Abstract
Resveratrol (RSV) is reported to extend life span1,2 and provide cardio-neuro-protective3, anti-diabetic4, and anti-cancer effects3,5 by initiating a stress response2 that induces survival genes. Because human tyrosyl tRNA synthetase (TyrRS) translocates to the nucleus under stress conditions6, we considered the possibility that the tyrosine-like phenolic ring of RSV might fit into the active site pocket to effect a nuclear role. Here we present a 2.1Å co-crystal structure of RSV bound to the active site of TyrRS. RSV nullified the catalytic activity and redirected TyrRS to a nuclear function, stimulating NAD+-dependent auto-poly-ADP-ribosylation of PARP-1. Downstream activation of key stress signaling pathways were causally connected to TyrRS-PARP-1-NAD+ collaboration. This collaboration was also demonstrated in the mouse, and was specifically blocked in vivo by a RSV-displacing tyrosyl adenylate analog. In contrast to functionally diverse tRNA synthetase catalytic nulls created by alternative splicing events that ablate active sites7, here a non-spliced TyrRS catalytic null reveals a new PARP-1- and NAD+-dependent dimension to the physiological mechanism of RSV.
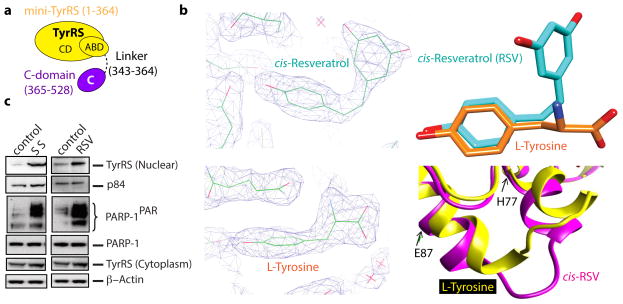
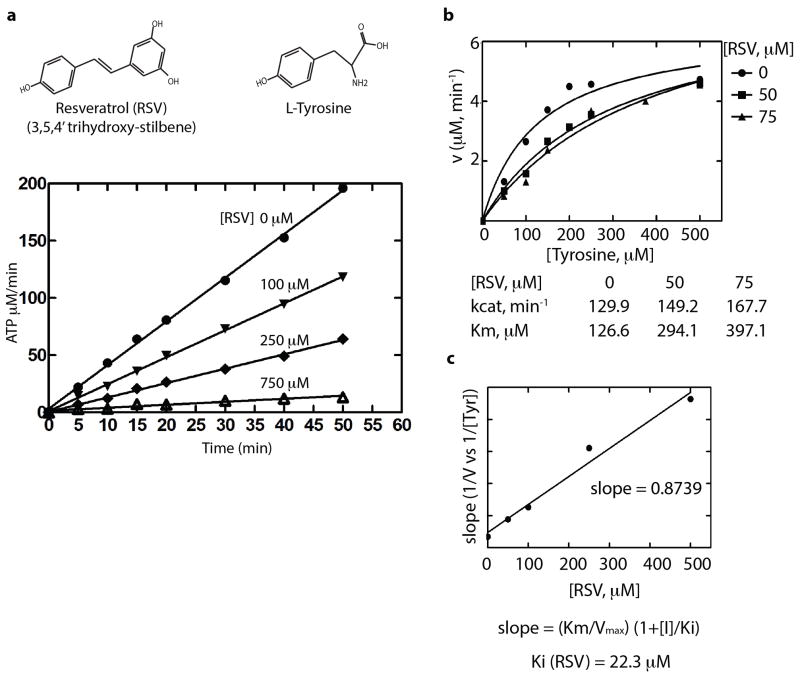
Human TyrRS is a homodimer of a 528 amino acid polypeptide that harbors an appended eukaryote-specific C-terminal EMAP-II domain (Fig. 1a)8,9. High-resolution structures of the catalytic unit of Hs TyrRS, known as mini-TyrRS, and the C-domain have been determined10,11. We found RSV strongly inhibited TyrRS with a Ki-value of 22 μM (Extended Data Fig. 1a–c). Crystallization of Hs mini-TyrRS with RSV and, separately, with tyrosine yielded co-crystal structures (at 2.1Å) (Fig. 1b and Extended Data Fig. 2a,b Extended Data Table 1, PDB ID code 4Q93 and 4QBT).
Figure 1. Resveratrol binds at the active site of TyrRS.
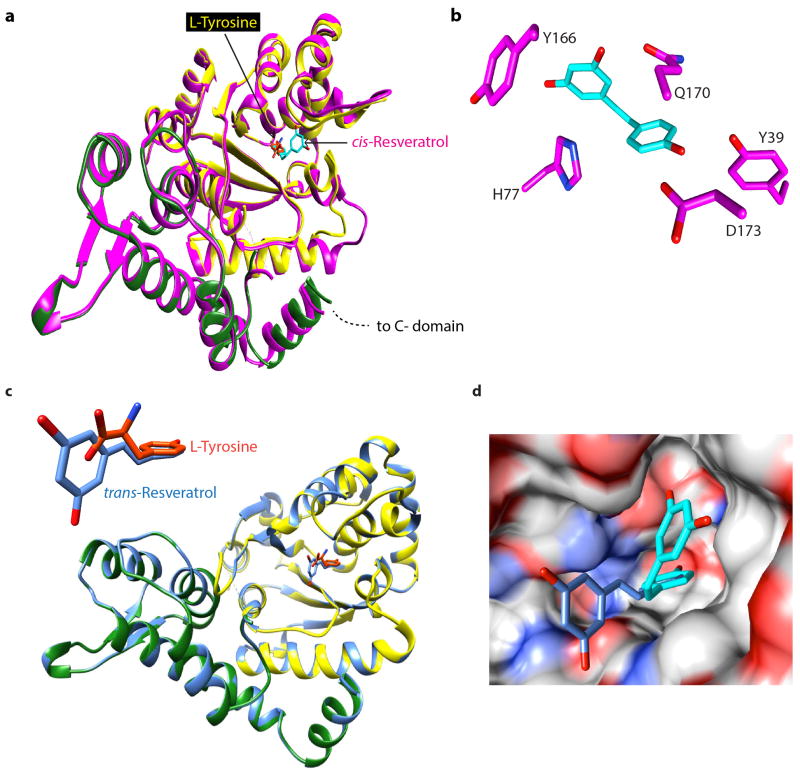
a, Cartoon illustration of the domain organization of Hs TyrRS. Both domains are connected by a linker of ~20 amino acids. b, Left, Electron density of co-crystal x-ray structures (2.1 A°) of TyrRS bound to cis-resveratrol and to L-tyrosine. Right, Resveratrol induced a local conformational change relative to bound tyrosine at the active site. c, Both serum starvation and resveratrol treatment (5 μM) facilitated the nuclear translocation of TyrRS with a concomitant increase in the PARylation of PARP-1.
While the phenolic ring of RSV and of the tyrosine have the same disposition in the respective co-crystals, accommodation of the cis-conformation of the dihydroxy ring of RSV forces a local structural change near the linker to the C-domain (Fig. 1b, Extended Data Fig. 2a). An RSV-promoted conformational change in TyrRS may drive the predominant trans RSV (in solution) into a cis conformation. (Extended Data Fig. 2c,d).
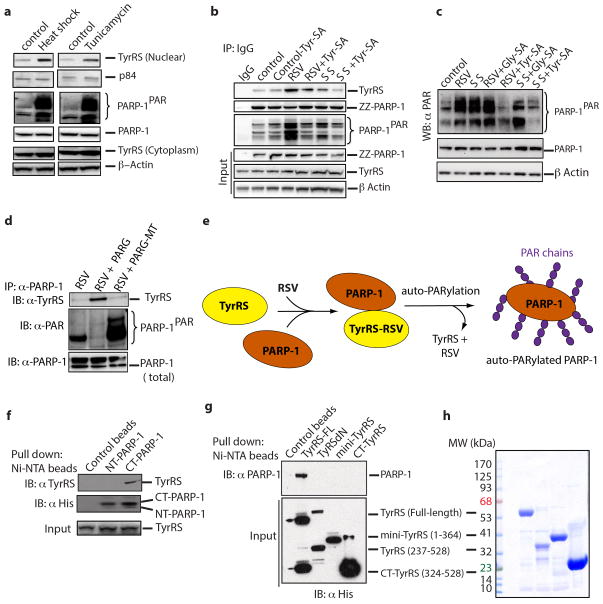
Associated with a previous study12, a distinct TyrRS-PARP-1 interaction was observed. PARP-1 is a major modulator of NAD+ metabolism and its related signaling13. Because RSV acts through NAD+-dependent proteins14, the TyrRS-PARP-1 interaction was further studied. Given that RSV treatment elicits a stress response2, serum starvation (SS) was used to mimic a general ‘stand-alone’ stress condition so that common signaling pathways, if any, between RSV treatment and a general stress condition, could be compared ex-vivo. Either serum starvation or RSV treatment promoted nuclear translocation of endogenous TyrRS in HeLa cells (Fig. 1c). Translocation was observed under different stress conditions (heat shock and ER stress, Extended Data Fig. 3a), suggesting that TyrRS is a general stress transducer. Nuclear translocation of endogenous TyrRS was concomitant with strong auto-PARylation of PARP-1 (PARP-1PAR) (Fig. 1c and Extended Data Fig. 3a).
Ex-vivo RSV also strongly promoted association of TyrRS with PARP-1, and robust auto-poly-ADP-ribosylation of PARP-1 (Extended Data Fig. 3b). Effects of RSV were blocked by a Tyr-AMP analog (Tyr-SA (5′-O-[N-(9L-tyrosyl) sulfamoyl] adenosine)), but not by Gly-SA (a control targeting GlyRS) (Extended Data Fig. 3b,c). Similar, but less pronounced, PARylation was seen with serum starvation. Enhanced PARylation correlated with increased amounts of TyrRS in the nucleus, which occurred upon serum starvation. Thus, both serum starvation and RSV promoted nuclear translocation of TyrRS and activation of PARP-1.
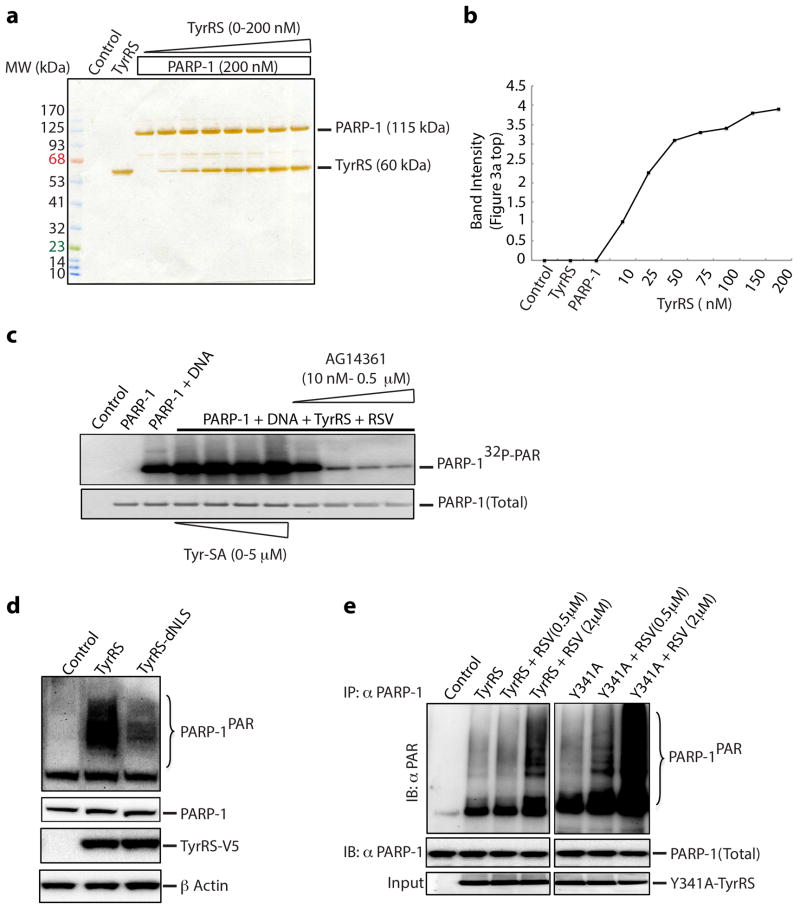
Cell lysates treated with the PARG hydrolyase and its hydrolase-inactive mutant supported that TyrRS preferentially bound to non-PARylated PARP-1 (Extended Data Fig. 3d,e). TyrRS interacted specifically with the C-domain of PARP-1 (CT-PARP-1) (Extended Data Fig. 3f). Neither mini-TyrRS nor the TyrRS C-domain interacted with PARP-1; only full-length native TyrRS bound PARP-1 (Extended Data Fig. 3g,h). In the absence of RSV, concentration-dependent activation of PARP-1 by TyrRS was observed in vitro (Fig. 2a, top, Extended Data Fig. 4a,b). RSV enhanced in-vitro auto-PARylation with the half-maximal effect at roughly 10 nM (Fig. 2a, middle), well below the Ki (about 22 μM) in Extended Data Figure 1a–c. Thus, PARP-1 may alter the apparent affinity of RSV for TyrRS. Also, concentration-dependent quenching of PARylation of PARP-1 by Tyr-SA was evident (Fig. 2a, bottom). Lastly, while broken DNA normally activates PARP-113, Tyr-SA did not interfere with this DNA-dependent-pathway of PARP-1 activation in vitro (Extended Data Fig. 4c). Therefore, TyrRS-RSV activation of PARP-1 is distinct.
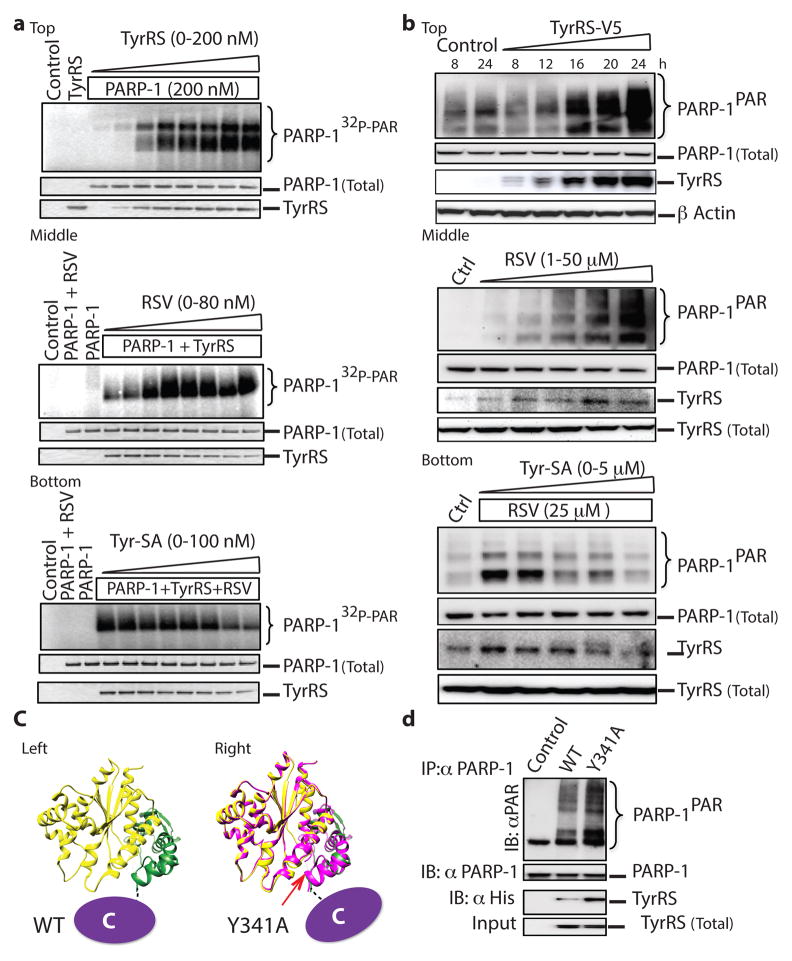
Figure 2. TyrRS facilitates the activation of PARP-1 in an active-site-dependent manner.
a, top, TyrRS activates PARP-1 in an in vitro assay. a, middle. Resveratrol potentiates TyrRS mediated activation of PARP-1. a, bottom. Tyr-SA blocks the resveratrol-mediated activation of PARP-1. b, top. TyrRS-V5 overexpression activates PARP-1 in HeLa cells in a concentration-dependent manner. b, middle. Resveratrol treatment activates PARP-1 in HeLa cells and enhances TyrRS interaction with PARP-1. b, bottom. Tyr-SA blocks the resveratrol-mediated interaction of TyrRS and activation of PARP-1. c, Cartoon illustration of the C-domain disposition in TyrRS (left) and Y341ATyrRS (right). d. Y341ATyrRS enhances its interaction and activates PARP-1 compared to WT.
Ectopically expressed TyrRS in HeLa cells for 0–24 h caused progressive increase in cellular concentrations of the synthetase (Fig. 2b, top) and a correlated progressive increase in PARP-1PAR (Fig. 2b, top). A TyrRS mutant (TyrRS-dNLS), with reduced nuclear localization6, reduced activation of PARP-1 (Extended Data Fig. 4d). Effects of RSV ex-vivo at various concentrations (Fig. 2b, middle) and of Tyr-SA (Fig. 2b, bottom) mirrored those seen in vitro (Fig. 2a, middle and bottom).
RSV and L-tyrosine produce different bound conformations of TyrRS, seen locally in the high-resolution structures of the two co-crystals (Fig. 1b and Extended Data Fig. 2a). These differences could affect the disposition of the C-domain needed for the interaction of TyrRS with PARP-1. This domain is tethered to the mini-TyrRS catalytic unit by a flexible linker (Fig. 2c, left). By breaking the Y341 OH--H-bond to a backbone carbonyl oxygen, a Y341A mutation releases tight tethering of the C-domain to the catalytic domain15 and its reorientation16 (Fig. 2c, right).
In the absence of RSV, Y341A-TyrRS showed robust interaction with and activation of PARP-1 in vitro (Fig. 2d). If Y341A is shifted more towards a conformation needed to bind to PARP-1, then it would be more sensitive than WT TyrRS to low concentrations of RSV. This expectation was fulfilled (Extended Data Fig. 4e). TyrRS-Tyr may thus have a conformation that prevents interaction with PARP-1, while TyrRS-RSV has a distinct conformation that binds PARP-1. In the absence of either ligand, a dynamic equilibrium populates both forms (Fig. 2c).
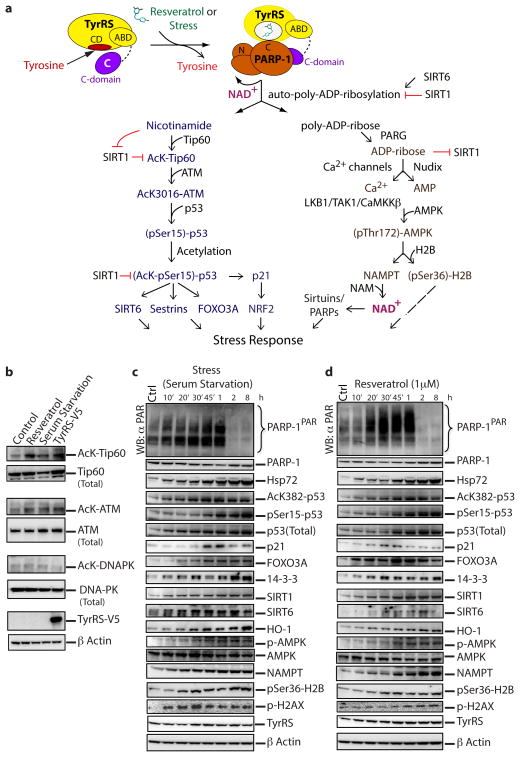
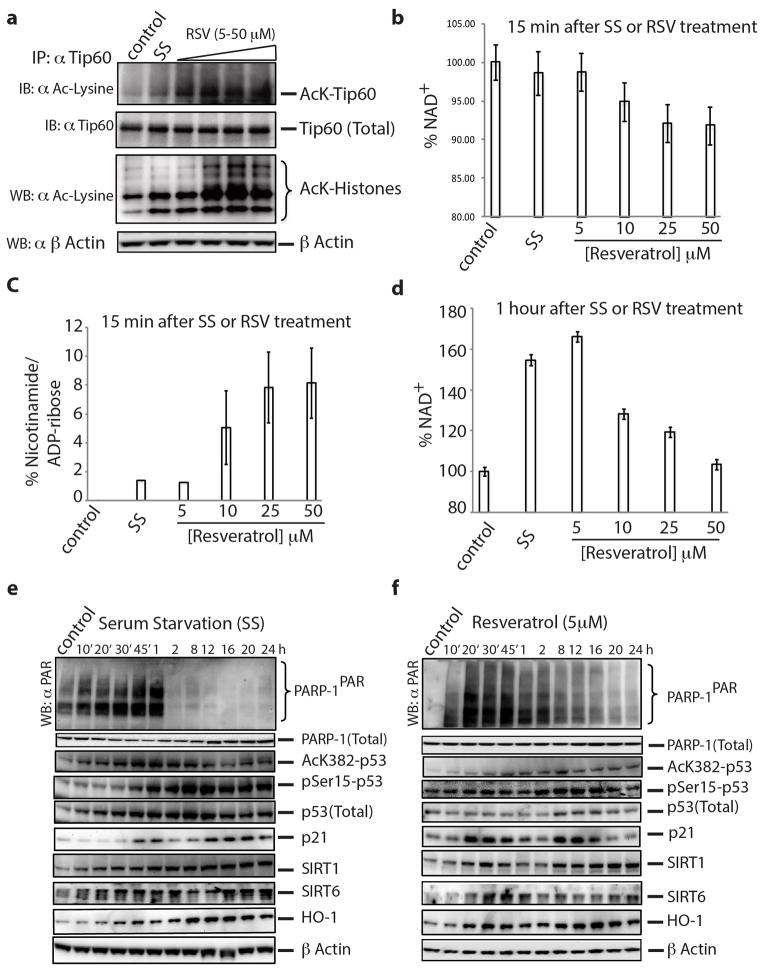
A working model of downstream signaling markers was developed as detailed in Methods (Fig. 3a). Effects from serum starvation and RSV administration involve cascades through protein acetylation and phosphorylation effects of serum starvation and of RSV, which have not been linked to TyrRS nor activation of PARP-1. Under conditions of serum starvation or RSV administration, the response in HeLa cells of PARylated and acetylated proteins, together with the aforementioned cell-signaling proteins, was monitored for up to 1h. Either RSV or serum-starvation promoted production of AcK-Tip60 and AcK-ATM (Fig. 3b). In a temporal response from 0 to 8 h, either or both of serum starvation or 1 μM RSV promoted increased levels of whole cell acetylated proteins, Hsp72, AcK382-p53, p-AMPK, pSer36-H2B, p21, FOXO3A, 14-3-3, HO-1, NAMPT, SIRT1 and SIRT6 (Fig. 3c,d). Also, RSV activated acetylation of Tip60 ex-vivo in a concentration-dependent manner (Extended Data Fig. 5a). As expected from previous reports that acetylation of p53 is indispensible for its activation17, a transient increase in acetylation of p53 (AcK382)1 was evident (Fig. 3c,d), possibly through a transient inhibition of SIRT1 by the nicotinamide18 being produced. Consistently, at 15 min, a RSV concentration-dependent reduction in total NAD+ with concomitant production of nicotinamide and ADP-ribose was observed (Extended Data Fig. 5b,c). Thus, serum starvation and RSV rapidly increased all monitored proteins in a way that is dependent on having active PARylation. This rapid increase is consistent with work showing rapid upregulation of PARP-1 target genes19,20, possibly due to a broader effect of PARP-1 on transcription and associated stress signaling13,21.
Figure 3. Resveratrol and serum starvation mimic similar downstream signaling events mediated by PARP-1 activation.
a, Cartoon illustrating the molecular basis and the integration of different signaling pathways to mediate a TyrRS/PARP-1 activated stress response evoked by either resveratrol or different stress conditions. b, PARP-1 activating conditions enhance Tip60-mediated activation of ATM. c, Time course of poly-ADP-ribosylation status and associated signaling events as depicted in figure 3a after serum starvation and (d) resveratrol (1 μM) treatment.
At longer times, and continuing at low concentrations (≤ 10 μM) of RSV, similarity between responses to RSV and serum starvation was ostensibly seen. By 1h, low concentrations of RSV elevated the NAD+ levels similar to those of serum-starvation conditions (Extended Data Fig. 5d). Responses of key stress-signaling markers in a more extended period of 8–24 hours at 5 μM RSV were mostly similar to those of serum starvation (Extended Data Fig. 5e,f). At 5 μM RSV, levels of PARP-1PAR were sustained longer when compared with what was observed at 1 μM RSV (compare 2, 8 h time points in Figure 3d with Extended Data Fig. 5f) and we thus continued further studies at 5 μM RSV.
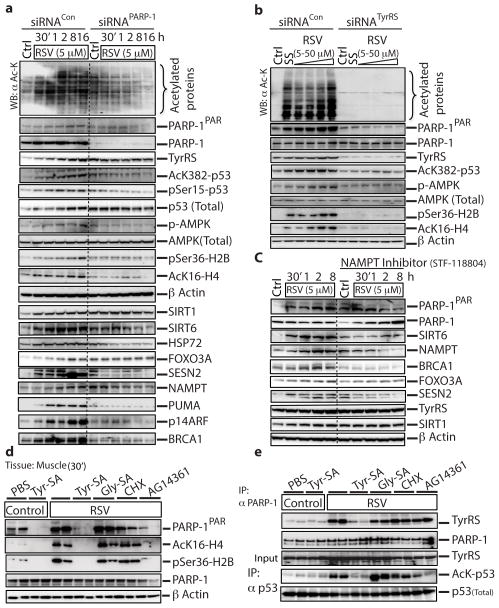
An si-RNA directed against PARP-1 (siRNAPARP-1) effectively abrogated the RSV-stimulated expression of Hsp72, p-AMPK, SIRT1, FOXO3A, SESN2, NAMPT, PUMA and SIRT6 (Fig. 4a). Also, 5 μM RSV promoted induction of BRAC1 and p14ARF (whose genes are directly regulated by PARP-122,23). Consistently, the RSV-stimulated induction of BRCA1 and p14ARF were prevented by siRNAPARP-1(Fig. 4a), further showing involvement of PARP-1 in RSV-mediated induction of these proteins.
Figure 4. Resveratrol treatment activates TyrRS-PARP-1 driven signaling events in mouse tissues.
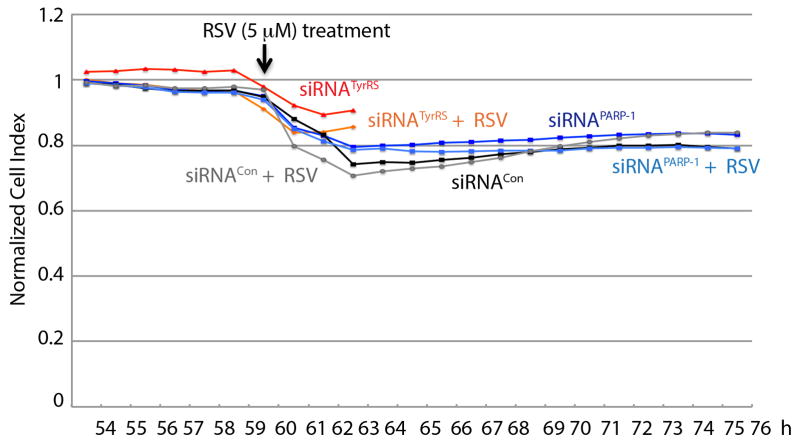
a, siRNA of PARP-1 or (b) siRNA of TyrRS abrogates resveratrol-mediated downstream signaling events. HeLa cells were treated with siRNAPARP-1 or siRNATyrRS for 60 h to knockdown PARP-1 or TyrRS expression levels by ~70–80%. Knockdown efficiency was monitored by with α-PARP-1 or α-TyrRS. For, siRNATyrRS HeLa cells were treated with either serum starvation or resveratrol (5, 10 25 and 50 μM) for 45 minutes. The dashed line in figure 4a represents the demarcation between siRNACon and siRNAPARP-1; all the samples were run on the same gel. c, NAMPT inhibition abrogates resveratrol-mediated downstream signaling events. HeLa cells were pre-treated with STF-118804 (100 nM), a NAMPT inhibitor, for 16 h and then treated with resveratrol (5 μM), with samples collected at intervals as indicated. Various signaling events, as depicted, were monitored using appropriate antibodies. Acetylation status was monitored using α-acetyl-lysine. The dashed line separates results without and with the inhibitor. d, Resveratrol mediated activation of PARP-1 is blocked by Tyr-SA. Activation of Tip60 and AMPK were monitored by using α-AcK16-H4 and α-pSer36-H2B, respectively. e, Resveratrol-mediated interaction of TyrRS with PARP-1 and acetylation of p53 are blocked by Tyr-SA. Immunoprecipitation of PARP-1 and p53 from muscle tissue demonstrated RSV-mediated TyrRS-PARP-1 interaction and p53 acetylation.
In figure 4a, RSV treatment led to enhanced activation of p53. However, in the absence of RSV, knockdown of PARP-1 resulted in a background increase in the products of these p53-regulated genes. Possibly p53 was activated by the knockdown of PARP-1, because removal of PARP-1 would enhance background levels of DNA damage. Indeed, activation of p53, as seen by enhanced acetylation and phosphorylation of p53, was observed (Fig. 4a).
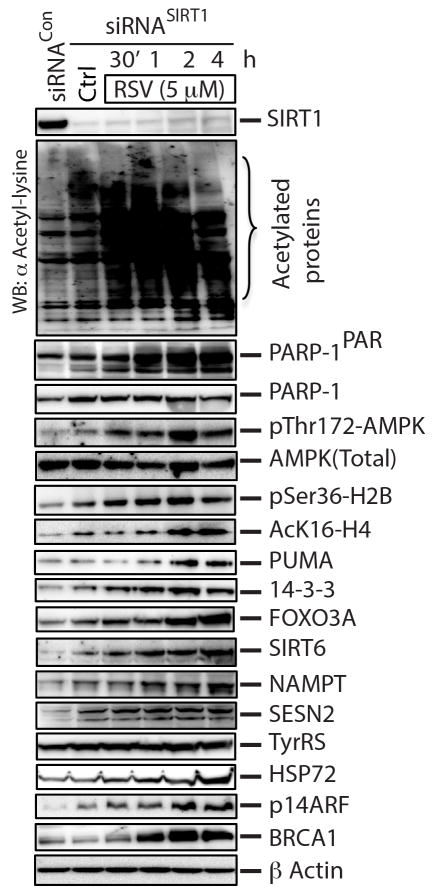
siRNATyrRS-knockdown at 5 μM RSV eliminated induction of, and dramatically reduced amounts of, PARylated PARP-1, of whole acetylated proteins, and of AcK382-p53, AcK16-H4 (Tip60 activation), p-AMPK, and pSer36-H2B (AMPK activation) (Fig. 4b). (Separately, RSV (5 μM) addition did not affect the viability of HeLa cells expressing either siRNATyrRS or siRNAPARP-1 (Extended Data Fig. 6)). In contrast, siRNASIRT1-knockdown did not affect RSV (5 μM)-mediated production of whole acetylated proteins or, among other downstream markers, activation of PARP-1 and induction of SIRT6, FOXO3A, NAMPT, AcK16-H4, p-AMPK24, and pSer36-H2B (Extended Data Fig. 7). Collectively, figures 4a, b showed that ex-vivo TyrRS and PARP-1 collaborate to activate RSV- and serum-starvation responses.
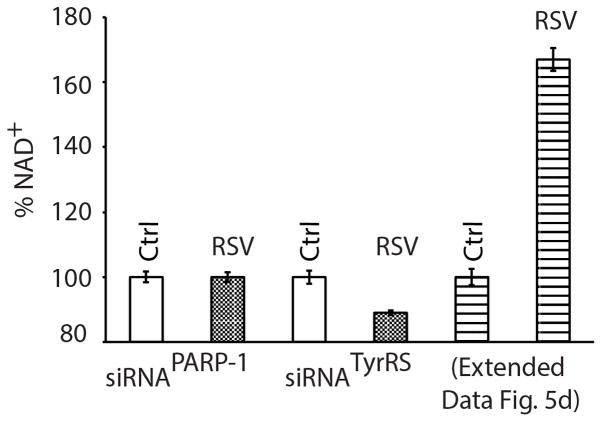
Although the RSV-TyrRS-PARP-1 axis initially consumes NAD+, at low RSV (5 μM), NAD+ levels are transiently raised after 1 h due to NAMPT activation (Extended Data Fig. 5d). At 1 h at low RSV, induction of NAD+ levels was abolished with either siRNATyrRS or siRNAPARP-1 (Extended Data Fig. 8). NAD+ depletion by NAMPT inhibition abolished RSV-mediated induction of BRCA1, FOXO3A, NAMPT, SESN2 and SIRT6 (Fig. 4c). These results are consistent with those using inhibitors against PLC-γ and the ryanodine receptor25, which inhibit PARP-1 activation26 and ADP-ribose-mediated calcium signaling25,27, and which prevented RSV-mediated AMPK activation. Similarly, a PARP-1 inhibitor prevented LKB1 activation by RSV28.
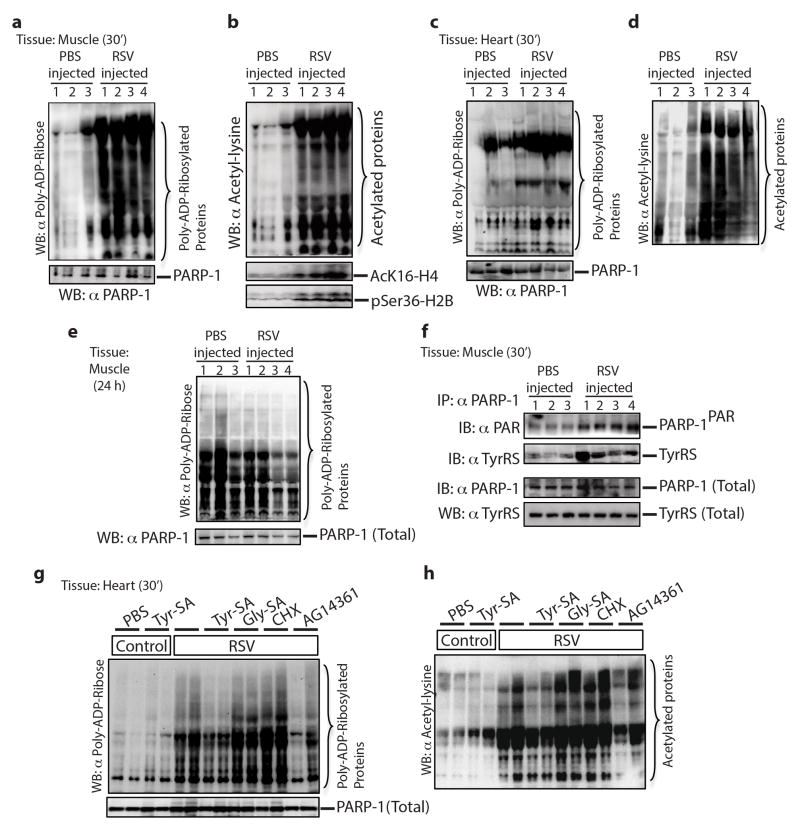
Mice were injected through tail vein-IV with 100 μl of 10 μM RSV and, after 30 min, were sacrificed. Compared to the PBS IV-injected control, PARylated and acetylated proteins, along with AcK16-H4 and pSer36-H2B, were significantly increased in skeletal muscle (Extended Data Fig. 9a,b). Results in cardiac tissue were similar (Extended Data Fig. 9c,d). In addition, PARP-1 activation diminished back to normal by 24 h (Extended Data Fig. 9e). Consistent with ex-vivo assays, tissue samples from RSV-injected mice showed higher levels of TyrRS-PARP-1 interaction together with increased auto-PARylation (Extended Data Fig. 9f).
Tyr-SA (5 μM) was added to the IV-injection with RSV. No interaction of TyrRS with PARP-1 could be detected in skeletal or cardiac muscle and levels of PARylated and acetylated proteins did not increase. In contrast, co-injection of Gly-SA or cycloheximide (CHX, protein synthesis inhibitor) did not block RSV/TyrRS-mediated activation of PARP-1 and its downstream signals (Fig. 4d (muscle), Extended Data Fig. 9g,h (cardiac)). Consistent with ex-vivo assays, immuno-precipitation of PARP-1 (from harvested muscle tissue) pulled-down TyrRS, and immuno-precipitation of p53 showed its increased acetylation. Pulldown of TyrRS and p53 acetylation were blocked in mice that had co-injections of Tyr-SA or the AG14361 (SelleckChem) PARP-1 inhibitor, but not of Gly-SA or cycloheximide (Fig. 4e). These responses in vivo parallel ex-vivo assays, and support that TyrRS is a major effector target for RSV that acts through PARP-1 as described in figure 3a.
The mechanism of action of RSV and of the stress response are both linked here to the activation of PARP-1 through TyrRS and NAD+. The interaction of RSV with TyrRS could be viewed as an example of xenohormesis through interactions of a natural ligand with a protein target29. In this instance, the natural ligand blocks the active site to create a tRNA synthetase catalytic null with a new, orthogonal function. This kind of catalytic null is in contradistinction to those created by alternative splicing events that specifically remove the active site7. The RSV-TyrRS activation of PARP-1 is readily observable in a functional in vitro assay even at sub-micromolar concentrations (e.g., 10 – 20 nM, Fig. 2a middle). Thus, TyrRS-RSV-induced PARP-1 activation appears at significantly lower RSV concentrations than seen with RSV functional binding to other targets1,25,30. As a consequence, the direct effects of RSV binding to these targets of RSV are layered over a pre-existing foundation that comes from the TyrRS-RSV-PARP-1-NAD+ connection.
METHODS
Construction of the working model as illustrated in figure 3a
Because both RSV and serum starvation activate PARP-1 leading to the metabolic breakdown of NAD+ into nicotinamide and ADP-ribose, downstream signaling markers associated with the NAD+ metabolic flux were investigated. Targets were selected if they were activated and/or inhibited under conditions of RSV-treatment, stress or NAD+ metabolic byproduct-treatment (nicotinamide and ADP-ribose). For example, nicotinamide activates Tip6031 through SIRT1 inhibition18 and Tip60 activates ATM32, the p53 kinase. p53 is a master regulator of NRF2 through p21 induction33 and inducible expression of survival genes like SIRT634, SIRT135, Sestrins36, and FOXO3A37. (SIRT6 promotes DNA repair under stress by activating PARP-138 and SIRT1 down-regulates the over-activation of PARP-139). Similarly, ADP-ribose (metabolic byproduct of NAD+ and an inhibitor of SIRT140) is a potent Ca2+ channel opener27,41 and a substrate for AMP production42 and hence an indispensible component for AMPK activation by resveratrol43 and the stress response by phosphorylating H2B44. AMPK is known to upregulate NAMPT expression45, the major regulator of NAD+ levels in the cell46. NAD+ is major substrate for sirtuins (deacetylation or mono-ADP-ribosylation38,47,48) and PARPs ((mono/poly)-ADP-ribosylation13,49). Although not depicted, p38 and NF-kB (major transducers of stress signaling) are also regulated by TAK1 in a PARP-1/ATM dependent manner50,51.
Antibodies
All antibodies were obtained from Cell Signaling Technology (Danvers, MA) unless and otherwise mentioned. α-ATM was from Abcam (Cambridge, MA) and GeneTex (San Antonio, TX). α-PARP-1 and α-poly(ADP-ribose) were from BD Biosciences (Franklin Lakes, NJ). α-p84 was from Sigma (St. Louis, MO). α-pSer36-H2B from ECM Biosciences. Rabbit α-human TyrRS polyclonal antibodies were homemade. α-Tip60 was from Millipore (Billerica, MA). α-AcK16-H4 was from Active Motif (Carlsbad, CA) and α-Hsp72 (cat # SPA-812) was from assay designs (Ann Arbor, MI).
Cell culture, transfection and resveratrol treatment
HeLa cells (from ATCC, mycoplasma free) were cultured in a humidified incubator with 5% CO2 in DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Invitrogen) and 1x penicillin/streptomycin. The cells were transfected with pcDNA6-TyrRS-V5 (wild type), or pcDNA3.1-ZZ-PARP-1 using Lipofectamine LTX (Invitrogen). Resveratrol (SantaCruz Biotechnologies), at a series of concentrations from 0 to 50 μM were used to treat HeLa cells 0–24 h in experiments described. As the effects of RSV are strongly influenced by the background level of PARP-1 activity, the experiments were done only after checking to verify that background PARP-1 activation was low. Our criterion was to see PARP-1 activation upon serum starvation, along with Hsp72 induction52. When serum starvation induces PARP-1 activation with Hsp72 induction within 30–45 min (from the same batch of a split of HeLa cells), we consistently see PARP-1 activation at lower concentrations of RSV (1μM). Cell culture medium was supplemented with Tyr-SA or Gly-SA (RNA-TEC, Coralville, IA) where mentioned. Silencer® Select Pre-designed siRNA against PARP-1 (siRNA ID#: s1099, 5′-GCAGCUUCAUAACCGAAGAtt-3′), SIRT1 (siRNA ID#: s23770, 5′-GGCUUGAUGGUAAUCAGUAtt-3′) and TyrRS (siRNA ID#: s443, 5′-GGACUUUGCUGCUGAGGUUtt-3′) were purchased from Invitrogen, Carlsbad, CA and was transfected into HeLa cells using Lipofectamine RNAiMAX (Invitrogen). NAMPT, the rate-limiting enzyme in the NAD+ salvage pathway, was inhibited using STF-11880453 according to manufactures’ protocol (SelleckChem). The effect of a concomitant increase in nicotinamide production with a reduction of NAD+ concentration (leading to SIRT1 inhibition) was monitored by immunoblotting with α-AcK382-p53, an acetylation site known to be specifically targeted by SIRT11. α-pSer15-p53 monitoring indicated if enhanced acetylation preceded the phosphorylation event54. Total p53 was blotted with α-p53. Transactivation of p53 was determined by monitoring its known targets, such as p21, PUMA, 14-3-3, FOXO3A, SESN2 and SIRT6. Activation of NRF2 was further confirmed by following HO-1 expression, using the cognate antibodies. Activation of AMPK (Thr172-phosphorylation) mediated through AMP and Ca2+ influx25 was monitored using α-pThr172-AMPK. Activation of AMPK on its targets was further determined by both α-pSer36-H2B and by the expression level of NAMPT.
Cell viability assay
HeLa (1×106) cells were reverse-transfected with siRNAs and viability was monitored using RTCA iCELLigence System (ACEA Biosciences). HeLa cells were cultured in a humidified incubator with 5% CO2 in DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Invitrogen) and 1x penicillin/streptomycin.
Preparation of cell fractions
Protocol for cell fractionation was followed as described previously12. Briefly, for whole cell lysate preparation, HeLa cells were dissolved in 1xSDS-PAGE loading buffer containing 300 mM NaCl. For cytoplasmic fraction preparation, HeLa cells were suspended in 0.1 ml of swelling buffer for 6 min on ice, and then incubated with 0.1 ml of plasma membrane lysis buffer for 5 min on ice. The cells were immediately passed through a 21-G needle 10 times and centrifuged for 10 min at 3220g, 4°C. The supernatant was harvested, while the pellets were used for the preparation of nuclear fractions. For this purpose, the pellet (nuclei) was incubated for 30 min at 4°C with 0.2 ml nuclear extraction buffer. Whenever required, PARP-1 (AG14361) and PARG inhibitors (ADP-HPD, Millipore) were added to the cell lysis buffer to ensure unwanted PARylation and removal of PAR chains with PARG.
Immuno-precipitation and Ni-NTA pull-down assays
Protocol for immuno-precipitation was followed as described previously12. Briefly, the supernatants were pre-cleared by incubation with protein G beads. The pre-cleared cell lysates were incubated at 4°C for 1 h with either α-PARP-1, α-ATM, α-DNA-PKcs, α-TyrRS, α-Tip60 or non-immune IgG, at a concentration of 5 μg/ml followed by incubation with 30 μl Protein G-beads (pretreated with 10 mg/ml BSA) at 4°C for 1h with rotation. PARP-1 (AG14361) and PARG inhibitors (ADP-HPD, Millipore) were added to the cell lysis buffer to ensure unwanted PARylation and removal of PAR chains with PARG. Immuno-precipitates were washed three times, subjected to SDS-PAGE and immunoblotted with specific antibodies. Whenever mentioned, the ZZ domain allowed immuno-precipitation of ectopically expressed ZZ-PARP-1, using anti-IgG. Whenever Ni-NTA pull-down was performed, proteins with a 6X-His tag were overexpressed in E. coli. Cells were lysed and the supernatant fractions containing the soluble proteins were mixed with HeLa cell lysates. For Ni-NTA pull-downs, normal procedures for immunoprecipitation were followed with 15–20 mM imidazole in the washing buffer.
Animal experiments
All experiments were approved by the Scripps Research Institute, IACUC (protocol number 13-0002) and conducted by the mice facility at The Scripps Research Institute. BALB/cByJ mice were originally purchased from Jackson Laboratories. Six week old male mice were kept with a 12 h light-dark cycle with free access to food and water for 3 days before conducting the experiment. For all studies, mice were dosed once and collected sample after either at 30 min or at 24 h. Briefly, activation of PARP-1 in mouse tissues treated with resveratrol was performed by the intravenous (IV) injection of a 100 μl sample of resveratrol (10 μM) in PBS into 8 mouse tails (0.012 mg/kg). A 100 μl PBS injection was used as a control in 6 mice. Tissue samples were collected from 4 resveratrol treated mice and 3 control mice after 30 min treatment. Tissue samples from the remaining mice were collected after 24h. In a different experiment, blocking of resveratrol mediated activation of PARP-1 by Tyr-SA was analyzed with 5 groups of two mice each, a 100 μl sample of resveratrol (10 μM ) in PBS alone and, separately, with Tyr-SA (5 μM), Gly-SA (5 μM), cycloheximide (5 μM) or the AG14361 PARP-1 inhibitor (10 μM) was IV-injected. A 100 μl PBS injection with Tyr-SA was used in a control group (2 mice) as indicated. Tissue samples were collected after 30 min treatment. Muscle or heart tissues samples were homogenized and analyzed by immunoblotting for the status of poly-ADP-ribosylation and associated signaling events.
Recombinant protein purification
Human TyrRS and PARP-1 and their variants were purified as previously described10,12. Briefly, DNA encoding either full-length TyrRS (aa 1-528), mini-TyrRS (aa 1-341), dN-TyrRS (aa 237-528), CT-TyrRS (aa 324-528), was cloned into Nde-I/Hind-III sites of pET-20b vector (Novagen, Gibstown, NJ). The expressed proteins include a 6-His tag from the vector sequence. The full-length PARP-1 and its variants (N-terminal domain (NTD) and C-terminal domain (CTD)) were cloned into pET 20b vector. All proteins having a C-terminal His-tag were expressed in E. coli strain BL21 (DE3) by induction for 4h with 1 mM isopropyl β-D-thiogalactopyranoside. Proteins were purified from the supernatants of lysed cells using Ni-NTA agarose (Qiagen, Chatsworth, CA) column chromatography according to the manufacturer’s instruction. We also included an additional high salt 2-column wash (1-1.5 M NaCl), to remove the endogenous DNA/RNA associated with the purified protein. The NaCl concentration was increased to 1.5M in increments and returned to final elution buffer having 250 mM NaCl in decrements. All the proteins were subjected to a gel-filtration (S-200) chromatography and collected the protein peak corresponding to homogenous protein. As endotoxin associated with purified protein interfered with the inhibitory/activating effect of RSV on TyrRS/PARP-1, the complete protein purification included an additional endotoxin removal step. The purified protein was further passed through a column containing Detoxi-Gel™ (Pierce-Thermo Scientific, IL, USA) and followed the manufactures instructions. The final endotoxin level (0.5 EU/mg/ml) was measured using The EndosafeR-PTS kit (Charles River Laboratories, Charleston, SC). The quality of each protein purification was validated by SDS-PAGE analysis.
Protein crystallization and data collection
All the steps were followed as mentioned in the previously report for mini-TyrRS crystallization10. Briefly, mini-TyrRS was mixed with either 1 mM RSV or 2 mM Tyrosine and incubated at 4°C for 16h. Before setting up the crystallization trials, the protein samples were subjected to high-speed centrifugation (13000 rpm for 15 min) to remove all the precipitants and transferred the clear soluble fraction to a new tube. The crystallization trials were done using 2.1 M (NH4)2SO4, 0.1 M NaH2PO4/K2HPO4 (pH 6–8), and 2% acetone at room temperature. Although, mini-TyrRS crystals with L-tyrosine were grown in 3 days (data collected at Stanford Synchrotron Radiation Lightsource), crystals with RSV were grown only after 3–4 months at around pH 7. X-ray data were collected at 2.1A° using the in-house x-ray diffraction facility (The Scripps Research Institute, La Jolla). Data were integrated and scaled using HKL2000. The electron density was refined by molecular replacement (CNS and CCP4/REFMAC suite and Coot) using the known structure of mini-TyrRS (Extended Data Table 1) and structures were deposited to PDB (ID code 4Q93 and 4QBT).
ATP-PPi exchange assay
Tyrosyl adenylate synthesis was measured by using the tyrosine-dependent ATP-pyrophosphate (PPi) exchange assay. A mixture containing 100 mM Hepes (pH 7.5), 20 mM KCl, 2 mM ATP, 1 mM NaPPi, 2 mM DTT, 250–500 μM L-tyrosine, 10 mM MgCl2, and 0.01 mCi/ml Na[32P]PPi was added to 50–100 nM purified TyrRS (endotoxin free, 0.5EU/mg/ml), pre-incubated with 0–1mM RSV at 4°C for 30 min. The ATP-PPi exchange reaction was incubated at room temperature, and aliquots were removed at specified time intervals and quenched in a mixture containing 40 mM NaPPi, 1.4% HClO4, 0.4% HCl, and 8% (wt/vol) of activated charcoal. After thoroughly mixing, the charcoal was filtered and washed with a solution of 7% HClO4 and 200 mM NaPPi using Spin-X Centrifuge Filters (Corning, Corning, NY) containing 0.45-μm pore-size cellulose acetate filters. After drying, the charcoal was punched into scintillation vials and the radioactivity of the ATP bound to the charcoal mixture was measured by scintillation counting.
In vitro PARylation assay
Protocol followed as described previously12. Briefly, 5 μl recombinant PARP-1 (2μg ) was mixed with 5 μl recombinant TyrRS (0–50 μM) and depending on the experiment either RSV, TyrSA, DNA or AG14361 were also added (5 μl) and adjusted the volume to 25 μl. This mixture was incubated at 4°C for 15 min, then incubated with 25 μl 2x PAR assay cocktail (50 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 40 nM NAD and 1 μCi [32P]-NAD (Perkin-Elmer, Boston, MA)) at 30°C for 30 min. [RSV] used in figure 2a middle is 0, 5, 10, 20, 30, 40, 60 and 80 nM and in figure 2a bottom is 40 nM. The reaction was stopped by addition of 50μl 2x SDS sample buffer (5% SDS,) and heated at 95°C for 1 min. The [32P]-PARylated PARP-1 was separated from free [32P]-NAD on SDS-PAGE, vacuum-dried and the poly-ADP-ribosylation activity of PARP-1 was analyzed using visualized by a phosphorimager (Typhoon™ FLA 7000, GE Healthcare).
Extended Data
Extended Data Figure 1. Resveratrol inhibits TyrRS activation.
a, The ATP-PPi exchange assay as described in methods demonstrated the inhibitory effect of resveratrol on TyrRS and (b), resveratrol shifts the Km for tyrosine. c, Resveratrol binds TyrRS better than tyrosine. The apparent Ki for resveratrol was deduced by varying the concentration of RSV and plotting the slope of (1/v vs 1/[Tyr]) versus [RSV] as indicated.
Extended Data Figure 2. Resveratrol induces a distinct conformational change upon binding to active site of TyrRS.
a, Comparison of the overall conformational change induced by resveratrol at the active site of TyrRS by structure based superposition (yellow-tyrosine-bound structure and magenta-resveratrol-bound structure). Note the conformational change near the helix region (P331-P342) that connects the linker region with the C-domain. b, Illustration of the extensive interactions of resveratrol with the active site. c, trans-resveratrol (dark blue) docks (manual) into TyrRS active site without significant structural disturbances. d, Generation of a new pocket through a RSV-induced conformational change in TyrRS accommodates the dihydroxy phenolic ring of RSV (otherwise exposed to the destabilizing aqueous environment in the trans form) and hence facilitates the trans (dark blue) to cis (light blue) conversion of RSV.
Extended Data Figure 3. Resveratrol facilitates the TyrRS/PARP-1 interaction in an active-site-dependent manner.
a, Both heat shock (42°C for 30 min) and tunicamycin-treatment (10 μg/ml, ER stress) facilitated the nuclear translocation of TyrRS and activation of PARP-1. b, Resveratrol or serum starvation facilitate TyrRS interaction with PARP-1 and Tyr-SA prevents this interaction. ZZ-PARP-1 was immunoprecipiated with IgG from HeLa cells treated with RSV or serum starvation alone or in combination with Tyr-SA. c, Resveratrol or serum starvation mediated PARP-1 activation is blocked only by Tyr-SA and not by Gly-SA. d, TyrRS interacts directly with PARP-1. HeLa cell lysate after RSV treatment (5 μM, 30 min) was divided into three parts and treated with PARG and catalytically inactive PARG-MT. PARP-1 was immunoprecipitated and analyzed for TyrRS interaction. e. Model illustrating the mechanism of RSV mediated TyrRS interaction with PARP-1 and subsequent release after auto-PARylation. f. Ni-NTA pull-down of N- and C-terminal fragments of PARP-1 overexpressed in E. coli demonstrated that TyrRS interacts with the C-terminal region of PARP-1. g, Only the full-length TyrRS (1-528) and none of the various fragments of TyrRS (mini-TyrRS (1-364), ΔN-TyrRS (228-528) or the C-domain (328-528)) interacts with PARP-1. h, Coomassie blue staining of a gel showing the total protein input in for the experiment of Extended Data Figure 3g.
Extended Data Figure 4. Tyrosyl-AMP analogue (Tyr-SA) does not affect DNA-dependent auto-PARylation of PARP-1.
a, Silver stained SDS-PAGE gel showing the purity and input of PARP-1 and TyrRS in the in vitro PARylation study of figure 2. b, Quantitation (Image J software) of the band intensity of PARylated PARP-1 in figure 2a top. c, Tyrosyl-AMP analogue (Tyr-SA) does not affect DNA-dependent auto-PARylation of PARP-1. d, Overexpression of nuclear translocation-weakened mutant of TyrRS6 is less effective in activating PARP-1. e. Y314A-TyrRS is more sensitive to RSV than is TyrRS in facilitating PARP-1 activation.
Extended Data Figure 5. Resveratrol enhances the acetylation of Tip60 and modulates [NAD+] in a dose and time dependent manner.
a, Treatment of HeLa cells (1 h) with increasing concentration of resveratrol enhances the acetylation level of Tip60. Activation of Tip60 was monitored by histone acetylation status. b, Total NAD+ content of serum starved cells or RSV treated samples were compared with untreated samples at 15 min using a commercially available BioVision NAD+/NADH quantitation colorimetric kit. c, Total nicotinamide or ADP-ribose produced was deduced from the difference in the amount of NAD+ in each sample with respect to the untreated sample (consumption of one mole of NAD+ would give rise one mole of nicotinamide and one mole of ADP-ribose). d, Total NAD+ content of the serum starved cells or RSV treated samples were compared with untreated samples at 1 hour. (Although the experiments were done in biological triplicates (all samples showing similar results), the error bars in the figure represent the deviations from the mean of the technical triplicates from one representative biological sample.) e,f Time course study of poly-ADP-ribosylation status and associated signaling events after (e) serum starvation (extended time course data of the same image shown in figure 3c) and (f) treatment with 5 μM RSV. Using the respective antibodies, Activation of p53 was monitored by the induction of p21 and SIRT6. Activation of NRF2 was monitored by HO-1 induction.
Extended Data Figure 6. siRNA ( siRNATyrRS or siRNAPARP-1), with and without low RSV (5 μM), does not affect cell viability.
HeLa cells (1× 106) were reverse-transfected with siRNA targeted to TyrRS or PARP-1. An siRNACon (a scrambled sequence of siRNAPARP-1) was used as a control. Viability was monitored using the RTCA iCELLigence System (ACEA Biosciences). Samples were treated with RSV (5μM) at 60h and monitoring was continued for another 2h for siRNATyrRS (total of 62h of monitoring) and for another 16h for siRNACon and siRNAPARP-1 (total 76h monitoring).
Extended Data Figure 7. siRNASIRT1 did not affect downstream signaling events at low RSV (5 μM).
HeLa cells were treated with siRNASIRT1 for 60 h to knockdown SIRT1. HeLa cells were treated with RSV (5 μM) for another 4 h and samples were collected intervals as indicated. Samples were analyzed for down stream signaling markers using appropriate antibodies.
Extended Data 8. | siRNA- (siRNATyrRS or siRNAPARP-1) treated cells did not upregulate the levels of NAD+ in response to RSV (5 μM) after 1h.
HeLa cells (1× 106) were reverse-transfected, separately, with siRNA targeted to PARP-1 or TyrRS. A scrambled sequence of target siRNA was used as a control. Total NAD+ content of RSV (5 μM)-treated samples were compared with untreated samples at 1h, using a commercially available BioVision NAD+/NADH quantitation colorimetric kit. (Although the experiments were done in biological triplicates (all samples showing similar results), the error bars in the figure represent the deviations from the mean of the technical triplicates from one reprentative biological sample.) The comparator (shown as a dashed bar) is taken from Extended Data Fig. 5d.
Extended Data Figure 9. Resveratrol treatment activates PARP-1 and associated signaling events in the mouse tissues.
a, Activation of PARP-1 in mouse muscle tissue treated with resveratrol monitored by increased PARylation and (b) by increased acetylation status. Activation of Tip60 and AMPK were monitored by using α-AcK16-H4 and α-pSer36-H2B, respectively. c, Activation of PARP-1 in mouse heart tissue treated with resveratrol monitored by increased PARylation and (d) by increased acetylation status. e, Resveratrol treatment cause only a transient activation on PARP-1. Immunoblotting of mouse muscle tissue samples after 24 hr of RSV treatment showed no significant difference in the level of PARP-1PAR with respect control. f, RSV treatment enhances TyrRS interaction with and activation of PARP-1 in the muscle tissue. g and h, Resveratrol-mediated activation of PARP-1 (g, monitored by PARylation status and h, monitored by acetylation status) is blocked by Tyr-SA in the mouse heart tissues.
Extended Data Table 1.
Data collection and refinement statistics.
Numerical parameters for the RSV-TyrRS and L-Tyr-TyrRS co-crystals are listed and compared side-by-side in separate columns.
| Resveratrol-TyrRS | L-Tyrosine-TyrRS | |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 73.97, 162.55, 35.0 | 73.97, 162.55, 35.0 |
| α, β, γ(°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 37 - 2.1(2.15–2.09)* | 34.63 - 2.1(2.15–2.09)* |
| Rsym or Rmerge | 27.1 | 44.7 |
| I/σI | 4.1 | 4.6 |
| Completeness (%) | 98.6 | 98 |
| Redundancy | 3.2 | 5.2 |
| Refinement | ||
| Resolution (Å) | 2.1 | 2.1 |
| No. reflections | 25438 | 24513 |
| Rwork/ Rfree | 0.19/0.24 | 0.22/0.27 |
| No. atoms | 2955 | 2858 |
| Protein | 2653 | 2636 |
| Ligand/ion | 16 | 12 |
| Water | 284 | 238 |
| B-factors | ||
| Protein | 37.664 | 38.199 |
| Ligand/ion | 43.2 | 42.3 |
| Water | 47.3 | 48.4 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.018 | 0.017 |
| Bond angles (°) | 2.017 | 2.008 |
Highest resolution shell is shown in parenthesis.
Acknowledgments
This work was supported in part the NCI grant CA92577, by a fellowship from the National Foundation for Cancer Research, and by aTyr Pharma through an agreement with The Scripps Research Institute. We thank the TSRI mouse facility for their efforts for this project. We also thank Prof. Paul Chang (Department of Biology, MIT, Cambridge, MA) for the ZZ-PARP-1 clone, and the PARG and PARG-MT proteins and Dr. Yi Shi (The Scripps Research Institute) for independently repeating some of the key experiments. We thank Professors Paul Chang (MIT, Cambridge, MA), Jay H. Chung (NHLBI, Bethesda, MD), Leonard Guarente (MIT, Cambridge, MA), Lee Krauss (UT Southwestern), David Sinclair (Harvard Medical School), and Dr. Christoph Westphal (Longwood Partners, Boston, MA) for helpful comments and suggestions on this work.
Footnotes
Author Contributions
M.S. and P.S. conceived the idea, designed the research, analyzed the data and wrote the manuscript. M.S. performed the experiments.
Although P.S. has a financial interest in aTyr Pharma (who partially supported this investigation), he and M. S. have no financial interest in this specific work. X-ray structure co-ordinates of resveratrol bound TyrRS (PDB ID code 4Q93) and L-Tyr bound TyrRS (PDB ID code 4QBT) were deposited to the Protein Data Bank.
References
- 1.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 2.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang M, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 6.Fu G, Xu T, Shi Y, Wei N, Yang XL. tRNA-controlled nuclear import of a human tRNA synthetase. The Journal of biological chemistry. 2012;287:9330–9334. doi: 10.1074/jbc.C111.325902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo WS, et al. Human tRNA synthetase catalytic nulls with diverse functions. Science. 2014;345:328–332. doi: 10.1126/science.1252943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 9.Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nature chemical biology. 2013;9:145–153. doi: 10.1038/Nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang XL, Skene RJ, McRee DE, Schimmel P. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XL, Liu FM, Skene RJ, McRee DE, Schimmel P. Crystal structure of an EMAP-II-like cytokine released from a human tRNA synthetase. Helv Chim Acta. 2003;86:1246–1257. doi: 10.1002/Hlca.200390107. [DOI] [Google Scholar]
- 12.Sajish M, et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nature chemical biology. 2012;8:547–554. doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes & development. 2012;26:417–432. doi: 10.1101/Gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinclair DA, Guarente L. Small-molecule allosteric activators of sirtuins. Annual review of pharmacology and toxicology. 2014;54:363–380. doi: 10.1146/annurev-pharmtox-010611-134657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapoor M, Otero FJ, Slike BM, Ewalt KL, Yang XL. Mutational separation of aminoacylation and cytokine activities of human tyrosyl-tRNA synthetase. Chemistry & biology. 2009;16:531–539. doi: 10.1016/j.chembiol.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee PS, Zhang HM, Marshall AG, Yang XL, Schimmel P. Uncovering of a short internal peptide activates a tRNA synthetase procytokine. The Journal of biological chemistry. 2012;287:20504–20508. doi: 10.1074/jbc.C112.369439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y, Zhao WH, Chen Y, Zhao YM, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/J.Cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. The Journal of biological chemistry. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 19.Petesch SJ, Lis JT. Activator-Induced Spread of Poly(ADP-Ribose) Polymerase Promotes Nucleosome Loss at Hsp70. Molecular cell. 2012;45:64–74. doi: 10.1016/J.Molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen-Armon M, et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Molecular cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Vyas S, Chang P. Dual roles for PARP1 during heat shock: transcriptional activator and posttranscriptional inhibitor of gene expression. Molecular cell. 2013;49:1–3. doi: 10.1016/j.molcel.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegan DC, et al. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlando G, Khoronenkova SV, Dianova, Parsons JL, Dianov GL. ARF induction in response to DNA strand breaks is regulated by PARP1. Nucleic acids research. 2013 doi: 10.1093/nar/gkt1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SJ, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butin-Israeli V, Drayman N, Oppenheim A. Simian Virus 40 Infection Triggers a Balanced Network That Includes Apoptotic, Survival, and Stress Pathways. J Virol. 2010;84:3431–3442. doi: 10.1128/Jvi.01735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bastide B, Snoeckx K, Mounier Y. ADP-ribose stimulates the calcium release channel RyR1 in skeletal muscle of rat. Biochemical and biophysical research communications. 2002;296:1267–1271. doi: 10.1016/s0006-291x(02)02073-9. [DOI] [PubMed] [Google Scholar]
- 28.Shin SM, Cho IJ, Kim SG. Resveratrol Protects Mitochondria against Oxidative Stress through AMP-Activated Protein Kinase-Mediated Glycogen Synthase Kinase-3 beta Inhibition Downstream of Poly(ADP-ribose)polymerase-LKB1 Pathway. Molecular pharmacology. 2009;76:884–895. doi: 10.1124/Mol.109.058479. [DOI] [PubMed] [Google Scholar]
- 29.Howitz KT, Sinclair DA. Xenohormesis: sensing the chemical cues of other species. Cell. 2008;133:387–391. doi: 10.1016/j.cell.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nwachukwu JC, et al. Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network. Elife. 2014;3:Artn 02057. doi: 10.7554/Elife.02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JD, Chen JJ. SIRT1 Regulates Autoacetylation and Histone Acetyltransferase Activity of TIP60. Journal of Biological Chemistry. 2010;285:11458–11464. doi: 10.1074/Jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen WM, et al. Direct Interaction between Nrf2 and p21(Cip1/WAF1) Upregulates the Nrf2-Mediated Antioxidant Response. Molecular cell. 2009;34:663–673. doi: 10.1016/J.Molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, et al. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/Science.1101731. [DOI] [PubMed] [Google Scholar]
- 36.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renault VM, et al. The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene. 2011;30:3207–3221. doi: 10.1038/Onc.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajamohan SB, et al. SIRT1 Promotes Cell Survival under Stress by Deacetylation-Dependent Deactivation of Poly(ADP-Ribose) Polymerase 1. Molecular and cellular biology. 2009;29:4116–4129. doi: 10.1128/Mcb.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith BC, Hallows WC, Denu JM. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Analytical biochemistry. 2009;394:101–109. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perraud AL, et al. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- 42.Formentini L, et al. Poly(ADP-ribose) catabolism triggers AMP-dependent mitochondrial energy failure. The Journal of biological chemistry. 2009;284:17668–17676. doi: 10.1074/jbc.M109.002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawley SA, et al. Use of Cells Expressing gamma Subunit Variants to Identify Diverse Mechanisms of AMPK Activation. Cell metabolism. 2010;11:554–565. doi: 10.1016/J.Cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bungard D, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/J.Devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey KM, et al. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD(+) Biosynthesis. Science. 2009;324:651–654. doi: 10.1126/Science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes & development. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 48.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–U416. doi: 10.1038/Nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vyas S, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nature communications. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, et al. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-kappaB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Molecular and cellular biology. 2011;31:2774–2786. doi: 10.1128/MCB.01139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stilmann M, et al. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Molecular cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 52.Martin N, et al. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. Embo Journal. 2009;28:3534–3548. doi: 10.1038/Emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matheny CJ, et al. Next-generation NAMPT inhibitors identified by sequential high-throughput phenotypic chemical and functional genomic screens. Chemistry & biology. 2013;20:1352–1363. doi: 10.1016/j.chembiol.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. The Journal of biological chemistry. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]