involved in the degradation of tyrosine. Microbiology. 1999; 145: 935–947. [DOI] [PubMed] [Google Scholar]
- 24. Serre L, Sailland A, Sy D, Boudec P, Rolland A, Pebay-Peyroula et al. Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway. Structure. 1999; 7: 977–988. [DOI] [PubMed] [Google Scholar]
- 25. Hernández-Romero D, Solano F, Sanchez-Amat A. Polyphenol oxidase activity expression in Ralstonia solanacearum . Appl Environ Microbiol. 2005; 71: 6808–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igbinosa IH, Igumbor EU, Aghdasi F, Tom M, Okoh AI. Emerging Aeromonas species infections and their significance in public health. The Scientific World J; 2012. [DOI] [PMC free article] [PubMed]
- 27. Aurstad K, Dahle H. The production and some properties of the brown pigment of Aeromonas liquefaciens . Acta Vet Scand. 1971; 13: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shieh H, Maclean J. Melanin biosynthesis by a fish pathogen, Aeromomas salmonicida . Int J Biochem. 1974; 5: 643–647. [Google Scholar]
- 29. Martinez-Murcia A, Esteve C, Garay E, Collins M. Aeromonas allosaccharophila sp. nov., a new mesophilic member of the genus Aeromonas . FEMS Microbiol Lett. 1992; 91: 199–205. [DOI] [PubMed] [Google Scholar]
- 30. Esteve C, Gutiérrez MC, Ventosa A. Aeromonas encheleia sp. nov., isolated from European eels. Int J Syst Bacteriol. 1995; 45: 462–466. [DOI] [PubMed] [Google Scholar]
- 31. Abbott SL, Cheung WK, Janda JM. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J Clin Microbiol. 2003; 41: 2348–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gibson LF, George AM. Melanin and novel melanin precursors from Aeromonas media . FEMS Microbiol Lett. 1998; 169: 261–268. [Google Scholar]
- 33. Wan X, Chai B, Liao Y, Su Y, Ye T, Shen P, et al. Molecular and biochemical characterization of a distinct tyrosinase involved in melanin production from Aeromonas media . Appl Environ Microbiol. 2009; 82: 261–269. [DOI] [PubMed] [Google Scholar]
- 34. Chai B, Wang H, Chen X. Draft genome sequence of high-melanin-yielding Aeromonas media strain WS. J Bacteriol. 2012; 194: 6693–6694. 10.1128/JB.01807-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wan X, Liu H, Liao Y, Su Y, Geng J, Yang M, et al. Isolation of a novel strain of Aeromonas media producing high levels of DOPA-melanin and assessment of the photoprotective role of the melanin in bioinsecticide applications. J Appl Microbiol. 2007; 103: 2533–2541. [DOI] [PubMed] [Google Scholar]
- 36. De Lorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990; 172: 6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dennis JJ, Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol. 1998; 64: 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983; 1: 784–791. [Google Scholar]
- 39. Green MR, Sambrook J. Molecular cloning: a laboratory manual Cold Spring Harbor Laboratory Press; New York; 2012. [Google Scholar]
- 40. Wang Y, Gu J. Influence of temperature, salinity and pH on the growth of environmental Aeromonas and Vibrio species isolated from Mai Po and the Inner Deep Bay Nature Reserve Ramsar Site of Hong Kong. J Basic Microb. 2005; 45: 83–93. [DOI] [PubMed] [Google Scholar]
- 41. Chatfield CH, Cianciotto NP. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect Immun. 2007; 75: 4062–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rholl DA, Trunck LA, Schweizer HP. In vivo Himar1 transposon mutagenesis of Burkholderia pseudomallei . Appl Environ Microbiol. 2008; 74: 7529–7535. 10.1128/AEM.01973-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995; 8: 457–463. [DOI] [PubMed] [Google Scholar]
- 44. Denoya CD, Skinner DD, Morgenstern MR. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli . J Bacteriol. 1994; 176: 5312–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum . J Bacteriol. 1996; 178: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995; 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 47. Gu W, Song J, Bonner CA, Xie G, Jensen RA. PhhC is an essential aminotransferase for aromatic amino acid catabolism in Pseudornonas aeruginosa . Microbiology. 1998; 144: 3127–3134. [DOI] [PubMed] [Google Scholar]
- 48. Zhao G, Xia T, Song J, Jensen RA. Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4 alpha-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc NatI Acad Sci USA. 1994; 91: 1366–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Flydal MI, Chatfield CH, Zheng H, Gunderson FF, Aubi O, Cianciotto NP, et al. Phenylalanine hydroxylase from Legionella pneumophila is a thermostable enzyme with a major functional role in pyomelanin synthesis. PloS one. 2012; 7: e46209 10.1371/journal.pone.0046209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rüetschi U, Odelhög B, Lindstedt S, Barros-Söderling J, Persson B, Jörnvall H. Characterization of 4-hydroxyphenylpyruvate dioxygenase. Eur J Biochem. 1992; 205: 459–466. [DOI] [PubMed] [Google Scholar]
- 51. Coon SL, Kotob S, Jarvis BB, Wang S, Fuqua W, Weiner RM, et al. Homogentisic acid is the product of MelA, which mediates melanogenesis in the marine bacterium Shewanella colwelliana D. Appl Environ Microbiol. 1994; 60: 3006–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song J, Xia T, Jensen RA. PhhB, a Pseudomonas aeruginosa homolog of mammalian pterin 4a-carbinolamine dehydratase/DCoH, does not regulate expression of phenylalanine hydroxylase at the transcriptional level. J Bacteriol. 1999; 181: 2789–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Griffin PJ, Snieszko S, Friddle S. Pigment formation by Bacterium salmonicida . J Bacteriol. 1953; 65: 652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McIntosh D, Austin B. Atypical characteristics of the salmonid pathogen Aeromonas salmonicida . J Gen Microbiol. 1991; 137: 1341–1343. [DOI] [PubMed] [Google Scholar]
- 55. Moran GR. 4-Hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys. 2005; 433: 117–128. [DOI] [PubMed] [Google Scholar]
- 56. Schmaler-Ripcke J, Sugareva V, Gebhardt P, Winkler R, Kniemeyer O, Heinekamp T, et al. Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus . Appl Environ Microbiol. 2009; 75: 493–503. 10.1128/AEM.02077-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hunter RC, Newman DK. A putative ABC transporter, hatABCDE, is among molecular determinants of pyomelanin production in Pseudomonas aeruginosa . J Bacteriol. 2010; 192: 5962–5971. 10.1128/JB.01021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rodríguez JM, Timm DE, Titus GP, De Bernabe DB-V, Criado O, Mueller HA, et al. Structural and functional analysis of mutations in alkaptonuria. Hum Mol Genet. 2000; 9: 2341–2350. [DOI] [PubMed] [Google Scholar]
- 59. Turick CE, Beliaev AS, Zakrajsek BA, Reardon CL, Lowy DA, Poppy TE, et al. The role of 4-hydroxyphenylpyruvate dioxygenase in enhancement of solid-phase electron transfer by Shewanella oneidensis MR-1. FEMS Microbiol Ecol. 2009; 68: 223–225. 10.1111/j.1574-6941.2009.00670.x [DOI] [PubMed] [Google Scholar]
- 60. Titus GP, Mueller HA, Burgner J, de Córdoba SR, Peñalva MA, Timm DE. Crystal structure of human homogentisate dioxygenase. Nat Struct Mol Biol. 2000; 7: 542–546. [DOI] [PubMed] [Google Scholar]
- 61. Borowski T, Georgiev V, Siegbahn PE. Catalytic reaction mechanism of homogentisate dioxygenase: a hybrid DFT study. J Amer Chem Soc. 2005; 127: 17303–17314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arrows and boxed text indicate the construction manipulations. The chloramphenicol resistance cassette from pBeloBAC11 is represented by an open arrow. All plasmids are drawn to scale. IR, inverted repeat.
(DOC)
(A) Photographs of cultures from 72 h LB cultures of wild-type A. media strain WS and the 14 transposon mutants (WS-M1, WS-M2, WS-M3, WS-M4, WS-M5, WS-M6, WS-M7, WS-M8, WS-M9, WS-M10, WS-M11, WS-M12, WS-M13, WS-M14). (B) Wild-type A. media strain WS and the 14 transposon mutants were cultured in LB, and then at 72 h post-inoculation, the OD400 of the cultures were determined. (C) Wild-type A. media strain WS and the 14 transposon mutants were cultured in LB, and then at 72 h post-inoculation, the OD600 of the cultures were determined.
(DOC)
Detection of HGA from cultures of wild-type A. media strain WS and non-pigmented mutants WS-M10 and WS-M13. Samples were taken at 12-h intervals until 72 h at 30°C in LB. (A) Analysis of the culture from wild-type A. media strain WS. (B) Analysis of the culture from non-pigmented mutant WS-M10. (C) Analysis of the culture from non-pigmented mutant WS-M13. The asterisk indicates the peak of HGA.
(DOC)
(A) MS analysis of an authentic L-DOPA sample. (B) MS analysis of the sample from culture of wild-type A. media strain WS. (C) MS analysis of an authentic HGA sample. (D) MS analysis of the sample from culture of wild-type A. media strain WS.
(DOC)
The primary structure and conservation of HppD from A. hydrophila_XS91-4-1 compared to that from A. hydrophila ML09-119, A. hydrophila ATCC7966, A. hydrophila 4AK4. Amino acids depicted in gray are little conserved.
(DOC)
Expression of phhA, phhB, tyrB, aspC and hppD transcripts in A. salmonicida_AB98041, A. salmonicida KACC14791, A. hydrophila_XS94-1-4 and A. media strain WS, which were cultivated in LB at 30°C for 12, 24, 48 and 72 h cultivation. And then RT-PCR was done using primers that amplify the specific transcripts. That the PCR products obtained resulted from mRNA templates was confirmed by the lack of product obtained when the PCR did not incorporate reverse transcriptase (- RT). PCR products obtained from genomic DNA appear in the left-most lane, indicating that the mRNAs observed are full-length. RT-PCR analysis of 16S rRNA served as a positive control.
(DOC)
(A) The hmgA gene in A. media strain WS is interrupted into two parts, hmgA1 and hmgA2. hmgA, gene encoding the homogentisate dioxygenase. B224_2888, gene encoding a transposase. (B) Wild-type A. media strain WS, WSΔhmgA1, WSΔhmgA2 were cultured in LB, and then at 72 h and 96 h post-inoculation, the OD400 of the cultures were determined. (C) Wild-type A. media strain WS, WS (pBBR1MCS-5), WS (pBBR1MCS-5-hmgA-AS), WS (pBBR1MCS-5-hmgA-KACC) and WS (pBBR1MCS-5-hmgA-AH) were cultured in LB, and then at 72 h and 96 h post-inoculation, the OD400 of the cultures were determined.
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.