Abstract
Objective
Data from animal models show that in utero exposure to a maternal high fat diet (HFD) renders susceptibility of these offspring to the adult onset of metabolic syndrome. We and others have previously shown that epigenetic modifications to histones may serve as a molecular memory of the in utero exposure, rendering risk of adult disease. Because mice heterozygous for GLUT4 (insulin sensitive glucose transporter) born to wild-type (WT) mothers demonstrate exacterbated metabolic syndrome when exposed to a high fat diet in utero, we sought to analyze the genome-wide epigenetic changes which occur in the fetal liver in susceptible offspring.
Study Design
WT and Glut4+/− (G4+/−) offspring of WT mothers exposed either to a control or a HF diet in utero were studied. Immunoblotting was used to measure hepatic histone modifications of fetal and 5 week animals. Chromatin immunoprecipitation (ChIP) followed by hybridization to chip arrays (ChIP on chip) was utilized to detect genome-wide changes of histone modifications with HFD exposure.
Results
We found that levels of hepatic H3K14ac and H3K9me3 significantly increased with HFD exposure in WT and G4+/− fetal and 5 week offspring. Pathway analysis of our ChIP on chip data reveal differential H3K14ac and H3K9me3 enrichment along pathways which regulate lipid metabolism, specifically in the promoter regions of Pparg, Ppara, Rxra and Rora.
Conclusion
We conclude that HFD exposure in utero is associated with functional alterations to fetal hepatic histone modifications in both WT and G4+/− offspring, some which persist up to 5 weeks of age.
Keywords: developmental origins of adult disease, GLUT4, H3K14ac, H3K9me3
INTRODUCTION
According to the developmental origins of health and disease (DOHaD) hypothesis, the in utero experience can have a profound effect on the individual. Studies have suggested that the effects of a suboptimal intrauterine milieu can persist into adulthood1. In utero exposure to either a maternal low protein diet, caloric restriction or a maternal high fat diet (HFD), is associated with an increased susceptibility to the adult onset of metabolic syndrome2, 3. In the current era of obesity, studies concerning how a mother’s HFD may influence the health of her offspring are of increasing relevance. High fat diet consumption during pregnancy is associated with gestational diabetes mellitus 4. Animal models of in utero HFD exposure have shown that offspring are more susceptible to fatty liver in early life as well as increased adiposity, diabetes and cardiovascular disease in adulthood 3, 5–8. The question therefore remains, how can the memory of an exposure only experienced during gestation be maintained over the lifetime of the individual?
The possibility that epigenetic modifications contribute to this memory is an intriguing one. Epigenetic modifications constitute changes to the local chromatin structure that do not change the underlying DNA sequence. The addition or removal of post-translational histone modifications alongside changes in DNA methylation patterns are potential mechanisms that could contribute to the memory of an in utero exposure. Some histone modifications are enriched within the promoters of transcriptionally active genes, such as acetylation of lysine 14 of histone H3 (H3K14ac)9, 10 while other modifications such as trimethylation of lysine 9 of histone H3 (H3K9me3) are enriched in promoters of repressed genes as well as within heterochromatin 11–13. It is also well established that activating and repressive marks are not mutually exclusive, even within the same promoter. During embryogenesis, chromatin domains which contain both repressive and activating motifs cluster throughout the genome 14. Our previous work in a non-human primate model has demonstrated that HFD exposure in utero alters the fetal hepatic histone code 15. Specifically, hepatic H3K14ac is increased in the HFD exposed fetal animals 15, 16. H3K14 appears particularly sensitive to the intrauterine milleu as it is also modified in a rat model of nutrient restriction in skeletal muscle 17. How these modifications are established and their relationship to the susceptibility to the adult onset of disease remains to be fully investigated.
Not every animal exposed to a HFD is equally susceptible, begging the question as to whether the offspring genotype may serve as a modifier of the in utero environment. We have studied offspring heterozygous for the Glut4 gene (G4+/−) from WT mothers fed a HFD 18. GLUT4 haploinsufficiency results in peripheral insulin resistance, altered lipid metabolism and type 2 diabetes 19–22. It has been shown that exposure to a HFD during critical periods of development leads to development of metabolic syndrome such as increased adiposity, impaired glucose tolerance, and insulin insensitivity in G4+/− and WT offspring. Interestingly, genotype-dependent differences were observed, suggesting that haploinsufficiency of GLUT4 may result in a different metabolic remodeling in response to the HFD 18. Thus, it is possible that an interaction between the in utero environment (exposure to a maternal HFD) and the offspring genotype (specifically a heterozygous deletion of GLUT4) may lead to different epigenetic changes either protecting against or increasing the risk of developing metabolic disease.
Based on our findings of an altered hepatic epigenome in a non-human primate model of maternal HFD consumption 15, 16, 23, we sought to determine whether (1) an increase in hepatic acetylation is similarly observed in a murine model of maternal HFD exposure and (2) whether offspring genetically susceptible to metabolic syndrome (the G4+/− offspring) 18 have a similarly altered hepatic epigenome with HFD exposure in order to study whether diet x genotype interactions may also contribute to offspring disease susceptibility. Because the paternal germline is the source of the Glut4 haploinsufficiency (and Glut4 is not expressed in the liver until late postnatal life 24), any intrauterine genotypic contribution would be attributable only to altered placental glucose and/or nutrient uptake in G4+/− offspring.
In this study we found that fetal hepatic H3K14ac and H3K9me3 increases with maternal HFD exposure. These alterations were also observed in animals at 5 weeks of age. Chromatin immunoprecipitation (ChIP) followed by hybridization to a promoter array chip (ChIP-on-chip) was used to determine which promoters on a genome-wide scale show differential enrichment for H3K14ac and H3K9me3 in response to maternal diet. We observed that these modifications are predominantly enriched among those gene promoters which relegate to lipid metabolism networks. We conclude that HFD exposure in fetal life is associated with significant alterations of distinct histone modifications, rendering enriched occupancy in the promoters of genes which regulate lipid metabolism.
MATERIALS AND METHODS
Murine model
All animal procedures were done in accordance with approved IRB protocols from both Baylor College of Medicine and Albert Einstein College of Medicine as previously described 8, 18. WT CD1 female mice were maintained on a control breeding chow (PicoLab® Mouse Diet #5058; 9% fat, 20% protein, 53% carbohydrate) or high fat (HF Bio-Serv Product #F3282; 35.5% fat as lard, 20% protein, 36.3% carbohydrate) diet two weeks prior to mating with G4+/− males throughout gestation and lactation. Offspring were weaned onto a low fat diet (Pico Lab #5053; 4.5% fat, 20% protein, 54.8% carbohydrate) at postnatal day 21. The animals used in this study were male WT and G4+/− offspring. For fetal tissue, pregnant mice were sacrificed at embryonic (e) day e18.5. Fetuses were sacrificed by decapitation and organs were immediately harvested and snap frozen using liquid nitrogen18.
Experimental methods can be found in the supplemental information.
RESULTS
Hepatic H3K14 acetylation and H3K9 trimethylation increase with HFD exposure in utero and during lactation
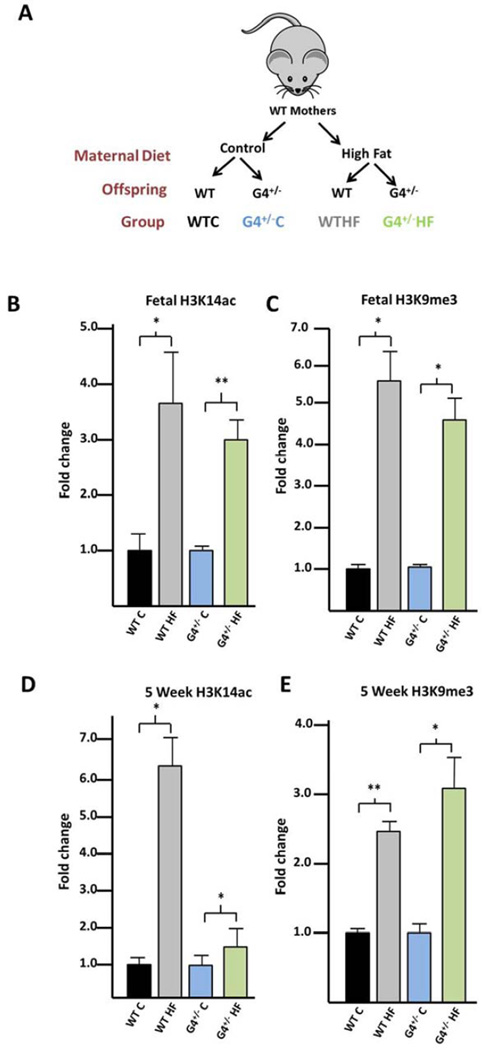
Immunoblotting was used to determine if hepatic histone modifications are altered in either WT or G4+/− offspring with HFD exposure (Figure 1A). In fetal liver at e18.5, H3K14ac is increased in both the WT (3.6-fold, p=0.002) and G4+/− (3.0-fold, p=0.002) offspring with HFD exposure (Figure 1B). H3K9me3 is also increased with HFD exposure in both the WT (5.7-fold, p=0.007) and G4+/− (4.6-fold, p=0.047) fetal offspring (Figure 1C).
Figure 1. H3K14ac and H3K9me3 are increased in the livers of HFD exposed WT and G4+/− offspring.
(A) WT mothers were exposed to either a control or HFD during pregnancy and lactation. Epigenetic changes in both WT and G4+/− offspring were characterized. (B) Immunoblotting with antibodies specific for each histone modification reveal that H3K14ac is significantly increased in the fetal liver in the WT and G4+/− offspring. (C) Similar results are seen for H3K9me3 in the fetal animals. (D,E) The increase in each modifiaction persists in the livers of 5 week old animals in both WT and G4+/− offspring. All results were normalized to total histone H3. Results are displayed as fold change compared with control diet exposed animals. P-values < 0.05 are designated *, and < 0.005 as **.
Immunobloting was similarly performed on livers from 5 week old animals exposed in utero to a maternal control or HF diet and weaned onto a low fat diet two weeks before harvesting the tissue. In these animals, both H3K14ac and H3K9me3 are significantly increased in HFD exposed animals compared with control diet in both WT and G4+/− offspring (Figures 1D and 1E). Relative levels of fetal hepatic H3K27me3, H4K20me3, H3K9ac, H3K18ac and H3K4me3 were found to be unchanged with HFD exposure in the fetal WT animals so were not further assessed (Supplementary Table 1).
Hepatic gene expression of histone modifying enzymes is reduced in WT 5 week old offspring with HFD exposure during lactation
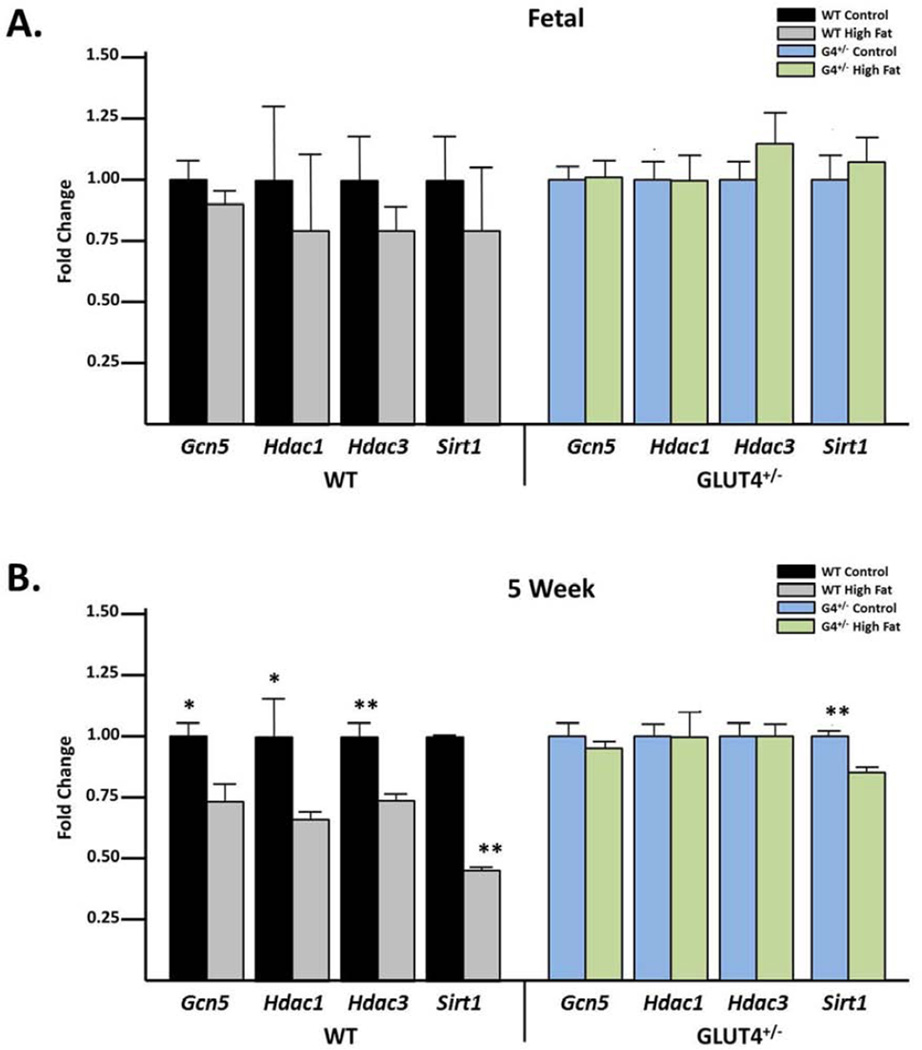
Because of the observed increase in histone acetylation in the fetal and 5 week old animals, we hypothesized that expression levels of histone acetyltransferases would be altered. GCN5 is a histone acetyltransferase of histone H3 25, 26. HDAC1, HDAC3 and SIRT1 deacetylate histone H3K14 27–30. Hepatic mRNA levels of these genes were measured in both fetal and 5 week old animals in the WT and G4+/− offspring. While we failed to observe a significant alteration in expression in the fetal animals (Figure 2A), at 5 weeks of age, Sirt1 expression was significantly decreased when compared to the control diet exposed cohort in both WT and G4+/− animals (Figure 2B). In WT offspring, gene expression of Gcn5, Hdac1 and Hdac3 were also significantly reduced in 5 week old offspring who experience both prenatal and postnatal HFD exposure (Figure 2B).
Figure 2. Hepatic SIRT1 levels are reduced in 5 week old animals exposed in utero to a maternal HFD.
(A) Using qRT-PCR levels of the histone acetyltransferase Gcn5 as well as the histone deacetylases Hdac1, Hdac3 and Sirt1 were analyzed. No significant changes with HFD exposure in WT and G4+/− animals were found. (B) Gcn5, Hdac1 and Hdac3 are all significantly decreased in the WT but not in the G4+/− animals. Sirt1 was significantly decreased with HFD exposure in both groups. P-values < 0.05 are designated *, and < 0.005 as **.
ChIP on chip reveals that global H3K14ac and H3K9me3 are both enriched at the transcription start site (TSS) regardless of genotype or diet exposure in the fetal liver
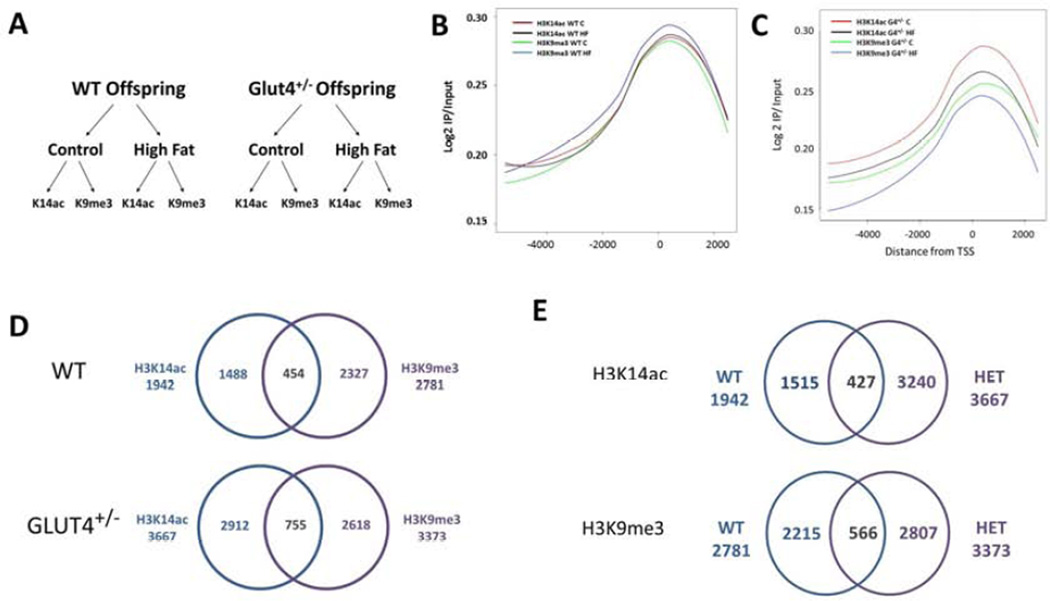
In order to determine in the localization of H3K14ac and H3K9me3, ChIP-on-chip was performed on the fetal liver from WT and G4+/− offspring exposed to either control or HFD in utero (Figure 3A). Enrichment (as measured by log 2 IP/ Input) of these modifications was plotted throughout the array as a means of internal validation 31, 32. Consistent with previous epigenome-wide characterizations 26, 33, we observed robust enrichment in a broad region surrounding the TSS in each of the eight groups studied (Figures 3B–C).
Figure 3. Chip on chip reveals a distinct profile for H3K14ac and H3K9me3 localization.
(A) This schemata shows an overview of the samples processed by ChIP for ChIP on chip analysis. Livers from fetal WT or G4+/− animals exposed to either a control or a HFD in utero were used for ChIP using either H3K14ac or H3K9me3 antibodies. Localization of both H3K14ac and H3K9me3 in WT (B) or G4+/− (C) animals surrounding the TSS is calculated using Log 2 IP/IN. (D) Partek was used to generate lists of genes with differential promoter occupancy between control and HFD exposed animals. In the WT animals, H3K14ac was differentially enriched in the promoters of 1942 genes with HFD exposure, and H3K9me3 n 2781 genes. There were 454 genes common to each group. In G4+/− animals, H3K14ac was differentially enriched in the promoters of 3667 genes, and H3K9me3 in 3373 genes. There were 755 genes common to each group. (E) To compare the overlap in genes marked by each modification in WT and G4+/−offspring a Venn Diagram was created. In the WT offspring, 1942 are differentially enriched with HFD exposure, in the G4+/− offspring 3667 genes show differential enrichment. Less than 10% (427 genes) are shared between the two groups. For H3K9me3, the WT offspring have 2781 and the G4+/− offspring have 3373 genes differentially enriched by virtue of HFD exposure. Only 566 genes are similar between the WT and G4+/− offspring.
H3K14ac and H3K9me3 are differentially enriched in genes involved in lipid metabolism in the fetal liver
In order to determine promoter specific changes of H3K14ac and H3K9me3 in the fetal animals, gene lists were generated from the ChIP-on-chip data which demonstrated differential enrichment over input (Supplementary Tables 2–9). We then determined which genes had differential enrichment between the control and HFD groups (Supplementary Tables 10–13). Of interest were the genes significantly altered for both H3K14ac and H3K9me3: , 454 genes in the WT animals and 755 genes in the G4+/− aimals (Figure 3D). To determine if there is an offspring genotype effect on histone modification localization, the generated gene lists were analyzed for overlap for each modification between WT and G4+/− offspring (Figure 3E). When comparing genes differentially enriched between control and HFD for H3K14ac, 8% (427 out of 5182 total) of genes were similarly enriched between WT and G4+/− liver while 10% (566 out of 5588 total) of the genes enriched for H3K9me3 were found in both WT and G4+/− liver.
For the genomic regions enriched for H3K14ac or H3K9me3 between control and HFD exposed animals (Supplementary Tables 10–13), HOMER 34 was used to determine which transcription factor binding sites were significantly (Benjamini q-value < 0.05) represented in the dataset. In the WT animals, there were no known motifs enriched in either the H3K14ac or H3K9me3 datasets. However, analysis of the G4+/− offspring revealed 9 significantly enriched motifs (Table 1). H3K14ac is differentially enriched in regions containing Gata1, 2, 4 and Myf5 binding motifs. H3K9me3 is differentially enriched in regions containing GFY, E2F, E2F4 and RUNX-AML binding motifs.
Table 1.
Known motifs differentially enriched by virtue of maternal high fat diet exposure
| Modification | Motif | Name | q-value |
|---|---|---|---|
| H3K14ac |  |
Gata1 | 0.0095 |
| H3K14ac |  |
Gata4 | 0.0436 |
| H3K14ac |  |
Gata2 | 0.0436 |
| H3K14ac |  |
Myf5 | 0.0436 |
| H3K9me3 |  |
E2F | 0.014 |
| H3K9me3 | GFY-Staf | 0.014 | |
| H3K9me3 | E2F4 | 0.0434 | |
| H3K9me3 | GFY | 0.0434 | |
| H3K9me3 | RUNX-AML | 0.0434 |
HOMER was used to determine known motifs enriched within the H3K14ac and H3K9me3 datasets.
Genes important for lipid metabolism are differentially enriched in H3K14ac and H3K9me3 in fetal livers of WT and G4+/− offspring
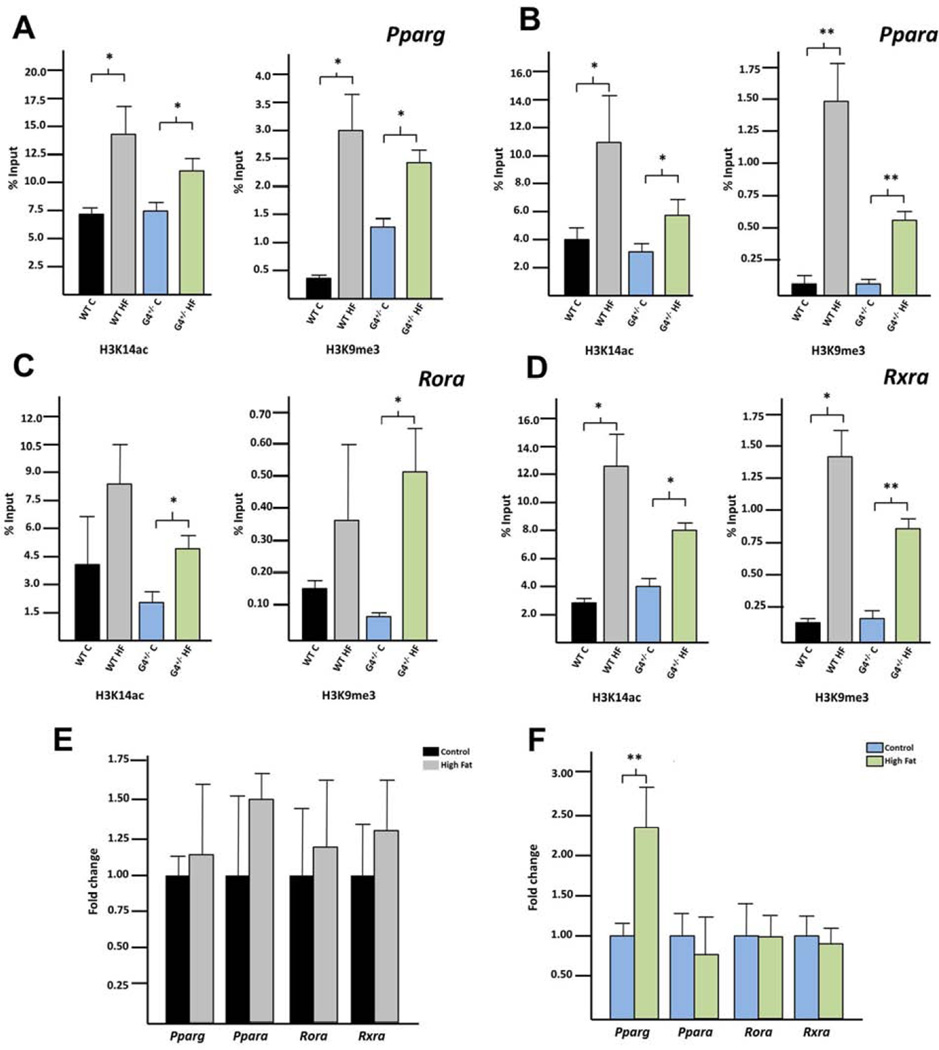
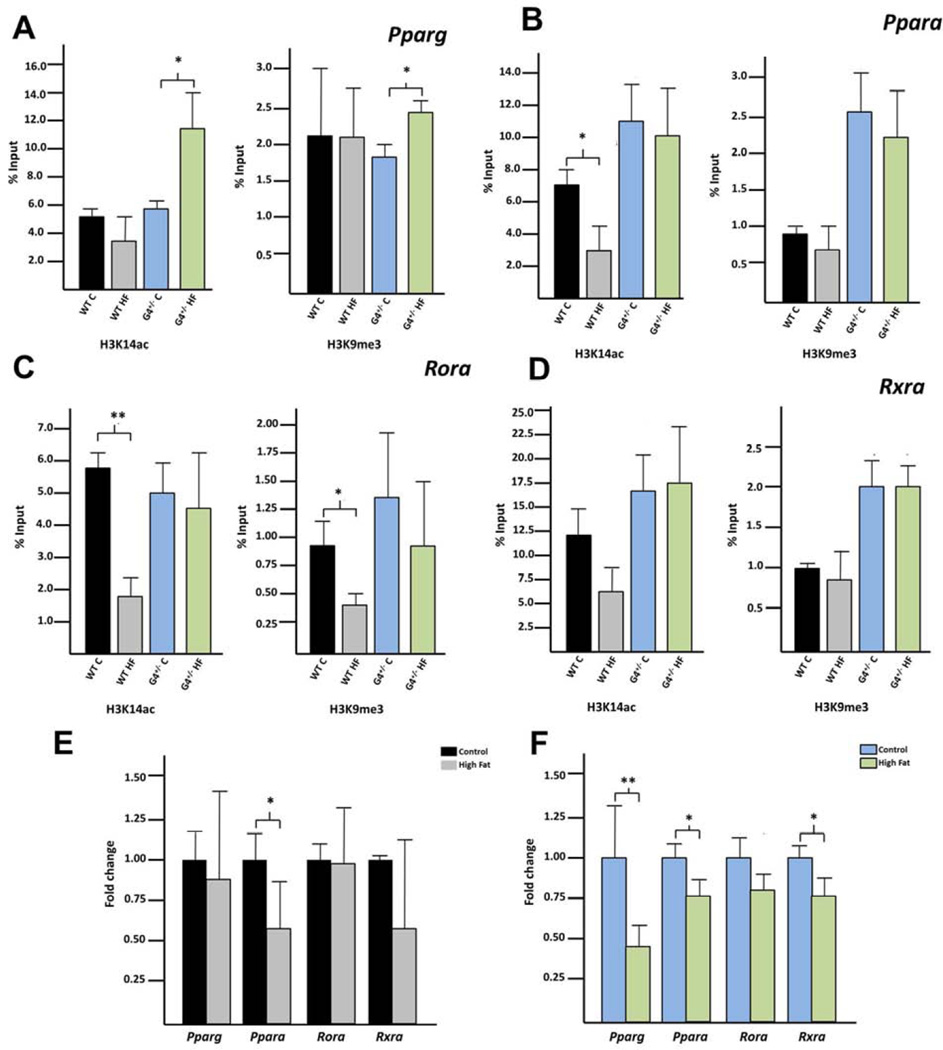
The generated gene lists were analyzed using Ingenuity Pathway Analysis (IPA) to determine which biological networks are differentially represented. For each group, lipid metabolism was the top network identified (Supplementary Table 14). In each analysis, four genes involved in lipid metabolism consistently emerged as central convergence nodes for the differentially represented pathways: Pparg, Ppara, Rora, and Rxra. Enrichment of H4K14ac and H3K9me3 within gene-specific promoters was interrogated employing qPCR on ChIP’ed DNA using primers proximal to the TSS (Supplementary Table 15). Indeed, both H3K14ac and H3K9me3 were enriched among both WT and G4+/− fetal liver following in utero HFD exposure compared to control diet (Figure 4 A–D).
Figure 4. Both H3K14ac and H3K9me3 are enriched at site specific promoters with HFD exposure in the fetal liver.
ChIP followed by site specific qPCR was used to determine enrichment (as calculated by percent input) of either H3K14ac or H3K9me3 within the promoters of (A) Pparg, (B) Ppara (C) Rora or (D) Rxra for WT and G4+/− offspring. qRT-PCR was used to determine if expression of each gene was altered in either the (E) WT or (F) G4+/−offspring. P-values < 0.05 are designated *, and < 0.005 as **.
We hypothesized that if there were an enrichment of these modifications with HFD exposure, this enrichment should correlate with significant alterations in gene-specific transcription. Pparg, Ppara, Rora and Rxra expression was quantified by qPCR (Figure 4 E+F). Pparg was increased in the G4+/− HFD exposed fetuses compared with the control diet group. Expression levels of Ppara, Rora and Rxra were not altered in HFD exposed fetuses in either the WT or G4+/− groups.
Promoter enrichment of H3K14ac and H3K9me3 is reversed at 5 weeks of age despite HFD exposure during lactation
We performed qPCR on H3K14ac and H3K9me3 ChIP’ed DNA using primers proximal to the TSS. At 5 weeks of age, both modifications are enriched with maternal HFD exposure in the Pparg promoter , but only among G4+/− offspring (Figure 5A). In HFD WT animals, there was a significant decrease in H3K14ac in the Ppara promoter (Figure 5B). Similarly, both H3K14ac and H3K9me3 were decreased in the Rora promoter with HFD exposure (Figure 5C). There were no significant changes in the Rxra promoter (Figure 5D). However, mRNA expression analysis demonstrated that Ppara expression was significantly decreased following maternal HFD exposure in both the WT and G4+/− 5 week old offspring (Figure 5 E+F), while mRNA expression of Pparg and Rxra were significantly decreased only in the G4+/− offspring (Figure 5F).
Figure 5. Enrichment of H3K14ac and H3K9me3 at site specific promoters in the livers of 5 week old animals.
ChIP followed by site specific qPCR was used to determine enrichment (as calculated by percent input) of either H3K14ac or H3K9me3 within the promoters of (A) Pparg, (B) Ppara (C) Rora or (D) Rxra for WT and G4+/− offspring. qPCR was used to determine if expression of each gene was altered in either the (E) WT or (F) G4+/− offspring. P-values < 0.05 are designated *, and < 0.005 as **.
COMMENT
In this study we undertook a discovery based, epigenome-wide approach to identifying which modifications were altered in the HFD exposed male offspring in fetal and postnatal life. Similar to our prior findings in a non-human primate model 15, 16, hepatic H3K14ac is increased in fetal life in the mouse. Hepatic H3K9me3 is also increased with HFD exposure in the fetal mouse, which was not observed in the non-human primate model. We conclude that K14ac and K9me3 are both modifiable in utero in response to maternal diet.
If epigenetic modifications contribute to a molecular memory of an in utero exposure, one would expect that changes which occur in utero would persist postnatally. Hepatic levels of both H3K14ac and K9me3 were measured in 5 week old offspring. Both the 5 week old WT and G4+/− offspring which were exposed to a HFD in utero showed an increase in both modifications compared with animals on a control diet. From this we conclude that in mice, an in utero exposure is sufficient to contribute to a change in the hepatic epigenome at 5 weeks of age.
Consistent with our work in primates, the Sirt1 deacetylase showed a significant decrease in expression in the 5 week old WT and G4+/− offspring following HFD exposure. Not only does Sirt1 have many reported functions in lipid metabolism and obesity 35, due to its dependence on NAD+, its activity is directly related to cellular energy levels. We have shown that Sirt1 functions as a histone deacetylase with preference for H3K14ac in vitro 16. Based on our collective findings, we speculate that hepatic Sirt1 is a crucial epigenomic modulator of fetal and juvenile histone acetylation.
Using ChIP on chip with a genome-wide promoter array we observed that H3K14ac and H3K9me3 are uniquely enriched within a 4 kb region surrounding the TSS and peaking directly at the start site (Figures 3 B+C). Although this was not surprising, this observation is crucial to our concluding that the increase in H3K14ac and K9me3 abundance by immunoblotting is not manifested through a delocalized spreading throughout the promoter regions.
We wanted to determine if both H3K14ac and H3K9me3 were potentially “marking” the same genes for reprogramming with HFD exposure. Such genes are potentially interesting because the mechanism(s) responsible for differential enrichment of each modification similarly targets these genes. Also, genes enriched for both repressive and permissive modifications in utero may be poised for rapid transcriptional activation when challenged ex utero 14. These histone marks may be key to understanding the susceptibility of offspring exposed to a maternal HFD to metabolic syndrome.
A surprising finding of this study was the difference in hepatic histone modification localization between the HFD exposed WT and G4+/− offspring. As evidenced in Figure 3E, only 10% of the genes which are differentially marked by either K14ac or K9me3 are similar between the WT and G4+/− animals. This was unexpected as Glut4 is not prominently expressed in fetal liver 24, and therefore it seemed unlikely that G4+/− offspring would have a different hepatic epigenome-wide pattern compared with WT offspring. However, it is important to note that Glut4 is expressed in mouse placenta 36 and therefore G4+/− offspring likely had a different intrauterine milieu resulting from a haploinsufficiency of the GLUT4 transporter in the placenta with altered placental nutrient uptake. It is also interesting to note the nearly two-fold increase in genes differentially enriched for H3K14ac in the G4+/− fetal liver when compared with the WT offspring (Figure 3D, 2912 genes vs 1488 genes). This may suggest that a diet x genotype interaction contributes to the observed phenotypic outcomes of offspring exposed to a maternal HFD. Consistent with this finding, we also determined that 9 transcription factor binding motifs are enriched within the G4+/− offspring dataset, but not that of the WT.
The promoter occupancy of four genes (Ppara, Pparg, Rora and Rxra) revealed differential enrichment of both modifications between control and HFD animals in both the WT and G4+/− groups. We hypothesize that these modifications may act as markers of in utero and lactation exposure, marking the genes for future transcription with continued metabolic stress of HFD consumption. However, we did not observe a similar pattern in the 5 week old animals. While we cannot conclude that the marks set in fetal life remain unaltered postnatally, it is important to note that the 5 week old animals were all weaned onto a low fat chow which they consumed for two weeks prior to sacrifice. We hypothesize that because both hepatic H3K14ac and H3K9me3 are both responsive to changes in the diet, the exposure to low fat chow may allow for a subtle altered enrichment of the modifications.
We conclude that the fetal hepatic epigenome is modified following maternal HFD exposure in utero and during lactation to enrich specific histone modifications in the promoters of genes well-characterized as key regulators of lipid overload. Whether these modifications contribute to the increased susceptibility of metabolic syndrome in adulthood remains to be determined, but this robust characterization of the fetal and postnatal epigenomic landscape lends crucial impetus to such studies.
MATERIALS AND METHODS
Immunoblotting
Immunoblots were performed as previously described 1. Briefly, acid extracted histones or whole cell lysates were run on 18% SDS-PAGE and transferred to polvinylidene fluoride (PVDF). Blots were incubated overnight at 4°C in primary antibody, washed three times and incubated for 45 minutes at room temperature in secondary antibody. Blots were visualized using Chemiluminescence. Bands were quantified using the UltraQuant system, and normalized to unmodified histone H3. The H3K9me3 antibody was purchased from Millipore (07–442); the H3K14ac antibody from Millipore (07–353) and total H3 from Cell Signaling (9715L). The HDAC1 and 3 antibodies were from Abcam (HDAC1 #46985; HDAC3 #28170). For the fetal animals an n of 5 per group was utilized and for the 5 week old animals, an n of 4 per group.
qPCR
RNA was extracted from liver using the Machery Nagel kit (740955.250) and converted to cDNA using Superscript III from Illumina (#18080-044). Commercially available TaqMan primers and probes were used for qPCR. Analysis was performed using the DDCT method 2. For the fetal animals an n of 5 per group was utilized, for the 5 week old animals there was an n of 4 per group.
ChIP
Chromatin immunoprecipitation was performed as previously described 3. Three fetal animals/ group were utilized. Histone modification antibodies were the same as from the western blots. Before the addition of antibody, 10% of the reaction was set aside as the input. Before genome wide amplification, samples were tested by qPCR for proper fold enrichment using previously published primers 4. Samples were amplified to approximately 7.5µg using the Sigma WGA kit (WGA-2) and submitted to the Baylor Microarray Core Facility for processing.
Array hybridization
Fragmentation of 7.5µg of double-stranded DNA was completed with the aid of 0.2U of DNase I, Amplification Grade, from Life Technologies (18068-015), producing fragment products 50–200bp in length. Fragment sizes were verified on the Agilent Bioanalyzer. Fragmentation was followed by terminal labeling with terminal deoxynucleotidyl transferase and the Affymetrix® proprietary DNA Labeling Reagent that is covalently linked to biotin. Hybridization cocktails containing Affymetrix spike-in controls and fragmented, labeled double-stranded DNA were loaded onto Affymetrix GeneChip® Mouse Promoter 1.0R arrays. The arrays were hybridized for 16 hours at 45°C with rotation at 60 rpm in the Affymetrix GeneChip® Hybridization Oven 640. The arrays were washed and stained with a streptavidin, R-phycoerythrin conjugate stain using the Affymetrix GeneChip® Fluidics Station 450. Signal amplification was done using biotinylated antistreptavidin. The stained arrays were scanned on the Affymetrix GeneChip® Scanner 3000. The images were analyzed and quality control metrics recorded using Affymetrix Command Console.
Array analysis
The analysis of ChIP-chip data was performed using Partek®, Genomic Suite™, Partek Inc. The CEL Files were imported and normalized by default method predefined in Partek, which includes Robust Multiarray Average (RMA) background correction, quantile normalization and Log 2 transformation. The difference of each comparison group was found by performing ANOVA, and then the MAT (Model based analysis of tiling-array) algorithm 5 was used to detect regions with enriched peaks. The p-value of the region is the empirical p-value of the most significant MAT score included within this region. MAT score of the region is the maximum MAT score for this region. The threshold for MAT score is 3.0. Enriched region with overlapping genes were annotated with Mus musculus NCBI Build 36.
Genes overlapping with significantly enriched regions were used to estimate the occupancy around TSS region. The probes corresponding to each region were identified and mapped into a window around transcription starting site (TSS) based on correct strand orientation. For each probe, the fold enrichment between the immunoprecipitated (IP) sample and negative control was calculated; the mean enrichment is the average of all the probes.
Transcription factor binding site motif analysis
Genomic regions identified by ChIP-chip as enriched for either H3K14ac or H3K9me3 in comparisons between control and HFD exposed animals were analyzed for transcription factor binding site motif enrichment using the findMotifsGenome.pl tool within the Hypergeometric Optimization of Motif EnRichment (HOMER) tool suite 6.
Statistical analysis
Statistical significance for immunoblotting and qPCR was calculated using a 2-tailed t-test in Excel with significance being a p-value of less than 0.05. The Known Motif Enrichment Results at a Benjamini multiple testing correction q-value < 0.05 were identified as being significantly enriched.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Aagaard, Hawkins and Charron labs for their input. We would like to thank the BCM Microarray Core Facility for their help in processing the arrays.
Financial support: This work was funded by NIH R01 DK089201 (K.A.), NIH DP2O D001500-01 (K.A.), a NIH REACH IRACDA K12 GM084897 (M.S.), NIH R21 DK081194 (MJC and PMV), Diabetes Research and Training Center NIH P60 DK020541, and the American Diabetes Association (MJC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors report no conflicts of interest
Results from this work were previously presented at the annual SMFM Meeting in 2009 and will be presented at the annual SMFM meeting in 2014.
REFERENCES
- 1.Hanson M, Gluckman P. Developmental origins of noncommunicable disease: population and public health implications. Am J Clin Nutr. 2011;94:1754S–1758S. doi: 10.3945/ajcn.110.001206. [DOI] [PubMed] [Google Scholar]
- 2.Suter MA, Aagaard-Tillery KM. Environmental influences on epigenetic profiles. Semin Reprod Med. 2009;27:380–390. doi: 10.1055/s-0029-1237426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seki Y, Williams L, Vuguin PM, Charron MJ. Minireview: Epigenetic programming of diabetes and obesity: animal models. Endocrinology. 2012;153:1031–1038. doi: 10.1210/en.2011-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saldana TM, Siega-Riz AM, Adair LS. Effect of macronutrient intake on the development of glucose intolerance during pregnancy. Am J Clin Nutr. 2004;79:479–486. doi: 10.1093/ajcn/79.3.479. [DOI] [PubMed] [Google Scholar]
- 5.Bertram CE, Hanson MA. Animal models and programming of the metabolic syndrome. Br Med Bull. 2001;60:103–121. doi: 10.1093/bmb/60.1.103. [DOI] [PubMed] [Google Scholar]
- 6.McCurdy CE, Bishop JM, Williams SM, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119:323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ainge H, Thompson C, Ozanne SE, Rooney KB. A systematic review on animal models of maternal high fat feeding and offspring glycaemic control. Int J Obes (Lond) 2011;35:325–335. doi: 10.1038/ijo.2010.149. [DOI] [PubMed] [Google Scholar]
- 8.Hartil K, Vuguin PM, Kruse M, et al. Maternal substrate utilization programs the development of the metabolic syndrome in male mice exposed to high fat in utero. Pediatr Res. 2009;66:368–373. doi: 10.1203/PDR.0b013e3181b33375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang G, Lin JC, Wei V, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Peters AH, Mermoud JE, O‘Carroll D, et al. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 12.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan RS, Zueva E, Cammas F, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487:249–253. doi: 10.1038/nature11173. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Aagaard-Tillery KM, Grove K, Bishop J, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suter MA, Chen A, Burdine MS, et al. A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. Faseb J. 2012;26:5106–5114. doi: 10.1096/fj.12-212878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri N, Raychaudhuri S, Thamotharan M, Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283:13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuguin PM, Hartil K, Kruse M, et al. Shared effects of genetic and intrauterine and perinatal environment on the development of metabolic syndrome. PLoS One. 2013;8:e63021. doi: 10.1371/journal.pone.0063021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenbit AE, Tsao TS, Li J, et al. GLUT4 heterozygous knockout mice develop muscle insulin resistance and diabetes. Nat Med. 1997;3:1096–1101. doi: 10.1038/nm1097-1096. [DOI] [PubMed] [Google Scholar]
- 20.Charron MJ, Kahn BB. Divergent molecular mechanisms for insulin-resistant glucose transport in muscle and adipose cells in vivo. J Biol Chem. 1990;265:7994–8000. [PubMed] [Google Scholar]
- 21.Li J, Houseknecht KL, Stenbit AE, Katz EB, Charron MJ. Reduced glucose uptake precedes insulin signaling defects in adipocytes from heterozygous GLUT4 knockout mice. Faseb J. 2000;14:1117–1125. doi: 10.1096/fasebj.14.9.1117. [DOI] [PubMed] [Google Scholar]
- 22.Rossetti L, Stenbit AE, Chen W, et al. Peripheral but not hepatic insulin resistance in mice with one disrupted allele of the glucose transporter type 4 (GLUT4) gene. J Clin Invest. 1997;100:1831–1839. doi: 10.1172/JCI119711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suter M, Bocock P, Showalter L, et al. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. Faseb J. 2011;25:714–726. doi: 10.1096/fj.10-172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim GH, Park EC, Yun SH, et al. Proteomic and bioinformatic analysis of membrane proteome in type 2 diabetic mouse liver. Proteomics. 2013;13:1164–1179. doi: 10.1002/pmic.201200210. [DOI] [PubMed] [Google Scholar]
- 25.Johnsson A, Durand-Dubief M, Xue-Franzen Y, Ronnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 2009;10:1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci U S A. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dangond F, Hafler DA, Tong JK, et al. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 29.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 30.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 31.Lindeman LC, Reiner AH, Mathavan S, Alestrom P, Collas P. Tiling histone H3 lysine 4 and 27 methylation in zebrafish using high-density microarrays. PLoS One. 2010;5:e15651. doi: 10.1371/journal.pone.0015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferraz-de-Souza B, Lin L, Shah S, et al. ChIP-on-chip analysis reveals angiopoietin 2 (Ang2, ANGPT2) as a novel target of steroidogenic factor-1 (SF-1, NR5A1) in the human adrenal gland. Faseb J. 2011;25:1166–1175. doi: 10.1096/fj.10-170522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xing AY, Challier JC, Lepercq J, et al. Unexpected expression of glucose transporter 4 in villous stromal cells of human placenta. J Clin Endocrinol Metab. 1998;83:4097–4101. doi: 10.1210/jcem.83.11.5290. [DOI] [PubMed] [Google Scholar]
References
- 1.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 3.Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R, Grove K, Lane R, Aagaard-Tillery K. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. Faseb J. 2011;25:714–726. doi: 10.1096/fj.10-172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.