Abstract
Background
Cytoplasmic dynein provides the main motor force for minus-end-directed transport of cargo on microtubules. Within the vertebrate central nervous system (CNS), proliferation, neuronal migration and retrograde axon transport are among the cellular functions known to require dynein. Accordingly, mutations of DYNC1H1, which encodes the heavy chain subunit of cytoplasmic dynein, have been linked to developmental brain malformations and axonal pathologies. Oligodendrocytes, the myelinating glial cell type of the CNS, migrate from their origins to their target axons and subsequently extend multiple long processes that ensheath axons with specialized insulating membrane. These processes are filled with microtubules, which facilitate molecular transport of myelin components. However, whether oligodendrocytes require cytoplasmic dynein to ensheath axons with myelin is not known.
Results
We identified a mutation of zebrafish dync1h1 in a forward genetic screen that caused a deficit of oligodendrocytes. Using in vivo imaging and gene expression analyses, we additionally found evidence that dync1h1 promotes axon ensheathment and myelin gene expression.
Conclusions
In addition to its well known roles in axon transport and neuronal migration, cytoplasmic dynein contributes to neural development by promoting myelination.
Keywords: Dynein, myelination, oligodendrocyte, axon, zebrafish
INTRODUCTION
Oligodendrocytes, the myelinating glial cell type of the central nervous system (CNS), extend numerous membrane processes that contact and wrap axons with myelin to facilitate rapid and efficient conduction of nerve impulses and maintain axon integrity. Myelin is a specialized membrane made up of specific lipids and proteins that forms only within the processes that contact axons and not elsewhere in the cell (Simons & Trotter 2007). Localized formation of myelin requires, at least in part, active transport of myelin components. For example, mRNAs encoding Myelin Basic Protein (MBP) accumulate in oligodendrocyte processes, where they are translated (Colman et al. 1982; Trapp et al. 1987). Experiments performed in cultured cells indicated that transport is dependent on microtubules, which are oriented with their plus ends in proximity to the cell membrane, and kinesins, motor proteins that move cargo on microtubule filaments toward the plus end (Carson et al. 1997; Song et al. 2003; Ainger et al. 1993). Consistent with this, mutation of zebrafish kif1b, which encodes a kinesin, disrupted mbp RNA transport and caused ectopic formation of myelin membrane (Lyons et al. 2009).
The major motor for minus end directed transport of cargo on microtubules is cytoplasmic dynein, a multi-subunit protein complex. Cytoplasmic dynein functions in numerous cellular processes including endosomal sorting, autophagy, cell division and cell migration (Eschbach & Dupuis 2011). Additionally, cytoplasmic dynein has a well-characterized role in transporting cargoes from the distal ends of axons toward neuronal cell bodies (Cosker et al. 2008; Zweifel et al. 2005). In principle, dynein could function similarly in oligodendrocytes, shuttling material from the distal tips of processes in contact with axons to the cell soma. Another possible role for dynein in oligodendrocytes is in promoting process extension or axon wrapping, because dynein at the cell cortex can stabilize microtubule plus ends and tether them to the cell periphery, potentially providing force on the cytoskeleton relative to the cell membrane (Hendricks et al. 2012). Consistent with both possibilities, immunolocalization experiments performed on cultured oligodendrocytes revealed concentration of cytoplasmic dynein within peripheral processes (King et al. 1996). Finally, our recent demonstration that Schwann cells require cytoplasmic dynein for myelination of peripheral nerves (Langworthy & Appel 2012) raised the possibility that dynein is also required for CNS myelination. However, no functional investigations of cytoplasmic dynein in oligodendrocytes have been reported and its potential roles in central nervous system myelination remain unknown.
To identify genes necessary for myelination, we carry out screens for mutations that disrupt glial development in zebrafish. Here we report analysis of a new mutant allele of dync1h1, which encodes the heavy chain subunit of cytoplasmic dynein. dync1h1 mutants have severe deficits of CNS myelin, which result from the combined effects of a deficit of oligodendrocyte progenitor cells (OPCs), failure to form normal myelin sheaths on axons and a deficit of myelin gene expression. Some human patients with dominant DYNC1H1 mutations have been diagnosed with intellectual disability and cognitive delay, which are associated with cortical malformations (Harms et al. 2012; Weedon et al. 2011; Willemsen et al. 2012; Vissers et al. 2010; Poirier et al. 2013). Our results raise the possibility that mutations that disrupt function of cytoplasmic dynein also contribute to abnormal brain development and cognitive deficits through disruption of myelination.
RESULTS
Oligodendrocyte Development is Abnormal in dync1h1vu76 Mutant Zebrafish Larvae
In a screen of zebrafish for ethyl nitrosourea (ENU)-induced mutations that alter the number and distribution of oligodendrocyte lineage cells, consisting of migrating and dividing oligodendrocyte progenitor cells (OPCs) and differentiating oligodendrocytes, we discovered one mutant allele, designated vu76, that caused small eyes, abnormal pigment deposition, uninflated swim bladders (Fig. 1A–F) and apparently fewer dorsal spinal cord OPCs and oligodendrocytes, marked by expression of the transgenic reporter Tg(olig2:EGFP) (Shin et al. 2003), than wild type (Fig. 1G,H). At higher magnification a dense network of oligodendrocyte membrane processes was evident in 5 day post fertilization (dpf) wild-type Tg(olig2:EGFP) larvae (Fig. 1I). vu76−/−;Tg(olig2:EGFP) larvae had substantially fewer processes and a deficit of cells with morphologies that are characteristic of myelinating glia (Fig. 1J).
Figure 1. The vu76 mutation causes defects in pigment deposition and oligodendrocyte development.
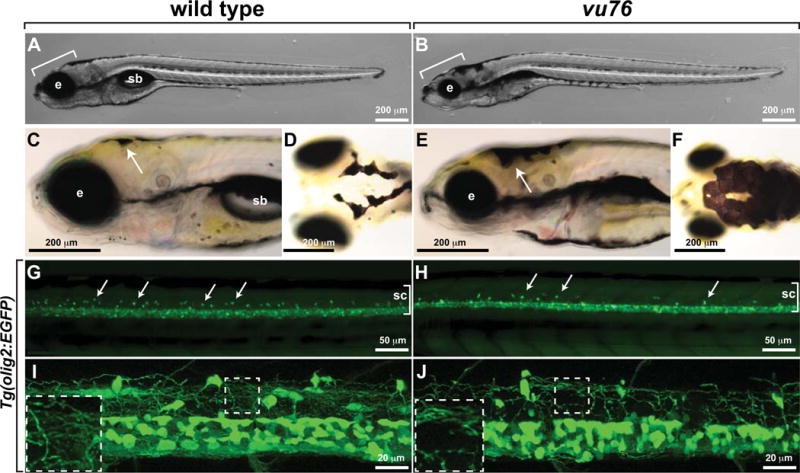
Images of living wild-type (A) and vu76 mutant (B) larvae at 6 dpf. The mutant has a smaller brain (bracket) and eye (e) than wild type and lacks a swim bladder (sb). Lateral (C, E) and dorsal (D, F) views of 5 dpf wild-type and vu76 mutant larvae to show pigment patterns. The dark pigment of the mutant is more broadly distributed than that of the wild type. Low (G, H) and high (I, J) magnification images of Tg(olig2:EGFP) reporter gene expression in wild-type and mutant larvae. Images are focused on the trunk spinal cord (sc, brackets) with dorsal up. In G and H dorsally migrated OPCs appear as green dots (arrows). Fewer dorsal OPCs are apparent in the vu76 mutant larva than in the wild-type larva. At 6 dpf OPC and oligodendrocyte processes form a dense meshwork (I, boxed area magnified as inset). Oligodendrocyte lineage cell processes are evident in the mutant larva, but the density of processes is less than in wild-type (J, boxed area magnified as inset).
To identify the gene that is disrupted by the vu76 mutation, we mapped the vu76 locus to zebrafish chromosome 17 using bulked segregant analysis with simple sequence length polymorphism (SSLP) markers (Knapik et al. 1998). In subsequent fine mapping we found one marker, z6010, which detected no recombination events in 2853 mutant larvae. Several genes are located near Z6010, including dync1h1. Larvae homozygous for the previously identified dync1h1hi3684Tg mutation have abnormally small eyes and brains and enlarged patches of black pigment (Amsterdam et al. 2005) similar to vu76 mutant larvae, raising the possibility that the vu76 mutation disrupts the dync1h1 locus. Therefore, we performed a complementation test by crossing vu76+/– and dync1h1hi3684Tg +/− adults. Approximately one quarter of the progeny had the morphological and pigment abnormalities common to larvae homozygous for either allele (Fig. 2A–C), indicating that the vu76 allele is a mutation of dync1h1. Consistent with this, injection of an antisense morpholino oligonucleotide (MO) designed to block translation of dync1h1 (Insinna et al. 2010) phenocopied the morphological (Fig. 2D) and myelination defects (see below) of vu76 mutant larvae. Western blotting revealed absence of full length Dync1h1 within protein extracted from homozygous mutant larvae at 5 dpf (Fig. 2E). Full length Dync1h1 detected in mutant extracts at 3 dpf likely represents maternal contribution. Sequencing of dync1h1 cDNA produced from vu76 mutant larvae revealed a G to A transition predicted to change a tryptophan residue to a premature stop codon at amino acid position 3561 (W3561X, Genbank accession NP_001036210), truncating the protein within the motor domain (Fig. 2F,G). Finally, whereas treatment of wild-type larvae with epinephrine, which stimulates rapid, Dync1h1-dependent retrograde transport of melanosomes (Clark & Rosenbaum 1982), caused aggregation of dark pigment (Fig. 2H–J), epinephrine had no effect on the pigment pattern of vu76 mutant larvae (Fig. 2K–M), consistent with loss of Dync1h1 motor function. We conclude that the vu76 mutation is a loss-of-function dync1h1 allele and hereafter refer to it as dync1h1vu76.
Figure 2. The vu76 allele is a mutation of dync1h1.
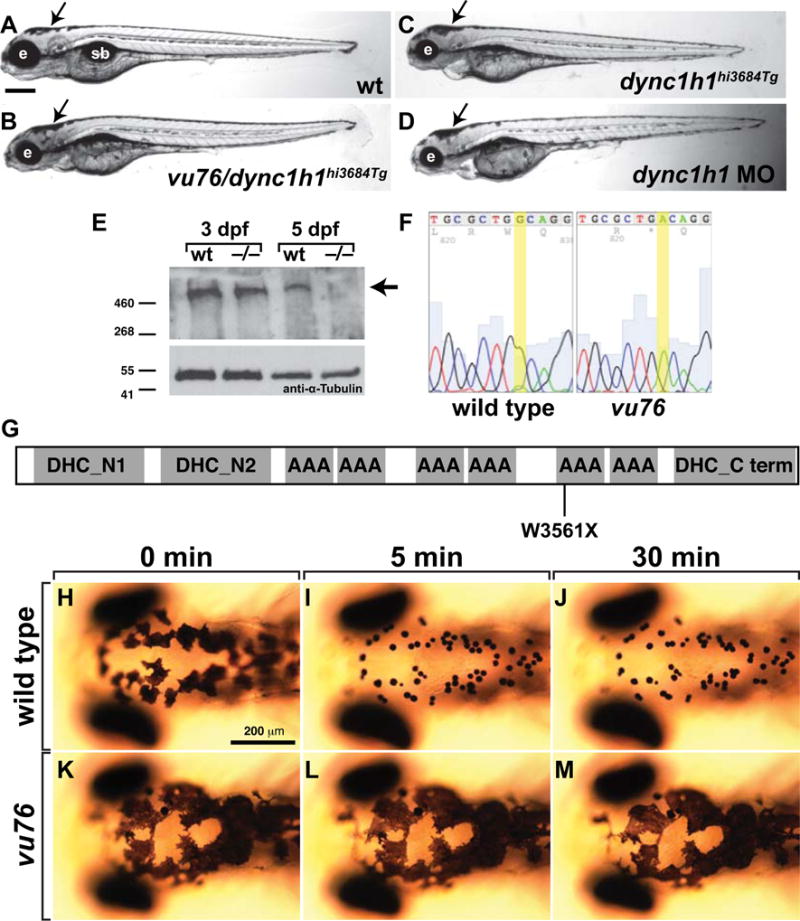
(A–D) Compound heterozygous vu76/dync1h1hi3684Tg, homozygous dync1h1hi3684Tg and dync1h1 MO-injected larvae have similar phenotypic characteristics including small heads and small eyes (e) and abnormally dispersed pigment (arrows). (E) Western blotting of protein extracts obtained from 3 and 5 dpf larvae with anti-Dync1h1 antibody reveals an approximately 500 kD protein present in 3 and 5 dpf wild-type larvae and 3 dpf mutant larvae but absent from 5 dpf mutant larvae (arrow). The blot was probed with anti-α-Tubulin antibody as a loading control. (F) Sequencing of dync1h1 cDNA from wild-type and vu76 mutant larvae revealed a G to A transition changing a tryptophan codon to a stop codon. This change is predicted to prematurely terminate translation at amino acid position 3561, within the motor domain of Dync1h1 (G). (H–M) Whereas 3 dpf wild-type larvae rapidly aggregate melanosomes into tight spots upon treatment with epinephrine, which reflects retrograde transport of melanosomes on microtubules, melanosome distribution in vu76 mutant larvae is not changed by epinephrine. Images in H-M show dorsal views of the head. Elapsed time following initiation of epinephrine treatment is indicated above the panels.
Dync1h1 Promotes Oligodendrocyte Number and Stable Axon Ensheathment by Oligodendrocyte Membrane
dync1h1vu76 mutant larvae have fewer dorsally migrated olig2:EGFP+ cells than wild type, indicative of a deficit of OPCs. To confirm this observation we used immunohistochemistry to dectect expression of Sox10, which specifically marks OPCs and oligodendrocytes (Park et al. 2005). This revealed that dync1h1vu76 mutant larvae had an approximately 2-fold reduction in the number of spinal cord oligodendrocyte lineage cells relative to wild type (Fig. 3A–C). To investigate the basis of the OPC deficit, we performed time-lapse imaging of Tg(olig2:EGFP) embryos injected at one-cell stage with dync1h1 MO. Fewer OPCs migrated from the pMN precursor domain and OPCs underwent fewer divisions in dync1h1 MO-injected larvae compared to control larvae (Fig. 3D,E). Furthermore, no OPC death was evident during the period of time-lapse imaging of dync1h1 MO-injected larvae. Therefore, the deficit of OPCs in dync1h1 deficient larvae appears to result both from production of fewer OPCs from neural precursors and failure to expand the OPC population by division.
Figure 3. Dync1h1 deficient zebrafish have a deficit of oligodendrocytes, each of which has fewer myelin sheaths.
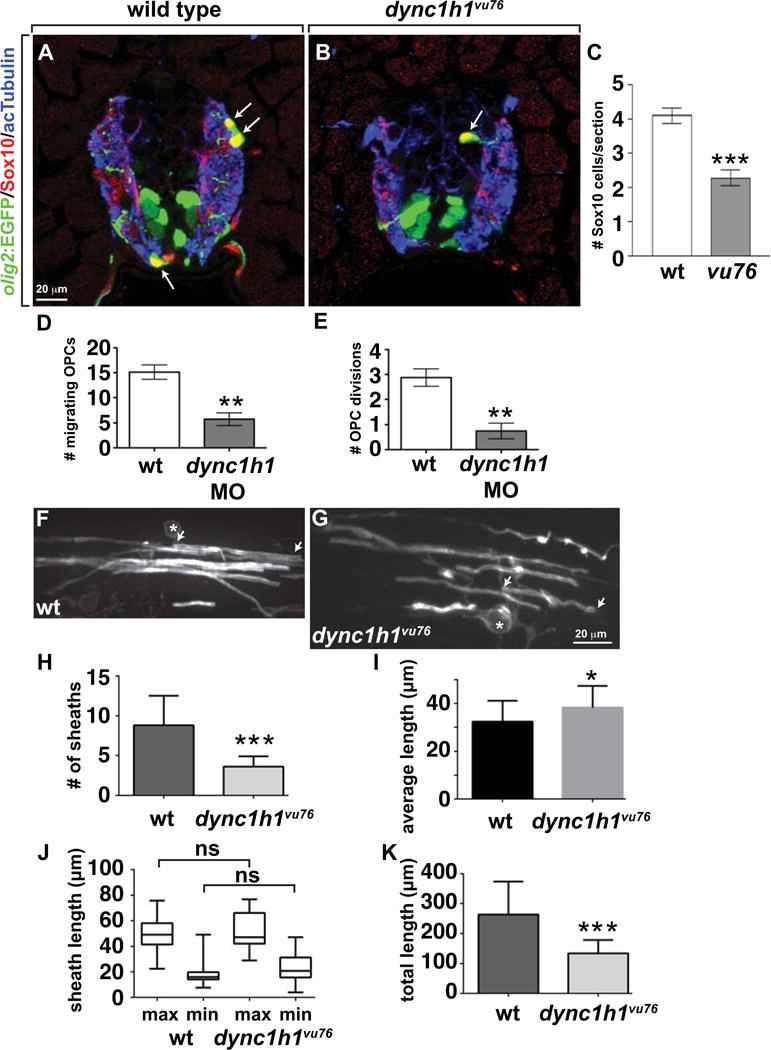
(A, B) Immunohistochemistry to detect Sox10 (red) in combination with olig2:EGFP (green) marks oligodendrocyte lineage cells in 4 dpf wild-type and dync1h1vu76 mutant larvae. Panels show representative images of transverse sections through the trunk spinal cord, with dorsal up. Anti-acetylated Tubulin staining (blue) marks axons. (C) Mutant larvae have about 50% the normal number of oligodendrocyte lineage cells. (D, E) Data, collected from time-lapse movies, showing fewer dorsally migrating OPCs and fewer OPC divisions in dync1h1 MO-injected larvae than in wild type. (F, G) Representative images of single, sox10:EGFP-CaaX+ oligodendrocytes in 6 dpf control (F) and dync1h1vu76 mutant (G) larvae. Asterisks mark cell bodies and arrows mark ends of single myelin sheaths. (H) Quantification revealed fewer sheaths in dync1h1vu76 mutant larvae than in control. (I) Graph showing that average myelin sheath length is slightly longer in dync1h1vu76 mutant larvae than in wild type. However, the statistical significance of the difference is weak (P=0.0414). (J) Box-and-whisker plot revealed that both the minimum (min) and maximum (max) lengths of sheaths in dync1h1vu76 mutant larvae were slightly more variable than in control larvae. However, the differences between the average maximum and minimum lengths were not significant (ns). (K) Graph showing the average total myelin sheath lengths of individual oligodendrocytes. Error bars represent +/− SEM. For panels H–K, wild-type data were collected from 28 cells in 11 larvae. Mutant data were collected from 15 cells in 9 larvae. Statistical significance was determined by Student’s t test for panels C–E and by nonparametric two tailed Mann-Whitney t test for panels H–K. *** P<0.0001; **P<0.001; * P<0.05.
The apparently decreased density of oligodendrocyte processes in dync1h1vu76 mutant larvae evident in Figure 1 likely reflects, at least in part, decreased number of oligodendrocytes. In addition, data showing that microtubules and cytoplasmic Dynein are enriched within oligodendrocyte processes (King et al. 1996) raised the possibility that loss of Dynein also reduces process activity and axon wrapping. To investigate this, we used a transgenic reporter system that reveals single oligodendrocytes, which permits visualization and quantification of oligodendrocyte sheath number and length. Specifically, we injected embryos at single cell stage with plasmid expressing membrane-tethered EGFP under control of sox10 regulatory DNA (sox10:EGFP-CaaX). In the spinal cords of control 6 dpf larvae, single oligodendrocytes formed multiple sheaths of variable length (Fig. 3F). Oligodendrocytes in 6 dpf dync1h1vu76 mutant larvae wrapped axons (Fig. 3G) indicating that Dync1h1 function is not necessary for axon ensheathment. However, oligodendrocytes in dync1h1vu76 mutant larvae formed fewer sheaths (Fig. 3J). The average length of sheaths was similar in control and dync1h1 MO-injected larvae (Fig. 3K) but sheath length was slightly more variable in mutant larvae than in control larvae (Fig. 3J). The total myelin sheath length for individual oligodendrocytes in dync1h1vu76 mutant larvae was approximately half that of wild type (Fig. 3K), reflecting the reduction in myelin sheath number. Together with the approximately two-fold reduction in oligodendrocyte lineage cell number, this deficit of myelin sheath formation likely results in a substantial deficit of myelination in Dync1h1-deficient larvae.
To investigate axon wrapping more closely we introduced the transgenic reporter Tg(nkx2.2a:mEGFP), which encodes membrane-tethered EGFP under control of nkx2.2a regulatory DNA (Ng et al. 2005), into the dync1h1vu76 mutant line and performed time-lapse imaging beginning at 2.5 dpf. In wild-type larvae, OPCs continuously extended and retracted multiple processes during migration and wrapped axons with tube-like sheaths of membrane (Fig. 4A; Video S1). Consistent with previous observations (Czopka et al. 2013), nascent sheaths rarely retracted from axons. We observed no obvious differences in OPC migration and process activity in dync1h1vu76 mutant larvae (Fig. 4B; Video S2). However, sheath formation appeared to be more variable in mutant larvae. Whereas some sheaths lengthened over time, others did not extend or became shorter (Fig. 4B; Video S2). Thus, the reduction in sheath number per oligodendrocyte resulting from dync1h1 knockdown may result from reduced sheath stability.
Figure 4. Axon wrapping by oligodendrocyte membrane is abnormal in dync1h1vu76 mutant larvae.
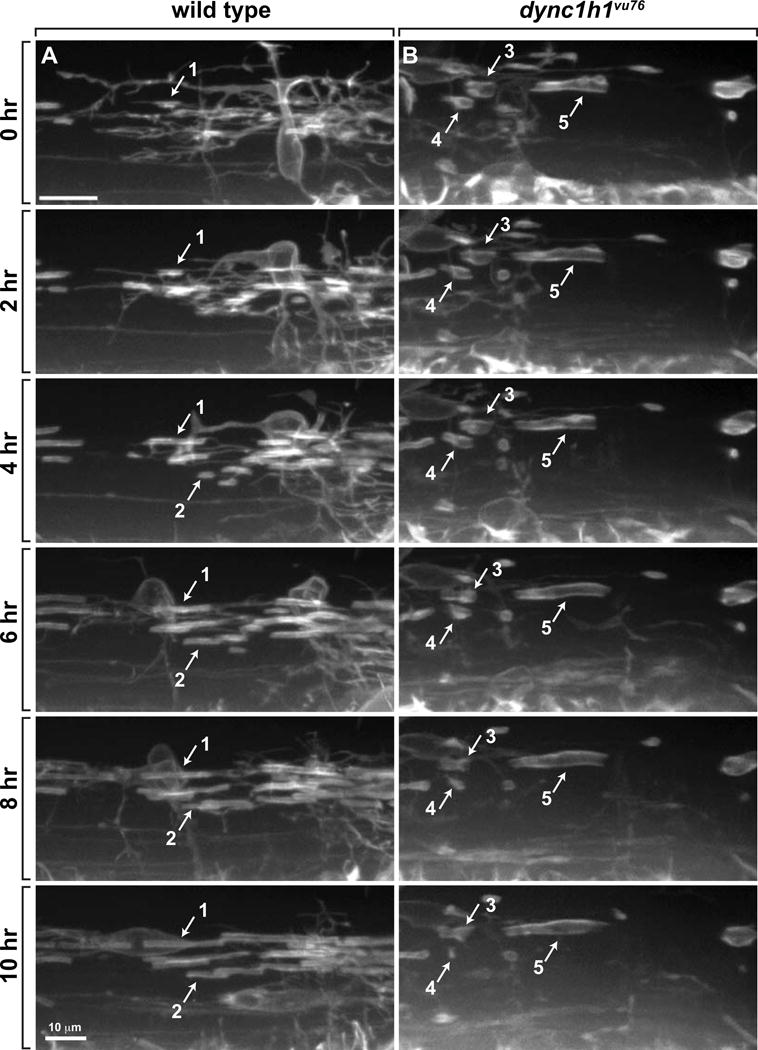
Panels show images captured from time-lapse movies of oligodendrocytes marked by Tg(nkx2.2:EGFP-CaaX) reporter expression beginning at 2.5 dpf. Time elapsed since beginning of movies is marked on the left. Images were obtained from lateral view of the trunk spinal cord, with dorsal up. (A) In wild-type larvae, oligodendrocyte membrane processes contact axons, initiate tight wrapping and lengthen bi-directionally to form myelin sheaths (see example sheaths 1 and 2). Once formed, most sheaths are stable and rarely lost. (B) In dync1h1vu76 mutant larvae, some sheaths do not lengthen (e.g. sheath 3) and some shorten and are subsequently lost (e.g. sheath 4). Sheath 5 forms a relatively long segment.
Dync1h1 Function is Required for Myelination
To determine if Dync1h1 is required for myelin gene expression in addition to promoting oligodendrocyte number and stable sheath formation, we first performed in situ RNA hybridization to detect transcripts encoded by myelin genes. Whereas wild-type larvae expressed robust levels of RNAs encoded by the myelin genes cldnk, plp1a and mbp (Fig. 5A–C), dync1h1vu76 mutant larvae expressed these genes at much lower levels (Fig. 5D–F), similar to our observations that Schwann cells express myelin genes at undetectable levels in the peripheral nervous system of dync1h1hi3684Tg mutant larvae (Langworthy & Appel 2012). Quantitative RT-PCR confirmed that transcripts encoding various myelin proteins were reduced both in mutant larvae and in larvae injected with dync1h1 MO relative to wild type (Fig. 5G,H). Consistent with these data, dync1h1 mutant larvae expressed lower levels of Mbp, detected by immunohistochemistry, than wild type (Fig. 5I,J).
Figure 5. Normal levels of CNS myelin gene expression require Dync1h1 function.
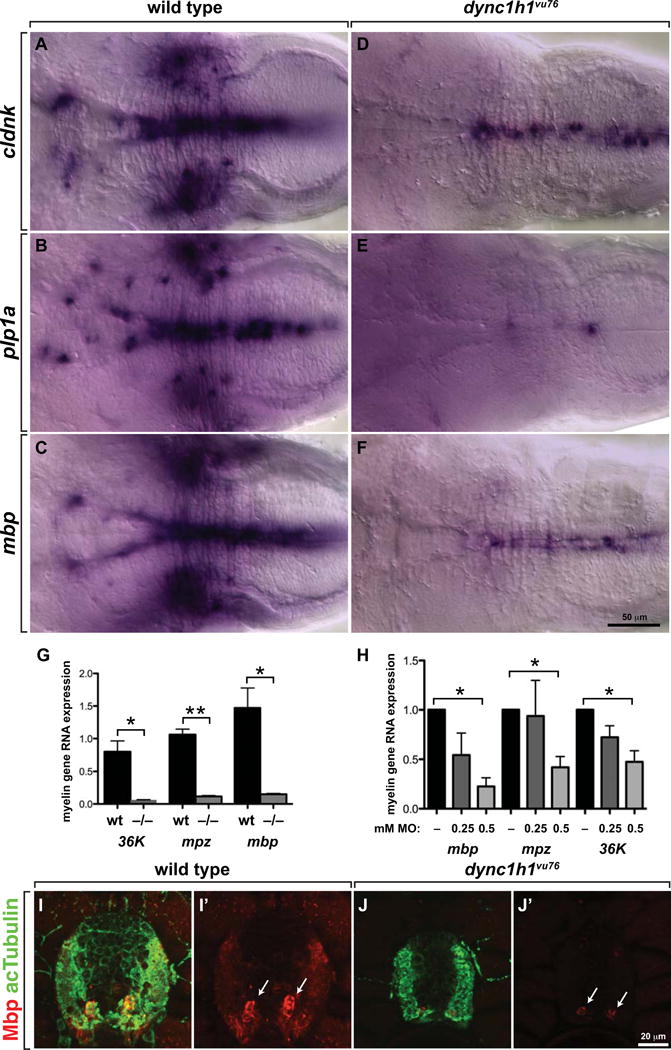
(A–F) Whereas 4 dpf wild-type larvae robustly express cldnk, plp1a and mbp RNA, only apparently low levels of transcripts are evident in similarly processed dync1h1vu76 mutant larvae. Images show dorsal views of whole larvae at the level of the midbrain and hindbrain, with anterior to the left. (G, H) Quantitative RT-PCR confirmed the deficit of myelin gene expression in dync1h1vu76 mutant larvae and dync1h1 MO-injected larvae. (I, J) Levels of Mbp (red), detected by immunohistochemistry, appear higher in wild-type spinal cord than in mutant spinal cord. Anti-acetylated Tubulin (green) reveals axons. Images show transverse sections, with dorsal up. Statistical analysis was performed using an unpaired t test with Welsh correction. * P<0.05; ** P<0.001. Error bars represent + SEM.
Finally, we investigated the effect of loss of dync1h1 function on CNS nerves using transmission electron microscopy. We focused our attention on the ventral spinal cord of 6 dpf larvae, where the large Mauthner axon serves as convenient landmark (Fig. 6A,B). By plotting the area of individual ventral spinal cord axons, we found that the average area of axons in dync1h1vu76 mutant larvae was smaller than that of wild-type larvae, due mostly to the presence of a relatively large number of small axons in dync1h1vu76 mutants (Fig. 6C). We next counted the number of myelinated and nonmyelinated axons in the intermediate size class, defined as axons having a cross sectional area of 0.201–3.99 μm. Consistent with our data showing that oligodendrocytes make fewer myelin sheaths in mutant larvae, the proportion of intermediate size axons that were myelinated was reduced from nearly 100% in wild type to approximately 63% in dync1h1vu76 mutant larvae (Fig. 6D). We then assessed myelin thickness by counting the number of myelin membrane wraps on intermediate size class axons that were clearly myelinated. Whereas there was no difference in the average cross sectional area of wild-type and mutant axons of this class (Fig. 6E), the average number of membrane wraps was reduced from about 4 in wild type to about 2 in dync1h1vu76 mutant larvae (Fig. 6F). Therefore, oligodendrocytes form fewer and thinner myelin sheaths in dync1h1vu76 mutant larvae, resulting in a substantial deficit of myelination.
Figure 6. Loss of Dync1h1 function causes CNS axon and myelin abnormalities.
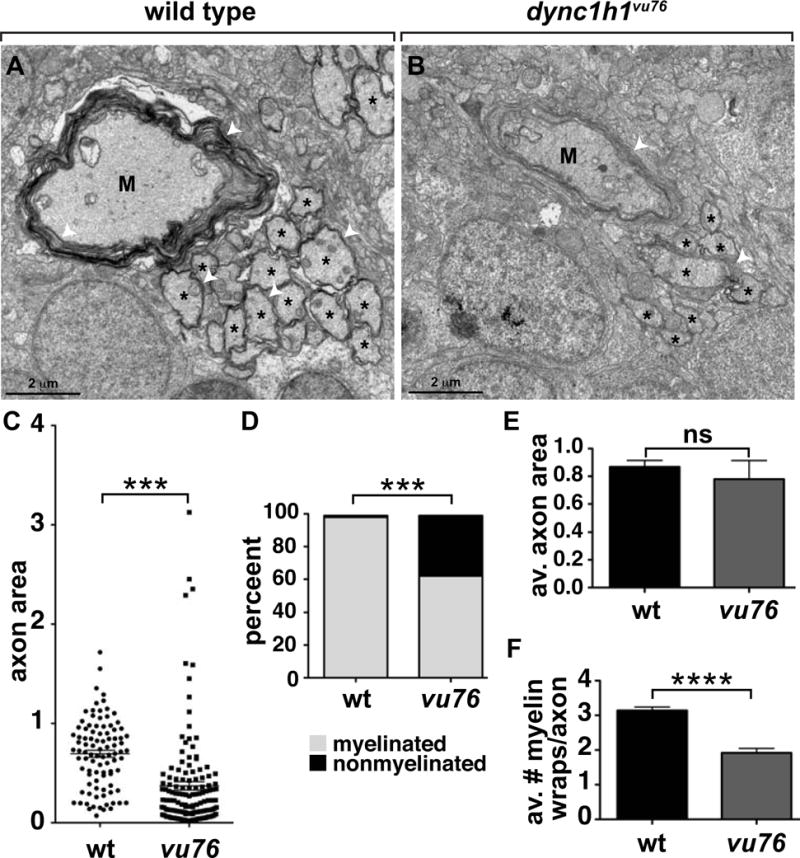
(A, B) EM micrographs of ventral spinal cords of 6 dpf wild-type and dync1h1vu76 mutant larvae. M indicates Mauthner axon, asterisks indicate intermediate sized axons and white arrowheads mark myelin membrane. (C) Scatter plot showing cross sectional area of axons. Mauthner axons were excluded from the plot. The average area of axons in mutant larvae is reduced relative to wild type. (D) Graph showing percentage of myelinated, intermediate size axons at 6 dpf. Intermediate was defined as axons having a cross sectional area between 0.201–3.99 μm. Loss of Dync1h1 function results in more nonmyelinated axons. (E) Graph showing the average axon cross sectional area of intermediate class myelinated axons. (F) Graph showing the average number of myelin wraps on intermediate class myelinated axons. Statistical analysis for the proportion of myelinated and nonmyelinated axons was performed using a two-sided Chi-square test. Statistical analysis for axon area and myelin wraps was performed using two-tailed, unpaired t test. *** P<0.0005, **** P<0.0001. ns, not significant. Error bars represent + SEM.
DISCUSSION
Cytoplasmic dynein is a multi-subunit protein complex that functions as the major motor for minus end directed transport of cargo on microtubules. Mutations of genes that encode dynein subunits have now been implicated in various pathologies, including neurological disease. For example, dominant, missense mutations of DYNC1H1 have been associated with an axonal form of Charot-Marie-Tooth peripheral neuropathy (Weedon et al. 2011) and spinal muscular atrophy with lower extremity predominance (SMA-LED) (Harms et al. 2012). Pathologies such as these might result from defects in axon transport (Millecamps & Julien 2013). Additionally, some individuals with dominant DYNC1H1 mutations have been diagnosed with intellectual disability and cognitive delay (Weedon et al. 2011; Willemsen et al. 2012; Vissers et al. 2010; Harms et al. 2012). Magnetic resonance (MR) and X-ray computed tomography (CT) imaging of two patients with severe intellectual disability revealed brain abnormalities suggestive of neuronal migration defects (Willemsen et al. 2012). This observation is consistent with interaction of DYNC1H1 with LIS1, mutations of which cause lissencephaly due to a failure of neuronal migration (Smith et al. 2000). Recently, recurrent mutations of DYNC1H1 were found in individuals with malformations of cortical development (Poirier et al. 2013), which are generally associated with defects in proliferation and neuronal migration (Barkovich et al. 2012). Our data now provide evidence that cytoplasmic dynein also is required for myelination.
Myelination of CNS axons requires formation of appropriate numbers of OPCs, migration of OPCs from their origins to their target axons, extension of OPC membrane to wrap axons, myelin gene expression and active transport of myelin membrane components. Our investigation reveals that, in zebrafish, several of these cellular functions depend on Dync1h1. We identified the dync1h1vu76 allele in our genetic screen because mutant larvae had too few OPCs. Time-lapse imaging revealed that formation of too few OPCs from ventral spinal cord precursors and subsequent failure of OPC division, and not cell death, caused the OPC deficit, indicating that Dync1h1 is required for oligodendrocyte lineage cells to progress through the cell cycle. Consistent with this, previous investigations showed that cytoplasmic dynein function is necessary for chromosome movements, organization and orientation of the spindle and check-point silencing (Raaijmakers et al. 2013; Howell et al. 2001; Busson et al. 1998; Varma et al. 2008; Sharp et al. 2000; Robinson et al. 1999; Schmidt et al. 2005). Cell division during early development of dync1h1vu76 mutant embryos is probably sustained by maternally contributed wild-type Dync1h1, which Western blotting revealed persists through 3 dpf.
Dync1h1 also is an important motor for cell migration. During development of the cerebral cortex, neurons born at the proliferative ventricular zone migrate radially to populate the upper layers of the cortex. These cells extend a leading process along radial glial fibers, followed by the cell soma, resulting in saltatory movements (Solecki et al. 2004). Microtubules form a cage around the nucleus and extend into the leading process of radially migrating neurons. Dynein is concentrated both in the leading process and the cell soma, and nuclear translocation to the leading process is disrupted by reduction of dynein function by RNA interference indicating that dynein activity pulls the nucleus forward during migration (Tsai et al. 2007; Vallee et al. 2009). Consistent with this, mice homozygous for the missense Loa allele of Dync1h1 have cortical lamination defects resulting from a reduction in radial migration of neurons (Ori-McKenney & Vallee 2011). Migrating OPCs also extend long leading processes and can exhibit saltatory movements (Kirby et al. 2006; Tsai et al. 2009), raising the possibility that similar motor mechanisms propel OPC and cortical neuron migration. However, in contrast to neuronal migration our time-lapse imaging revealed no obvious OPC migration defects in dync1h1vu76 mutant larvae, indicating that cytoplasmic dynein may not play a major role in OPC migration.
Upon reaching their target locations, OPCs extend numerous processes that wrap axons. Microtubules fill the main oligodendrocyte processes, oriented so that their plus ends face the membrane, whereas microfilaments fill finer processes (Lunn et al. 1997; Kachar et al. 1986; Wilson & Brophy 1989). In principle, loss of dynein function could disrupt process activity by destabilizing microtubules or disrupting motor forces required to extend processes, but our time-lapse imaging revealed no evidence for this. Process number, length and remodeling appeared normal, indicating that these activities can operate in the absence of Dync1h1 function. However, individual oligodendrocytes formed fewer sheaths on axons in dync1h1 deficient larvae and fewer intermediate size axons in the ventral spinal cord were myelinated. Time-lapse imaging revealed that some sheaths were less stable in dync1h1vu76 mutant larvae than in wild-type, whereby they became shorter in length and sometimes lost contact with axons. Thus, Dync1h1 helps maintain sheaths on axons, promoting effective myelination.
Perhaps the most surprising feature of the dync1h1vu76 mutant phenotype was the dramatic reduction of myelin gene expression. We showed previously that the deficit of myelin gene expression in Schwann cells of dync1h1 mutant larvae could be rescued by elevation of cAMP levels, raising the possibility that Dync1h1 is required for signal transduction necessary for gene expression (Langworthy & Appel 2012). Cytoplasmic dynein can transport transcription factors such as CREB (Cox et al. 2008) and NF-κB (Shrum et al. 2009; Mikenberg et al. 2007) from cytoplasm to the nucleus in response to extracellular signals and cytoplasmic dynein knockdown reduced NF-κB-dependent gene expression in various types of cells (Shrum et al. 2009; Mikenberg et al. 2007). Several signal transduction pathways are implicated in myelination by oligodendrocytes (Krämer-Albers & White 2011; Wood et al. 2013; Ishii et al. 2012; Furusho et al. 2011), but whether cytoplasmic dynein is necessary for signaling by one or more of these pathways is not known. Our data raise the possibility that one or more of these pathways requires intracellular transport by cytoplasmic dynein. One possible result of a deficit in myelin production is that myelin sheaths are less stable, as revealed by our time-lapse imaging, resulting in formation of fewer sheaths by individual oligodendrocytes.
Loss of Dync1h1 function also affected CNS axons. In particular, the average size of axons, as determined by cross sectional area, was reduced in mutant larvae relative to wild type. In principle, a reduction of axon size might account for a reduction in axon wrapping by myelin membrane. However, the size range of our intermediate class of axons exceeds the minimum diameter necessary for wrapping (Lee et al. 2012) and the average cross sectional area of axons assessed for myelin wrapping in wild-type and mutant larvae did not differ. Axons might also require Dync1h1 function to deliver molecules that promote myelination to oligodendrocytes. Because oligodendrocytes can express myelin genes in the absence of axons, this possibility seems unlikely to explain the dramatic deficit of myelination in dync1h1 mutant larvae. Nevertheless, discriminating between potential roles of Dync1h1 in axons and oligodendrocytes that are important for myelination will require cell-type specific mutation.
In summary, our investigation reveals that dync1h1-deficient zebrafish larvae have a substantial deficiency of myelination, resulting from the combined effects of fewer oligodendrocytes, fewer axon sheaths and abnormally low myelin gene expression. Our work should now call attention to the possibility that myelin abnormalities contribute to the intellectual disability associated with DYNC1H1 mutations.
EXPERIMENTAL PROCEDURES
Ethics Statement
The animal work in this study was approved by the Institutional Animal Care and Use Committees of Vanderbilt University and the University of Colorado School of Medicine.
Zebrafish Lines and Husbandry
Embryos were raised at 28.5°C in egg water of embryo medium (EM) and staged according to hours post fertilization (hpf), days post fertilization (dpf) and morphological criteria (Kimmel et al. 1995). The dync1h1vu76 mutation was uncovered in an ENU mutagenesis screen. The dync1h1hi3684Tg was uncovered in an insertional mutagenesis screen (Amsterdam et al. 2005) and kindly provided by Adam Amsterdam and Nancy Hopkins. Tg(olig2:EGFP)vu12 (Shin et al. 2003), Tg(nkx2.2a:EGFP-CaaX)vu16 (Kirby et al. 2006; Ng et al. 2005) and Tg(olig2:DsRed2)vu19 (Kucenas et al. 2008) fish were used for this study.
Positional Cloning of dync1h1vu76
We created a mapping cross by mating vu76+/− fish, which were from the AB strain, to WIK strain fish and raising the progeny to adulthood. 24 each of 5 dpf wild-type and vu76 mutant larvae were collected from crosses of identified vu76+/− map cross fish and mixed genomic DNA pools were prepared. By bulked segregant analysis using 223 simple sequence length polymorphism (SSLPs) markers, we linked the vu76 mutation to markers z6010, z1990, z34756, G45581, G39834, z7295 and G45067, located on chromosome 17. We next tested 2853 individual mutant larvae for linkage with these markers. z6010, located at 17: 29478–29606, was most tightly linked, with 0 recombinants. The entire coding region of dync1h1 was sequenced from PCR products amplified in seven overlapping fragments from cDNA prepared from 4 dpf vu76 mutant and wild-type larvae.
Melanosome Aggregation Assay
At 3 dpf, wild-type and dync1h1vu76 larvae were dark adapted for maximal melanophore dispersion and mounted, dorsal side upwards, in 4% methyl cellulose (1500 centipoises, Sigma-Aldrich, St. Louis, MO, USA) submerged in EM on a depression slide. After taking images of melanocytes in the head region, we discarded EM on the slide and applied epinephrine (0.5 mg/ml, Sigma-Aldrich, St. Louis, MO, USA) dissolved in EM (Yen et al. 2006). Images were taken every 5 min after epinephrine treatment.
Morpholino Injections
We purchased a previously described antisense MO (designed to block transcription of dync1h1 and consisting of the sequence 5′-CGCCGCTGTCAGACATTTCCTACAC-3′ (Insinna et al. 2010) from Gene tools, LLC (Corvallis, OR, USA). To prevent off-target effects, we performed co-injections with a p53 translational blocking MO consisting of the sequence 5′-GCGCCATTGCTTTGCAAGAATTG-3′ (Robu et al. 2007). MOs were dissolved in water and diluted prior to injections. We injected 1–2 nL of the MO into the yolk at the 1 cell stage. All MO-injected embryos were raised in EM at 28.5°C.
In situ RNA hybridization, Immunohistochemistry and Western Blotting
plp1a, mbp (Brosamle & Halpern 2002) and cldnk (Roberts & Appel 2009) RNA probes were generated using digoxigenin RNA labeling kits (Roche). In situ RNA hybridization was performed as described previously (Hauptmann & Gerster 2000). For immunohistochemistry, larvae were fixed using 4% paraformaldehyde, embedded, frozen and sectioned using a cryostat microtome as previously described (Park & Appel 2003). We used rabbit anti-Sox10 (1:1000) (Park et al. 2005), rabbit anti-Mbp (1:200) (Kucenas et al. 2009) and mouse anti-acetylated Tubulin (T6793, 1:1000, Sigma-Aldrich, St. Louis, MO, USA) as primary antibodies. For fluorescent detection of antibody labeling, we used Alexa Fluor 568 goat anti-rabbit conjugate and Alexa Fluor 647 goat anti-mouse conjugate (1:200, Life Technologies, Grand Island, NY, USA). In situ hybridization images were collected using a QImaging Retiga Exi color CCD camera mounted on a compound microscope and imported into Adobe Photoshop. Image manipulations were limited to levels, curve and contrast adjustments. Fluorescence images were collected using a Zeiss LSM510 laser scanning confocal microscope and imported into Adobe Photoshop.
For Western blotting 50 larvae of each genotype were homogenized in sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.2% bromophenol blue) and the equivalent of 2 larvae were loaded per lane on a 4%–15% Tris-HCl ready SDS polyacrylamide gel (Bio-Rad Life Science, Hercules, CA, USA). After transfer the blot was cut and incubated with rabbit anti-Dync1h1 antibody (a generous gift from Richard Vallee) and anti α-Tubulin (Sigma-Aldrich, St. Louis, MO, USA). Blots were developed using an ECL kit (Bio-Rad Life Science, Hercules, CA, USA).
Time-lapse Imaging
For analysis of axon wrapping, 2.5 dpf wild-type Tg(nkx2.2a:mEGFP) and dync1h1vu76;Tg(nkx2.2a:mgfp) larvae were anesthetized using 3-aminobenzoic acid ethyl ester (Tricaine), immersed in 0.8% low melting temperature agarose, mounted on their sides and covered with EM in glass bottomed 35 mm Petri dishes. Time-lapse images were captured using a 40× oil immersion objective mounted on a motorized Zeiss Axiovert 200 microscope equipped with a PerkinElmer spinning disk confocal system and heated stage and chamber to maintain larve at 28.5°C. Z image stacks were collected every 5 minutes and 3D data sets compiled using Sorenson 3 video compression and exported using QuickTime to create movies. For analysis of OPC migration and division, 2 dpf wild-type Tg(olig2:EGFP) and dync1h1 MO-injected Tg(olig2:EGFP) embryos were anesthetized using 3-aminobenzoic acid ethyl ester, immersed in 0.8% lower melting temperature agarose, mounted on their sides, and covered with EM in a glass bottomed 35mm Petri dish. Time-lapse images were captured using a 20× objective on the imaging system as above. Z image stacks were collected every 12 minutes from 48 hpf to 58 hpf and 3D data sets compiled as above.
Myelin Sheath Length Measurements
We measured sheath length in larvae injected with approximately 10 pg of the plasmid pEXPR-Tol2-sox10:EGFP-CaaX at the one cell stage. Embryos were screen for fluorescent reporter expression at 6 dpf, anesthetized using 3-aminobenzoic acid ethyl ester, immersed in 0.8% lower melting temperature agarose, mounted on their sides, and covered with EM in a glass bottomed 35mm Petri dish. Z stack images of well-isolated oligodendrocytes were captured using a 40× oil immersion objective mounted on a motorized Zeiss Axiovert 200 microscope equipped with a PerkinElmer spinning disk confocal system. Volocity software was used to measure the length of each sheath. P values for the average number of sheaths per cell and average length of sheaths were calculated using nonparametric Mann-Whitney U-test statistical analysis. The minimum and maximum sheath length for each cell was graphically depicted in a box-and-whiskers plot to indicate the mean and extrema using Prism 6.0 software. The total sheath length per cell was calculated by the addition of all sheath lengths associated with a single cell.
Quantitative PCR
RNA was isolated from 20 pooled larvae for each control or experimental condition. RNA isolation for each experiment was performed in triplicate. Reverse transcription was performed using iScript Reverse Transcriptase Supermix (#170-8840, Bio-Rad Life Science, Hercules, CA, USA). Real-time qPCR was performed in triplicate for each cDNA sample using an Applied Biosystems StepOne Plus machine and software version 2.1. Taqman Gene Expression Assays were used to detect mpz (Dr03131917_m1), plp1a (Dr03433493_g1), 36k (Dr03438574_g1) and rpl13a (Dr03101115_g1) as an endogenous control. A custom designed assay to detect mbp consisted of the primers mbp-“A” forward, 5′-GTTCTTCGGAGGAGACAAGAAGAG-3′; mbp-“A” reverse, 5′-GTCTCTGTGGAGAGGAGGATAGATGA-3′; mbp-“a” probe, 5′-AAGGGAAAGGGTTCATT-3′.
Electron Microscopy
At 6 dpf, zebrafish larvae were anesthetized with Tricaine and fixed in a solution of paraformaldehyde, glutaraldehyde and sodium cacodylate followed by osmium fixation using osmium tetroxide and sodium cacodylate as previously described (Langworthy & Appel 2012). Reagents were purchased from Electron Microscopy Sciences (Hatfield, PA, USA). 50–70 nm sections were imaged on a FEI Technai Biotwin microscope with a Gatan Ultrascan camera. Axon area was measured using ImageJ software (NIH). Large axons were defined as measuring >4 μm, intermediate as 0.201–3.99 μm and small as <0.2 μm.
Key findings.
Loss of cytoplasmic dynein function results in fewer oligodendrocytes and fewer myelin sheaths on axons in zebrafish.
Loss of cytoplasmic dynein results in a substantial deficit of myelin gene expression.
Axons of similar size have fewer myelin membrane wraps in dynein mutant zebrafish larvae than in wild type.
Acknowledgments
We thank Adam Amsterdam and Nancy Hopkins for providing the dync1h1hi3684Tg allele, Richard Vallee for providing anti-Dync1h1 antibody, Mary Goll, Michael Nonet, and Chi-Bin Chien for Gateway plasmids, Dot Dill for electron microscopy support and Sarah Casper for performing Western blotting. The University of Colorado Anschutz Medical Campus Zebrafish Core Facility was supported by the Rocky Mountain Neurological Disorders Core Center (NIH P30 NS048154). The electron micrographs were generated in the EM core facility of the Department of Cell and Developmental Biology, supported by NIH grant P30 NS08154.
Grant sponsor and number: This work was supported by NIH Grant RO1 NS046668 and the Gates Frontiers Fund. M.L.Y. was supported by a Research Award from the Children’s Hospital Colorado Research Institute and a Colorado Clinical and Translational Sciences Institute Neuroscience Co-Pilot award from the University of Colorado Denver. J.H.H. was supported by a National Institutes of Mental Health T32 MN015442 Fellowship and a National Multiple Sclerosis Postdoctoral Fellowship (FG 2024-A-1).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Author contributions: B.A. designed the study and wrote the manuscript. M.L.Y. performed the complementation tests, time-lapse imaging and myelin sheath length experiments. J.S. identified the vu76 mutant allele, performed genetic mapping and initial phenotype characterizations including the time-lapse imaging shown in Figure 4. H.S. provided OPC quantitative data shown in Figure 3. M.L.L. performed the in situ RNA hybridizations shown in Figure 5. C.A.K. performed qPCR and electron microscopy experiments. M.B.W. sequenced dync1h1 cDNA.
References
- Ainger K, et al. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. The Journal of cell biology. 1993;123(2):431–41. doi: 10.1083/jcb.123.2.431. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2119827&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, et al. Submission and curation of data from an insertional mutagenesis screen. ZFIN Direct Data Submission 2005 [Google Scholar]
- Barkovich aJ, et al. A developmental and genetic classification for malformations of cortical development: update 2012. Brain : a journal of neurology. 2012;135(Pt 5):1348–69. doi: 10.1093/brain/aws019. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3338922&tool=pmcentrez&rendertype=abstract [Accessed May 23, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39(1):47–57. doi: 10.1002/glia.10088. Available at: http://www.ncbi.nlm.nih.gov/htbin-post/Entrez/query?db=m&form=6&dopt=r&uid=12112375. [DOI] [PubMed] [Google Scholar]
- Busson S, et al. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Current biology : CB. 1998;8(9):541–4. doi: 10.1016/s0960-9822(98)70208-8. Available at: http://www.ncbi.nlm.nih.gov/pubmed/9560347 [Accessed May 20, 2013] [DOI] [PubMed] [Google Scholar]
- Carson JH, et al. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil Cytoskeleton. 1997;38(4):318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9415374. [DOI] [PubMed] [Google Scholar]
- Clark TG, Rosenbaum JL. Pigment particle translocation in detergent-permeabilized melanophores of Fundulus heteroclitus. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(15):4655–9. doi: 10.1073/pnas.79.15.4655. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=346734&tool=pmcentrez&rendertype=abstract [Accessed December 30, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman DR, et al. Synthesis and incorporation of myelin polypeptides into CNS myelin. J Cell Biol. 1982;95(2 Pt 1):598–608. doi: 10.1083/jcb.95.2.598. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=6183276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Courchesne SL, Segal RA. Action in the axon: generation and transport of signaling endosomes. Curr Opin Neurobiol. 2008;18(3):270–275. doi: 10.1016/j.conb.2008.08.005. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18778772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, et al. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10(2):149–159. doi: 10.1038/ncb1677. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18193038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T, Ffrench-Constant C, Lyons Da. Individual Oligodendrocytes Have Only a Few Hours in which to Generate New Myelin Sheaths In Vivo. Developmental cell. 2013;25(6):599–609. doi: 10.1016/j.devcel.2013.05.013. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23806617 [Accessed August 7, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach J, Dupuis L. Cytoplasmic dynein in neurodegeneration. Pharmacology & therapeutics. 2011;130(3):348–63. doi: 10.1016/j.pharmthera.2011.03.004. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21420428 [Accessed November 28, 2012] [DOI] [PubMed] [Google Scholar]
- Furusho M, et al. Fibroblast growth factor signaling is required for the generation of oligodendrocyte progenitors from the embryonic forebrain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(13):5055–66. doi: 10.1523/JNEUROSCI.4800-10.2011. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21451043 [Accessed April 4, 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology. 2012;78(22):1714–20. doi: 10.1212/WNL.0b013e3182556c05. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22459677 [Accessed November 28, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Multicolor whole-mount in situ hybridization. Methods Mol Biol. 2000;137:139–148. doi: 10.1385/1-59259-066-7:139. Available at: http://www.ncbi.nlm.nih.gov/htbin-post/Entrez/query?db=m&form=6&dopt=r&uid=10948532. [DOI] [PubMed] [Google Scholar]
- Hendricks AG, et al. Dynein tethers and stabilizes dynamic microtubule plus ends. Current biology : CB. 2012;22(7):632–7. doi: 10.1016/j.cub.2012.02.023. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22445300 [Accessed March 9, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. The Journal of cell biology. 2001;155(7):1159–72. doi: 10.1083/jcb.200105093. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2199338&tool=pmcentrez&rendertype=abstract [Accessed March 11, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, et al. Analysis of a zebrafish dync1h1 mutant reveals multiple functions for cytoplasmic dynein 1 during retinal photoreceptor development. Neural Dev. 2010;5:12. doi: 10.1186/1749-8104-5-12. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20412557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, et al. ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(26):8855–64. doi: 10.1523/JNEUROSCI.0137-12.2012. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3521511&tool=pmcentrez&rendertype=abstract [Accessed June 2, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachar B, Behar T, Dubois-Dalcq M. Cell shape and motility of oligodendrocytes cultured without neurons. Cell Tissue Res. 1986;244(1):27–38. doi: 10.1007/BF00218378. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3516402. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, et al. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- King SM, et al. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. The Journal of biological chemistry. 1996;271(32):19358–66. doi: 10.1074/jbc.271.32.19358. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8702622. [DOI] [PubMed] [Google Scholar]
- Kirby BB, et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9(12):1506–1511. doi: 10.1038/nn1803. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17099706. [DOI] [PubMed] [Google Scholar]
- Knapik EW, et al. A microsatellite genetic linkage map for zebrafish (Danio rerio) Nature genetics. 1998;18(4):338–43. doi: 10.1038/ng0498-338. Available at: http://www.ncbi.nlm.nih.gov/pubmed/9537415 [Accessed November 15, 2012] [DOI] [PubMed] [Google Scholar]
- Krämer-Albers EM, White R. From axon-glial signalling to myelination: the integrating role of oligodendroglial Fyn kinase. Cellular and molecular life sciences : CMLS. 2011;68(12):2003–12. doi: 10.1007/s00018-010-0616-z. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21207100 [Accessed November 13, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S, et al. A selective glial barrier at motor axon exit points prevents oligodendrocyte migration from the spinal cord. J Neurosci. 2009;29(48):15187–15194. doi: 10.1523/JNEUROSCI.4193-09.2009. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19955371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S, et al. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat Neurosci. 2008;11(2):143–151. doi: 10.1038/nn2025. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18176560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langworthy MM, Appel B. Schwann cell myelination requires Dynein function. Neural development. 2012;7:37. doi: 10.1186/1749-8104-7-37. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3520773&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, et al. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nature methods. 2012;9(9):917–22. doi: 10.1038/nmeth.2105. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3433633&tool=pmcentrez&rendertype=abstract [Accessed January 27, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn KF, Baas PW, Duncan ID. Microtubule organization and stability in the oligodendrocyte. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17(13):4921–32. doi: 10.1523/JNEUROSCI.17-13-04921.1997. Available at: http://www.ncbi.nlm.nih.gov/pubmed/9185530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons Da, et al. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nature genetics. 2009;41(7):854–8. doi: 10.1038/ng.376. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2702462&tool=pmcentrez&rendertype=abstract [Accessed November 14, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikenberg I, et al. Transcription factor NF-kappaB is transported to the nucleus via cytoplasmic dynein/dynactin motor complex in hippocampal neurons. PloS one. 2007;2(7):e589. doi: 10.1371/journal.pone.0000589. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1899224&tool=pmcentrez&rendertype=abstract [Accessed May 26, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nature reviews Neuroscience. 2013;14(3):161–176. doi: 10.1038/nrn3380. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23361386 [Accessed February 15, 2013] [DOI] [PubMed] [Google Scholar]
- Ng AN, et al. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286(1):114–135. doi: 10.1016/j.ydbio.2005.07.013. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16125164. [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Vallee RB. Neuronal migration defects in the Loa dynein mutant mouse. Neural development. 2011;6(1):26. doi: 10.1186/1749-8104-6-26. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3127822&tool=pmcentrez&rendertype=abstract [Accessed March 14, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Appel B. Delta-Notch signaling regulates oligodendrocyte specification. Development. 2003;130:3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- Park HC, et al. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J Neurosci. 2005;25(29):6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16033893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nature genetics. 2013;45(6):639–647. doi: 10.1038/ng.2613. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23603762 [Accessed May 23, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers Ja, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. The Journal of Cell Biology. 2013;201(2):201–215. doi: 10.1083/jcb.201208098. Available at: http://www.jcb.org/cgi/doi/10.1083/jcb.201208098 [Accessed April 15, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RK, Appel B. Apical polarity protein PrkCi is necessary for maintenance of spinal cord precursors in zebrafish. Dev Dyn. 2009;238(7):1638–1648. doi: 10.1002/dvdy.21970. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19449304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. The Journal of cell biology. 1999;146(3):597–608. doi: 10.1083/jcb.146.3.597. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2150560&tool=pmcentrez&rendertype=abstract [Accessed April 13, 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3(5):e78. doi: 10.1371/journal.pgen.0030078. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17530925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt DJ, et al. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Molecular biology of the cell. 2005;16(3):1200–12. doi: 10.1091/mbc.E04-06-0523. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=551485&tool=pmcentrez&rendertype=abstract [Accessed April 13, 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nature cell biology. 2000;2(12):922–30. doi: 10.1038/35046574. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11146657 [Accessed May 20, 2013] [DOI] [PubMed] [Google Scholar]
- Shin J, et al. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003;25(1–2):7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14739582. [DOI] [PubMed] [Google Scholar]
- Shrum CK, Defrancisco D, Meffert MK. Stimulated nuclear translocation of NF-kappaB and shuttling differentially depend on dynein and the dynactin complex. Proc Natl Acad Sci U S A. 2009;106(8):2647–2652. doi: 10.1073/pnas.0806677106. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19196984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Trotter J. Wrapping it up: the cell biology of myelination. Current opinion in neurobiology. 2007;17(5):533–40. doi: 10.1016/j.conb.2007.08.003. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17923405 [Accessed March 14, 2013] [DOI] [PubMed] [Google Scholar]
- Smith DS, et al. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nature cell biology. 2000;2(11):767–75. doi: 10.1038/35041000. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11056530 [Accessed December 12, 2012] [DOI] [PubMed] [Google Scholar]
- Solecki DJ, et al. Par6alpha signaling controls glial-guided neuronal migration. Nature neuroscience. 2004;7(11):1195–203. doi: 10.1038/nn1332. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15475953 [Accessed May 25, 2013] [DOI] [PubMed] [Google Scholar]
- Song J, et al. RNA transport in oligodendrocytes from the taiep mutant rat. Molecular and Cellular Neuroscience. 2003;24(4):926–938. doi: 10.1016/s1044-7431(03)00254-9. Available at: http://linkinghub.elsevier.com/retrieve/pii/S1044743103002549 [Accessed November 28, 2012] [DOI] [PubMed] [Google Scholar]
- Trapp BD, et al. Spatial segregation of mRNA encoding myelin-specific proteins. Proc Natl Acad Sci U S A. 1987;84(21):7773–7777. doi: 10.1073/pnas.84.21.7773. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3478726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Macklin WB, Miller RH. Distinct modes of migration position oligodendrocyte precursors for localized cell division in the developing spinal cord. Journal of neuroscience research. 2009;87(15):3320–30. doi: 10.1002/jnr.22058. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2861839&tool=pmcentrez&rendertype=abstract [Accessed May 25, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nature neuroscience. 2007;10(8):970–9. doi: 10.1038/nn1934. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17618279 [Accessed May 22, 2013] [DOI] [PubMed] [Google Scholar]
- Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends in cell biology. 2009;19(7):347–55. doi: 10.1016/j.tcb.2009.03.009. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2844727&tool=pmcentrez&rendertype=abstract [Accessed March 30, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, et al. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. The Journal of cell biology. 2008;182(6):1045–54. doi: 10.1083/jcb.200710106. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2542467&tool=pmcentrez&rendertype=abstract [Accessed March 6, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, et al. A de novo paradigm for mental retardation. Nature genetics. 2010;42(12):1109–12. doi: 10.1038/ng.712. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21076407 [Accessed November 4, 2012] [DOI] [PubMed] [Google Scholar]
- Weedon MN, et al. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. American journal of human genetics. 2011;89(2):308–12. doi: 10.1016/j.ajhg.2011.07.002. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3155164&tool=pmcentrez&rendertype=abstract [Accessed July 17, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, et al. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. Journal of medical genetics. 2012;49(3):179–83. doi: 10.1136/jmedgenet-2011-100542. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22368300 [Accessed November 8, 2012] [DOI] [PubMed] [Google Scholar]
- Wilson R, Brophy PJ. Role for the oligodendrocyte cytoskeleton in myelination. J Neurosci Res. 1989;22(4):439–448. doi: 10.1002/jnr.490220409. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=2474666. [DOI] [PubMed] [Google Scholar]
- Wood TL, et al. mTOR: a link from the extracellular milieu to transcriptional regulation of oligodendrocyte development. Asn Neuro. 2013;5(1):63–79. doi: 10.1042/AN20120092. Available at: http://www.asnneuro.org/an/005/an005e108.htm [Accessed April 22, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HJ, et al. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Human molecular genetics. 2006;15(5):667–77. doi: 10.1093/hmg/ddi468. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16399798 [Accessed March 18, 2013] [DOI] [PubMed] [Google Scholar]
- Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6(8):615–625. doi: 10.1038/nrn1727. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16062170. [DOI] [PubMed] [Google Scholar]


