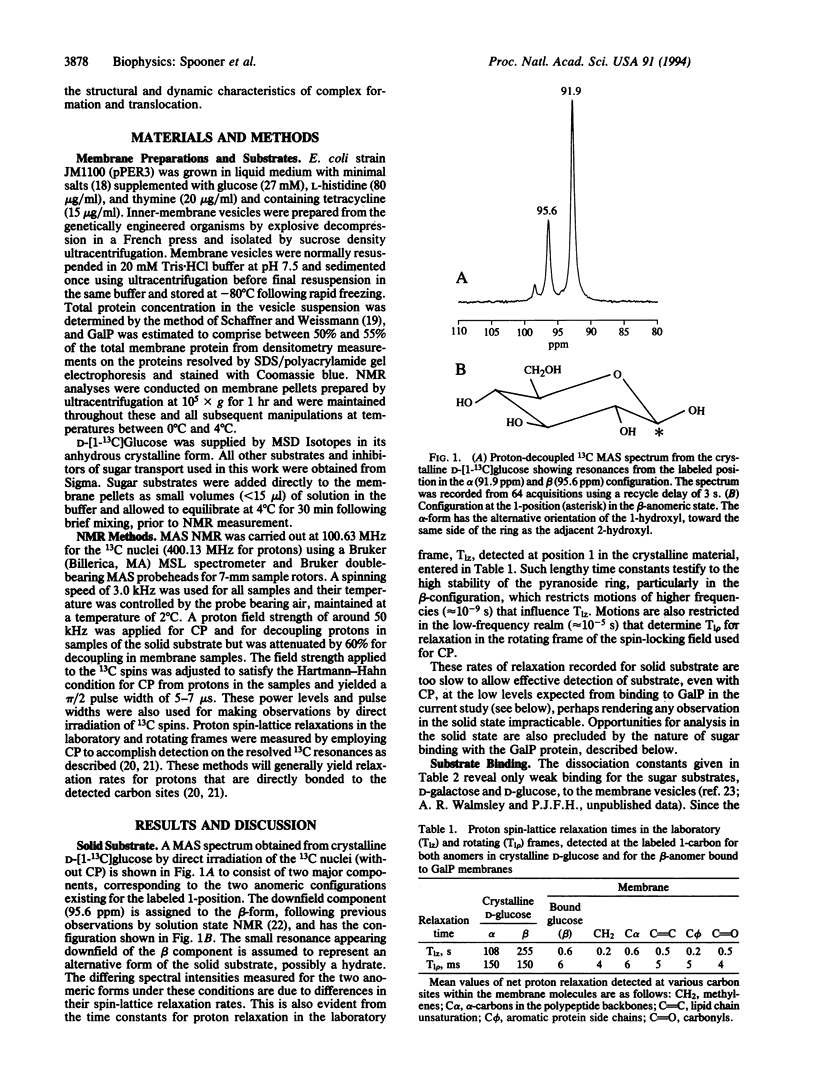
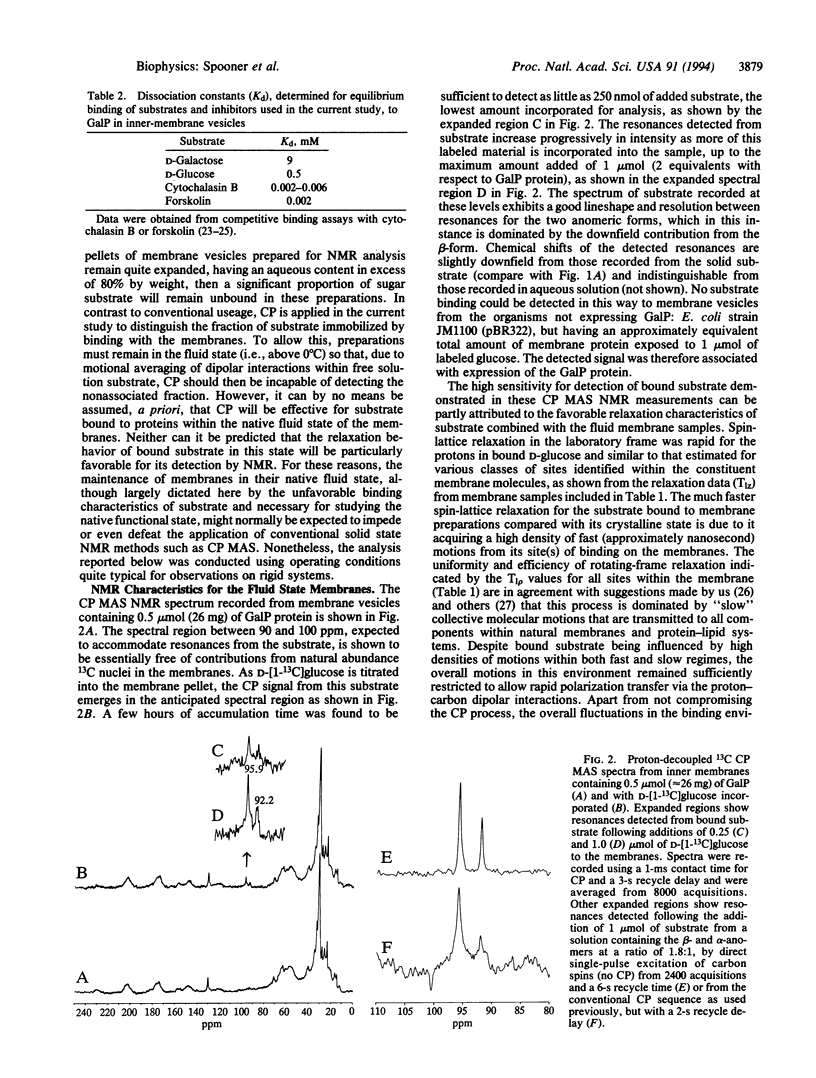
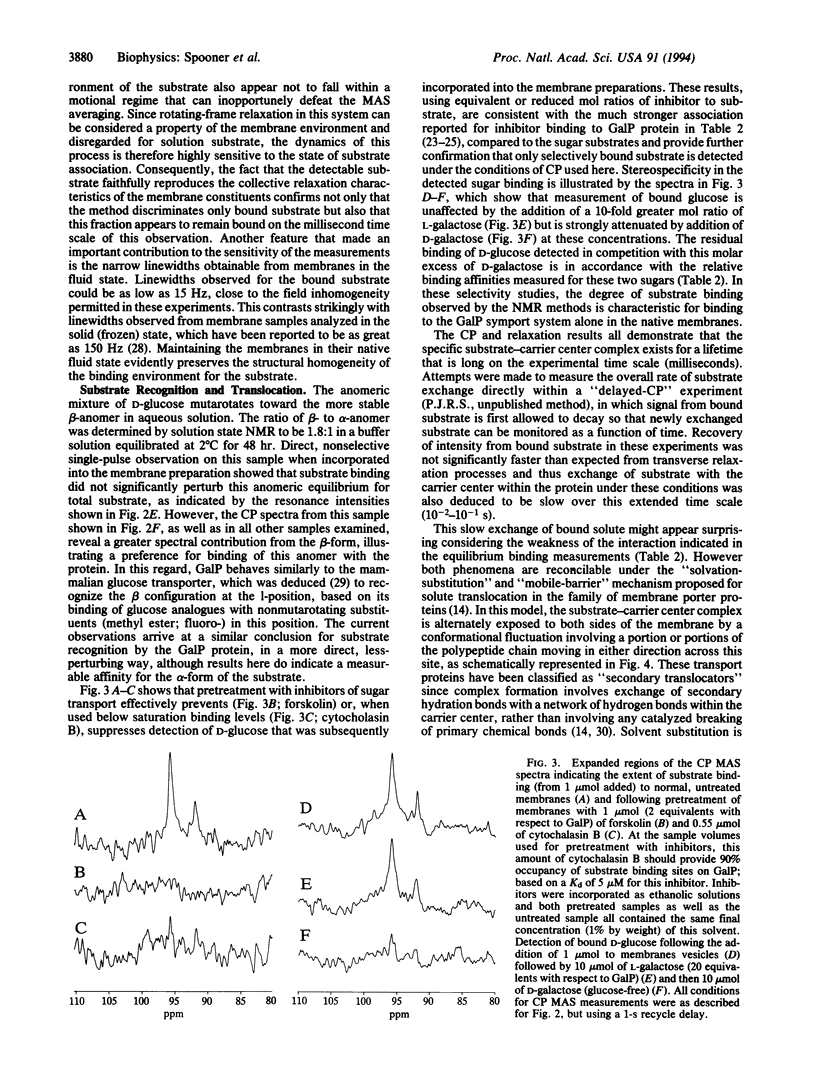
Abstract
NMR methods have been adopted to observe directly the characteristics of substrate binding to the galactose-H+ symport protein GalP, in its native environment, the inner membranes of Escherichia coli. Sedimented inner-membrane vesicles containing the GalP protein, overexpressed to levels above 50% of total protein, were analyzed by 13C magic-angle spinning NMR, when in their normal "fluid" state and with incorporated D-[1-13C]glucose. Using conditions of cross-polarization intended to discriminate bound substrate alone, it was possible to detect as little as 250 nmol of substrate added to the membranes containing about 0.5 mumol (approximately 26 mg) of GalP protein. Such high measuring sensitivity was possible from the fluid membranes by virtue of their motional contributions to rapid relaxation recovery of the observed nuclei and due to a high-resolution response that approached the static field inhomogeneity in these experiments. This good spectral resolution showed that the native state of the membranes presents a substrate binding environment with high structural homogeneity. Inhibitors of the GalP protein, cytochalasin B and forskolin, which are specific, and D-galactose, but not L-galactose, prevent or suppress detection of the 13C-labeled glucose substrate, confirming that the observed signal was due to specific interactions with the GalP protein. This specific substrate binding exhibits a preference for the beta-anomer of D-glucose and substrate translocation is determined to be slow, on the 10(-2) s time scale. The work describes a straightforward NMR approach, which achieves high sensitivity, selectivity, and resolution for nuclei associated with complex membrane proteins and which may be combined with other NMR methodologies to yield additional structural information on the binding site for the current transport system without isolating it from its native membrane environment.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the cofactors. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5730–5734. doi: 10.1073/pnas.84.16.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin S. A., Henderson P. J. Homologies between sugar transporters from eukaryotes and prokaryotes. Annu Rev Physiol. 1989;51:459–471. doi: 10.1146/annurev.ph.51.030189.002331. [DOI] [PubMed] [Google Scholar]
- Barnett J. E., Holman G. D., Munday K. A. Structural requirements for binding to the sugar-transport system of the human erythrocyte. Biochem J. 1973 Feb;131(2):211–221. doi: 10.1042/bj1310211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns M. T., McDonald T. P., Horne P., Henderson P. J., Baldwin S. A. Cytochalasin B as a probe of protein structure and substrate recognition by the galactose/H+ transporter of Escherichia coli. J Biol Chem. 1991 May 5;266(13):8176–8183. [PubMed] [Google Scholar]
- Cornell B. A., Hiller R. G., Raison J., Separovic F., Smith R., Vary J. C., Morris C. Biological membranes are rich in low-frequency motion. Biochim Biophys Acta. 1983 Jul 27;732(2):473–478. doi: 10.1016/0005-2736(83)90065-2. [DOI] [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Creuzet F., McDermott A., Gebhard R., van der Hoef K., Spijker-Assink M. B., Herzfeld J., Lugtenburg J., Levitt M. H., Griffin R. G. Determination of membrane protein structure by rotational resonance NMR: bacteriorhodopsin. Science. 1991 Feb 15;251(4995):783–786. doi: 10.1126/science.1990439. [DOI] [PubMed] [Google Scholar]
- Cross T. A. Anisotropy and NMR of macromolecules. Biophys J. 1993 Feb;64(2):301–302. doi: 10.1016/S0006-3495(93)81369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. J., Giddens R. A., Jones-Mortimer M. C. Transport of galactose, glucose and their molecular analogues by Escherichia coli K12. Biochem J. 1977 Feb 15;162(2):309–320. doi: 10.1042/bj1620309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. J. Proton-linked sugar transport systems in bacteria. J Bioenerg Biomembr. 1990 Aug;22(4):525–569. doi: 10.1007/BF00762961. [DOI] [PubMed] [Google Scholar]
- MITCHELL P. A general theory of membrane transport from studies of bacteria. Nature. 1957 Jul 20;180(4577):134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- Maiden M. C., Davis E. O., Baldwin S. A., Moore D. C., Henderson P. J. Mammalian and bacterial sugar transport proteins are homologous. Nature. 1987 Feb 12;325(6105):641–643. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Osmochemistry of solute translocation. Res Microbiol. 1990 Mar-Apr;141(3):286–289. doi: 10.1016/0923-2508(90)90002-8. [DOI] [PubMed] [Google Scholar]
- Nikaido H., Saier M. H., Jr Transport proteins in bacteria: common themes in their design. Science. 1992 Nov 6;258(5084):936–942. doi: 10.1126/science.1279804. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973 Dec;56(2):502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- Spooner P. J., Duralski A. A., Rankin S. E., Pinheiro T. J., Watts A. Dynamics in a protein-lipid complex: nuclear magnetic resonance measurements on the headgroup of cardiolipin when bound to cytochrome c. Biophys J. 1993 Jul;65(1):106–112. doi: 10.1016/S0006-3495(93)81048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner P. J., Watts A. Cytochrome c interactions with cardiolipin in bilayers: a multinuclear magic-angle spinning NMR study. Biochemistry. 1992 Oct 20;31(41):10129–10138. doi: 10.1021/bi00156a037. [DOI] [PubMed] [Google Scholar]
- Thompson L. K., McDermott A. E., Raap J., van der Wielen C. M., Lugtenburg J., Herzfeld J., Griffin R. G. Rotational resonance NMR study of the active site structure in bacteriorhodopsin: conformation of the Schiff base linkage. Biochemistry. 1992 Sep 1;31(34):7931–7938. doi: 10.1021/bi00149a026. [DOI] [PubMed] [Google Scholar]
- Weiss M. S., Wacker T., Weckesser J., Welte W., Schulz G. E. The three-dimensional structure of porin from Rhodobacter capsulatus at 3 A resolution. FEBS Lett. 1990 Jul 16;267(2):268–272. doi: 10.1016/0014-5793(90)80942-c. [DOI] [PubMed] [Google Scholar]



