Abstract
Background
Antibodies to citrullinated proteins (ACPAs) are a hallmark of rheumatoid arthritis (RA). Porphyromonas gingivalis peptidylarginine deiminase (PPAD) has been implicated in the initiation of RA by generating citrullinated neoantigens and due to its ability to autocitrullinate.
Objectives
To define the citrullination status and biology of PPAD in P gingivalis and to characterize the anti-PPAD antibody response in RA and associated periodontal disease (PD).
Methods
PPAD in P gingivalis cells and culture supernatant was analyzed by immunoblotting and mass spectrometry to detect citrullination. Recombinant PPAD (rPPAD), inactive mutant PPAD (rPPADC351S), and N-terminal truncated PPAD (rPPADNtx) were cloned and expressed inE coli. Patients with RA and healthy controls were assayed for IgG antibodies to citrullinated rPPAD and unmodified rPPADC351S by ELISA. Anti-PPAD antibodies were correlated with anti-CCP3 antibody levels, RA disease activity, and PD status.
Results
PPAD from P gingivalis is truncated at the N- and C-terminal domains and not citrullinated. Only when artificially expressed in E coli, full-length rPPAD, but not truncated (fully active) rPPADNtx, is autocitrullinated. Anti-PPAD antibodies show no heightened reactivity to citrullinated rPPAD, but are exclusively directed against the unmodified enzyme. Antibodies against PPAD do not correlate with anti-CCP levels and disease activity in RA. By contrast, anti-PPAD antibody levels are significantly decreased in RA patients with PD.
Conclusions
PPAD autocitrullination is not the underlying mechanism linking PD and RA. N-terminal processing protects PPAD from autocitrullination and enhances enzyme activity. Anti-PPAD antibodies may have a protective role for the development of PD in RA patients.
Keywords: Rheumatoid arthritis, periodontal disease, Porphyromonas gingivalis peptidylarginine deiminase, anti-PPAD antibodies, ACPA
INTRODUCTION
Rheumatoid arthritis (RA) is a systemic autoimmune disease of unknown etiology characterized by synovial inflammation and joint destruction.1,2 Loss of tolerance to citrullinated proteins is a hallmark of RA pathogenesis.3 Autoantibodies to citrullinated proteins (ACPA) are highly specific for RA and precede the onset of clinical disease by years.4–6 Peptidylarginine deiminases (PAD) are enzymes that catalyze the posttranslational modification reaction of arginine residues to citrulline.7,8 By modifying autoantigens in synovial tissue and alternative sites of inflammation, PAD activity may be a sine qua non in initiating and sustaining autoimmunity in ACPA-positive RA. Porphyromonas gingivalis peptidylarginine deiminase (PPAD), a bacterial deiminase evolutionarily unrelated to human PAD,8,9 has received considerable attention by investigators trying to identify a mechanism linking periodontal disease (PD), bacterial citrullination and RA.10–13
PD is a chronic inflammatory disease caused by infection of the supporting tissues of the teeth, ultimately resulting in alveolar bone destruction and tooth loss.14 An increased prevalence of PD has repeatedly been reported in RA.15–19 Among the periodontal pathogens, P gingivalis, an anaerobic bacterium strongly associated with chronic periodontitis,20–22 is unique in its expression of a bacterial PAD.9 PPAD is believed to be a major virulence factor of P gingivalis due to its capacity to generate ammonia in the deimination reaction of arginine to citrulline.8 Ammonia may protect P gingivalis during acidic cleansing cycles in the mouth,9,23,24 and promote periodontal infection via inhibitory effects on neutrophil function.25,26
PPAD is almost exclusively detected in outer membrane (OM) fractions of P gingivalis,12 and as a secreted enzyme.9 Interestingly, OM-associated PPAD and secreted PPAD were only recently shown to be truncated both at the C- and N-terminal domains.9,27 While C-terminal cleavage is required for cell surface translocation of PPAD,27–29 N-terminal processing appears to maintain enzyme activity and stability.30
By citrullinating C-terminal peptidylarginine in the context of periodontal infection, P gingivalis has been hypothesized to play a primary role in RA pathogenesis.11,31,32 Moreover, the recent finding that recombinant PPAD (rPPAD) is autocitrullinated and preferentially recognized by antibodies in RA suggests that loss of tolerance to citrullinated proteins in RA may originate from an antimicrobial immune response directed against citrullinated PPAD.12
This study defines the structure and citrullination status of the cellular and secreted forms of PPAD (cPPAD and sPPAD, respectively) in P gingivalis. The relevance of the anti-PPAD antibody response for RA and associated PD is examined in the context of PPAD biology.
MATERIAL AND METHODS
Patients and controls
Serum was obtained from 83 RA patients (1987 revised ARA criteria)33 and 39 healthy controls recruited under the protocol of the Comprehensive Oral Health Assessment in Patients with Arthritis and Autoimmune Inflammatory Diseases (OHARA). The protocol was approved by the Johns Hopkins Medicine Institutional Review Board, and informed consent was obtained from all participants. Study participants underwent a comprehensive dental evaluation. PD was defined using the 2007 CDC/ AAP standard cases definitions for surveillance of moderate and severe periodontitis based on assessments of pocket depth and attachment loss;14,34 subjects not fulfilling these definitions were considered periodontally healthy.
Bacterial samples and immunoblotting
P gingivalis strain W83 was obtained from the University of Maryland (courtesy of Mark A. Reynolds). Bacterial cells were pelleted by centrifugation, lysed in ice-cold NP-40 lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 Alternative), sonicated, and boiled in SDS sample buffer. P gingivalis culture supernatant was concentrated using Amicon Ultra-15 units (EMD Millipore) and buffer exchanged with 20 mM Tris pH 7.6, 150 mM NaCl, 10% glycerol. Samples were resolved by SDS-PAGE and analyzed by anti-modified citrulline (AMC) immunoblotting,35 or using a polyclonal antibody raised in rabbits by immunization with recombinant PPAD (Covance).
Cloning and purification of recombinant PPAD and alpha-enolase
The full-length PPAD coding sequence was amplified from P gingivalis W83 DNA (ATCC) and cloned into pET-28a(+) (Novagen), generating a fusion protein with N-terminal His-tag. Enzymatically inactive PPAD was generated by site-directed mutagenesis to replace cysteine 351 at the active site of the protein with serine (PPADC351S).36 N-terminal truncated PPAD (rPPADNtx) was generated by amplifying the coding sequence for amino acids 44 to 556 of full-length PPAD with primers containing a 5’ Kozak sequence. The PCR product was cloned into pET-28a(+) to encode for a C-terminal His-tagged fusion protein. Alpha-enolase encoding cDNA was cloned into pET-28a(+). Recombinant PPAD, PPADC351S, PPADNtx, and enolase were expressed in E coli BL21 (DE3) (Agilent), and purified from the soluble fraction of cell lysates prepared in 20 mM Tris pH 7.6, 400 mM NaCl, 5 mM imidazole, 20 mM β-mercaptoethanol, 1% Triton X by Ni-NTA affinity chromatography (Qiagen). Purity of rPPAD and rPPADC351S used in ELISA assays exceeded 95% (see online supplementary figure S1A). Enolase was citrullinated in vitro using human rPAD4 as previously described.37
PPAD ELISA
Polystyrene plates (Costar) were coated with 100ng/well citrullinated rPPAD (cit-rPPAD), rPPADC351S or PBS pH 7.4 alone (used as a negative control). Coated plates were washed with PBS 0.05% Tween 20 (PBS–T), and unoccupied binding sites blocked with PBS–T 3% non-fat dry milk (PBS–TM). Sera were diluted at 1:1000 in PBS–TM 1% and assayed in duplicate. HRP-conjugated anti-human IgG (Jackson ImmunoResearch) was used as a secondary antibody. A serial dilution of rabbit anti-PPAD was used as a standard. Arbitrary units (AU) were calculated from standard dilutions, and individual values corrected for background by subtracting the reactivity of PBS-coated wells.
RESULTS
PPAD from P gingivalis is truncated and not citrullinated
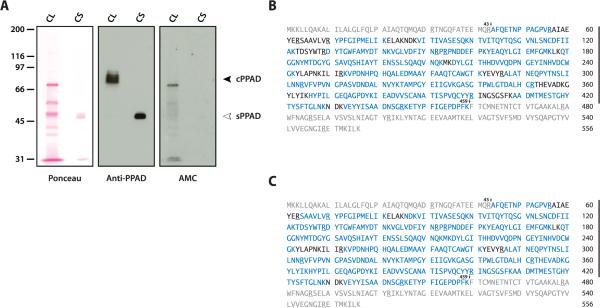
Cell lysates and supernatant from P gingivalis were initially studied by immunoblotting to characterize bacterial PPAD. Two distinct patterns of PPAD were identified. In bacterial cells, anti-PPAD immunoblotting detected bands of approximately 75-85 kDa (cPPAD), while a single band of 47 kDa (sPPAD) was identified in P gingivalis supernatant (figure 1A, middle panel). The observed size of sPPAD is consistent with the truncated form of PPAD.9,27 In contrast to a previous report,11 only low levels of protein citrullination were detected by AMC immunoblotting in bacteria lysed in NP-40 buffer (figure 1A, right panel), and no citrullination was found when P gingivalis was directly lysed and boiled in SDS sample buffer (see online supplementary figure S1B). Bacterial citrullination may therefore occur in vitro during cell lysis. Importantly, cPPAD and sPPAD are not citrullinated in bacterial samples (figure 1A, right panel and online supplementary figure S1B).
Figure 1.
Cellular PPAD (cPPAD) and secreted PPAD (sPPAD) from P gingivalis are truncated and not citrullinated. (A) P gingivalis cell lysate (CL) and culture supernatant (CS) were analyzed by SDS-PAGE. Ponceau staining before antibody probing is shown to visualize loading (left panel). Anti-PPAD immunoblotting shows cPPAD (black arrowhead) and sPPAD (white arrowhead) in P gingivalis CL and CS, respectively (middle panel). AMC immunoblotting was used to detect citrullination in bacterial samples (right panel). (B and C) Mass spectrometry sequence coverage map of P gingivalis cPPAD (B) and sPPAD (C). Peptide sequences identified are shown in blue; the N-terminal and C-terminal domains missing in cPPAD and sPPAD are shown in grey. Arginine residues are underlined. Numbered arrows indicate possible PPAD truncation sites at R43 and K459.
Mass spectrometry (MS) analysis of cPPAD and sPPAD invariably identified a truncated protein missing the first 43 N-terminal and 97 C-terminal amino acids of full-length PPAD (figure 1B and 1C, respectively). Peptides outside these terminal domains showed sequence coverage of 82.9% (cPPAD) and 92.5% (sPPAD). In contrast to a previous study of full-length rPPAD expressed in E coli,12 we found that PPAD in P gingivalis is not citrullinated (figure 1B,C). Lack of autocitrullination was not attributable to loss of enzymatic function as confirmed using the BAEE assay (data not shown).
N-terminal truncation prevents PPAD autocitrullination and amplifies enzyme function
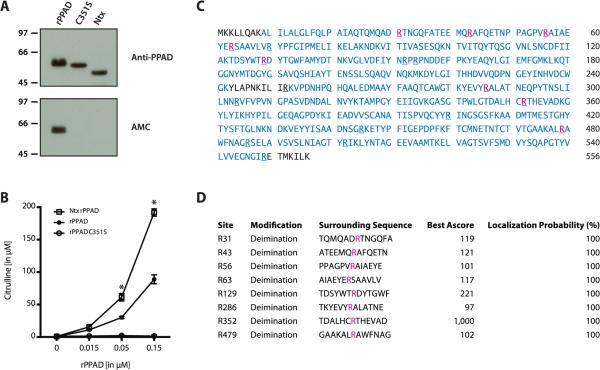
Previous studies of recombinant full-length PPAD and N-terminal truncated PPAD have shown conflicting results with regard to enzyme autocitrullination.12,30 While full-length PPAD is strongly citrullinated during expression in E coli,12 the truncated enzyme is not autocitrullinated.30 We hypothesized that N-terminal processing of PPAD may protect the enzyme from autocitrullination in P gingivalis. Indeed, when we expressed full-length PPAD (rPPAD) and N-terminal truncated PPAD (rPPADNtx) in E coli, only the full-length enzyme was citrullinated (figure 2A). Analogously, enzymatically inactive full-length PPAD (PPADC351S) was not citrullinated when expressed in E coli (figure 2A, lower panel), confirming that PPAD autocitrullination is dependent on both the catalytic activity of the enzyme and its N-terminal domain. Importantly, we confirmed that the absence of autocitrullination in rPPADNtx was not due to loss of enzymatic function. Indeed, N-terminal truncated PPAD showed a significant increase in enzyme activity as compared to full-length rPPAD (mean citrulline production in μM: 191.9 vs. 88.9; *p<0.0001) (figure 2B).
Figure 2.
Citrullination of rPPAD expressed in E coli is dependent on the N-terminal domain. (A) Purified rPPAD, mutant rPPADC351S (C351S) and truncated rPPAD missing the N-terminal domain (Ntx) were analyzed by SDS-PAGE and visualized by anti-PPAD immunoblotting (top panel). AMC immunoblotting (bottom panel) was used to detect citrullination. (B) Enzymatic activity of rPPAD, rPPADC351S and rPPADNtx was measured in the BAEE assay. Average citrulline concentrations (in μM) from two independent experiments are shown (error bars indicate standard deviation). Enzyme activities were analyzed by two-way ANOVA (rPPADNtx vs. rPPAD; *p<0.0001). (C) Mass spectrometry sequence coverage map of full-length rPPAD expressed in E coli. Peptide sequences identified by MS are shown in blue; confirmed citrullination sites in pink. Arginine residues are underlined. (D) Localization probabilities for individual arginine deimination sites in rPPAD were calculated using the Ascore algorithm. An Ascore of 20 (99% certainty; p=0.01) was considered significant.38
Mass spectrometry of full-length rPPAD achieved 96% protein sequence coverage, which included 17 of the 18 arginines found in the enzyme. Among these, 8 arginine residues were citrullinated (figure 2C). The probability of correct citrullination site localization approached 100% for all eight sites (figure 2D). These findings are in general accordance with the citrullination sites previously reported for rPPAD,12 with two additional sites identified at R129 and R286. In peptides covering R63 and R70, citrullination was located to R63, but not confirmed at R70 as previously reported (figure 2D).12 While this data confirms that rPPAD is autocitrullinated in E coli, this process appears to be unique to artificially expressed PPAD in the absence of physiological enzyme processing and regulators found in P gingivalis.
Anti-PPAD antibodies in RA are not directed against citrullinated PPAD
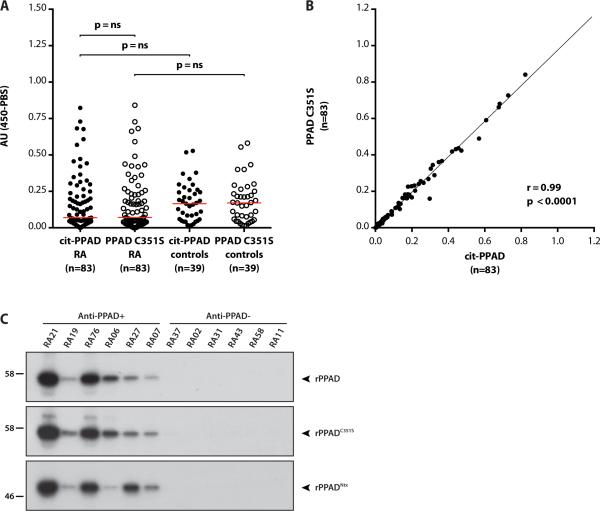
While we found no evidence that PPAD is citrullinated in P gingivalis, antibodies specific for citrullinated rPPAD have previously been reported in patients with RA.12 To dissect the role of citrullination for anti-PPAD antibody recognition, we assayed serum samples of RA patients and healthy controls for antibodies to cit-rPPAD and uncitrullinated rPPADC351S by ELISA. IgG anti-PPAD antibodies were common in patients with RA and in healthy controls (figure 3A). Strikingly, citrullination did not heighten antibody reactivity to PPAD. Antibody levels to citrPPAD vs. uncitrullinated rPPADC351S did not differ significantly among RA patients (median 0.069 vs. 0.070; median of differences: 0.0; p=0.14) or controls (median 0.165 vs. 0.170; median of differences: -0.001; p=0.87) (figure 3A and online supplementary figure S2). Indeed, anti-cit-rPPAD and anti-rPPADC351S antibody levels in RA were highly correlated (r=0.99, *p<0.0001) (figure 3B). Control subjects demonstrated a similar degree of correlation (r=0.99, *p<0.0001).
Figure 3.
Antibodies to citrullinated rPPAD (cit-rPPAD) and uncitrullinated PPAD (rPPADC351S) in serum of patients with RA and controls. (A) RA patients (n=83) and controls (n=39) were assayed for antibodies to cit-rPPAD (full circles) and rPPADC351S (empty circles) by ELISA. Antibody reactivity is expressed as arbitrary units (AU). Statistical analysis was performed using the Wilcoxon matched-pairs signed-rank test (anti-citPPAD vs. anti-PPADC351S) and Mann-Whitney test (RA vs. control subjects). The red line represents the median reactivity for individual groups. ns: not significant. (B) Correlation of anti-cit-rPPAD and anti-rPPADC351S antibody levels in patients with RA (r=0.99, p<0.0001). (C) 35S-methionine-labeled PPAD (top panel), PPADC351S (middle panel) and truncated PPADNtx (bottom panel) generated by IVTT were immunoprecipitated using RA patient serum. Immunoprecipitates were electrophoresed on SDS-PAGE and visualized by fluorography. Representative results for anti-rPPAD positive (anti-PPAD+) and anti-rPPAD negative (anti-PPAD-) patients, as determined by ELISA, are shown.
Importantly, levels of anti-cit-rPPAD were not significantly different between patients with RA and controls (median 0.069 vs. 0.165; p=0.085). Although the highest antibody reactivity was observed in the RA group (5/83 above the 95th percentile) compared to controls (1/39), statistical significance to support this association was not attained (Fisher's exact test; p=0.66). In fact, the overall trend observed was towards higher anti-PPAD antibody levels in the control group (figure 3A). To confirm the specificity of the ELISA assay for anti-PPAD, antibodies were additionally demonstrated by immunoprecipitation (IP) using radiolabeled PPAD generated by in vitro transcription/translation (figure 3C). In agreement with the ELISA assay, RA patient sera showed similar reactivity against PPAD and mutant PPADC351S by IP (figure 3C, top and middle panel, respectively). Importantly, only sera reactive by ELISA immunoprecipitated radiolabeled PPAD (figure 3C, anti-PPAD+ vs. anti-PPAD-). Similarly, radiolabeled truncated PPADNtx (figure 3C, bottom panel), a protein that better mimics PPAD in P gingivalis, was only precipitated by sera positive by ELISA. These data support the specificity of the ELISA assay and strongly suggest that anti-PPAD antibody binding in RA is independent of PPAD citrullination.
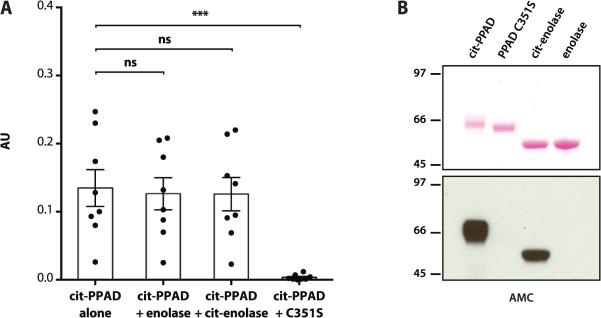
To confirm that citrullination is not a determinant in the antibody recognition of rPPAD, we further performed competition experiments. Antibody binding to cit-rPPAD was completely abrogated by preincubation of anti-cit-rPPAD positive RA sera with uncitrullinated rPPADC351S, but not by preincubation with unmodified or citrullinated enolase (figure 4A,B). This demonstrates that anti-PPAD antibodies in RA are exclusively directed against unmodified PPAD.
Figure 4.
Anti-PPAD antibodies in RA are exclusively directed against uncitrullinated PPAD. (A) Antibody reactivity against cit-rPPAD was measured after preincubation of anti-cit-rPPAD positive RA sera (n=8) with buffer alone (column 1), purified enolase (column 2), citrullinated enolase (cit-enolase) (column 3) or uncitrullinated rPPADC351S (column 4; ***p<0.001). Comparisons were made using Friedman's test/ Dunn's post test for matched groups. Column height shows mean antibody reactivity expressed as arbitrary units (AU); bars indicate SEM. ns: not significant. (B) AMC immunoblotting to demonstrate the presence or absence of citrullination in proteins used in A. Purified cit-rPPAD, rPPADC351S, enolase and cit-enolase were resolved by SDS-PAGE, transferred onto nitrocellulose and visualized by Ponceau staining as a loading control (top panel). Protein citrullination in the same membrane was detected by AMC immunoblotting (bottom panel).
Anti-PPAD antibodies do not correlate with anti-CCP levels or RA disease activity
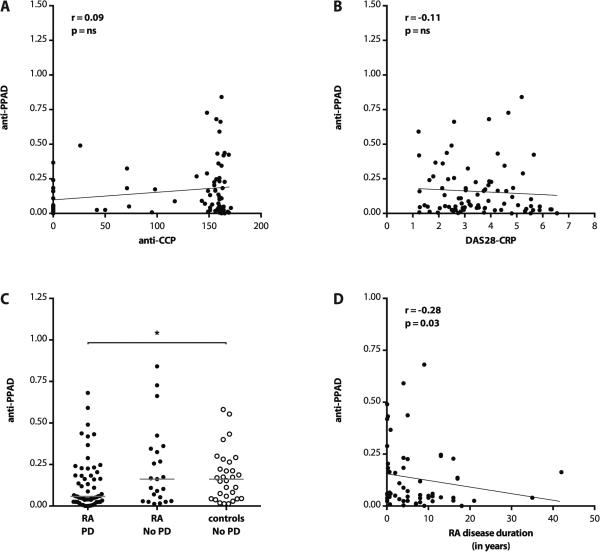
To further characterize their relevance in RA, we inquired how anti-PPAD antibodies relate to anti-CCP levels and disease activity measures. Anti-CCP3 antibodies in RA did not correlate with anti-cit-rPPAD or anti-rPPADC351S levels (r=0.07, p=0.56; r=0.09, p=0.44; respectively) (figure 5A). Moreover, we found no significant association between anti-rPPAD antibody levels and RA disease activity as measured using DAS28-CRP (r=−0.11, p=0.30) (figure 5B).
Figure 5.
Anti-PPAD levels do not correlate with anti-CCP antibodies or RA disease activity, but are negatively associated with periodontal disease in RA. (A) Correlation of anti-PPAD antibodies with anti-CCP3 levels in RA. Antibody levels to CCP3 were measured by ELISA (QUANTA Lite IgG, INOVA). (B) Correlation of anti-PPAD levels with RA disease activity as measured using DAS28-CRP. ns: not significant. (C) Anti-PPAD antibody levels in RA patients with and without periodontal disease (RA PD and RA No PD, respectively; full circles) compared to periodontally healthy controls (empty circles). Antibody levels are expressed in arbitrary units (red line shows median reactivity; *p<0.05). (D) Antibody levels in PD-positive RA patients as a function of RA disease duration in years.
Anti-PPAD antibody levels are decreased in RA patients with PD
To elucidate the role of anti-PPAD antibodies in RA-associated PD, we investigated the PD status in RA and healthy controls (clinical characteristics summarized in Table 1). PD was more common among patients with RA compared to age and sex-matched controls (60/83; 72.3% vs. 9/39; 23.1%). Interestingly, anti-rPPAD antibody levels were significantly lower in RA patients with PD compared to periodontally healthy controls (median 0.058 vs. 0.162; *p=0.046). Analogously, anti-rPPAD levels were decreased in PD-positive RA when compared to PD-negative RA patients (median 0.058 vs. 0.162). However, this analysis was underpowered and did not reach statistical significance (p=0.073) (figure 5C). Moreover, a significant decline in antibody levels with RA disease duration was observed in PD-positive RA (r=-0.28, *p=0.03) (figure 5D), but not in the PD-negative RA group (r=−0.01, p=0.97; data not shown). Although the duration of PD in these groups is unknown, the negative association with RA-associated PD may suggest a protective role of anti-PPAD antibodies for periodontal health.
Table 1.
Demographic and clinical characteristics of rheumatoid arthritis patients and controls
| Parameter | RA (83) | Controls (39) | p* |
|---|---|---|---|
| Female (%) | 79.5 (66) | 82.1 (32) | 0.74 |
| Mean age in years (± SD) | 50.7 (± 12.9) | 48.1 (± 13.3) | 0.31 |
| Race / ethnicity | 0.00 | ||
| Caucasian (%) | 74.7 (62) | 33.3 (13) | |
| African-American (%) | 16.9 (14) | 35.9 (14) | |
| Smoking (%)† | 16.9 (14) | 12.8 (5) | 0.56 |
| Periodontal disease (%) | 72.3 (60) | 23.1 (9) | 0.00 |
| Mean RA disease duration in years (± SD) | 6.3 (± 7.9) | ||
| Seropositive for IgM RF (%) | 68.5 (50) | ||
| Seropositive for IgG anti-CCP3 (%) | 75.9 (63) | ||
| Mean CRP in mg/L (± SD) | 6.9 (± 11.3) | ||
| Mean DAS28-CRP (± SD) | 3.5 (± 1.4) | ||
| Mean CDAI (± SD) | 15.6 (± 14.0) |
Comparison of parameters reflected in percentages: chi-square test for proportions, comparison of means: independent-samples t-test; SD, standard deviation.
Current cigarette smoking
RF, rheumatoid factor, cut-off level for positivity 20 IU; anti-CCP3, anti-cyclic citrullinated peptide antibody (third-generation), cut-off level for positivity 20 units; CRP, C-reactive protein; DAS28-CRP, Disease Activity Score 28-CRP; CDAI, Clinical Disease Activity Index.
DISCUSSION
PPAD autocitrullination has provided an intriguing framework to explain the loss of tolerance to citrullinated proteins in RA. In this study, we show that PPAD expressed by P gingivalis is not citrullinated, and demonstrate that PPAD citrullination is not recognized by anti-PPAD antibodies in RA. These findings have critical implications for the biology of PPAD and redefine the relevance of the anti-PPAD response in RA.
Despite the difference in molecular weight, cellular PPAD (75-85 kDa) and secreted PPAD (47 kDa) were indistinguishable in protein sequence. Mass spectrometry identified both as a truncated form of PPAD missing the first 43 N-terminal and 97 C-terminal amino acids. Full-length PPAD was not detected in our analysis, suggesting that similar to proforms of other P gingivalis virulence factors, this pro-enzyme is rapidly processed by truncation and concomitantly translocated to the bacterial cell surface.27,39,40 As such, glycosylation of cPPAD for membrane attachment may be sufficient to explain the striking difference in molecular weight to sPPAD,29 as previously shown for arginine gingipain.41 The truncation sites identified in this study may indicate specific processing by P gingivalis envelope-associated cysteine proteases with arginine-X (at R43) and lysine-X activity (at K459).42 Truncation at R43 has previously been shown for sPPAD by N-terminal sequencing,9 and MS.27 While we cannot exclude that truncation at K459 is an artifact of tryptic digestion, semitryptic peptides indicative of alternative cleavage sites were not detected.
Considering full-length rPPAD is readily autocitrullinated when expressed in E coli,12 the lack of citrullinated PPAD in P gingivalis is astounding. Unique strategies, which are absent in E coli, may have evolved in P gingivalis to prevent PPAD autocitrullination. In this study, we provide evidence that N-terminal processing may be the primary mechanism by which PPAD is protected from autocitrullination. Indeed, PPAD citrullination may be biologically unfavorable for P gingivalis. Rodríguez et al reported that PPAD activity declines with enzyme autocitrullination,30 and citrullination of R352 adjacent to the active site of PPAD (C351) has been hypothesized to explain this reduction in enzyme activity.12,30,36 Mechanisms that block PPAD autocitrullination in P gingivalis may therefore conserve PPAD activity. The molecular basis by which N-terminal truncation abrogates PPAD autocitrullination is unclear. While PPAD is generally restricted to modify only free arginine and C-terminal arginine residues in cleaved proteins,9,11,12 PPAD autocitrullination involves citrullination of internal arginine residues in the intact enzyme. The N-terminal domain may therefore facilitate peptidylarginine autocitrullination by uniquely expanding the catalytic function of full-length PPAD. Rapid removal of the N-terminal domain by P gingivalis may render autocitrullination insufficient and aid bacterial survival in the periodontal pocket.
The finding that anti-PPAD antibodies in RA do not target citrullinated PPAD is consistent with the biology of PPAD identified in this study. However, our findings markedly contrast the conclusions reached by another study in which antibodies specific for citrullinated full-length rPPAD were identified in 38% of RA patients.12 The discrepancy between the two studies may be explained by differences in ELISA methodology, in patient cohorts studied, or in the rPPAD fusion proteins used. We tried to minimize error and exclude plate variability by measuring antibodies to cit-rPPAD, rPPADC351S and background reactivity on the same plate. Using competition assays, we show that anti-PPAD antibodies in patients with RA are exclusively directed against the unmodified enzyme. We cannot fully exclude major differences in the populations studied. Lastly, there is a significant difference in the recombinant proteins used in our study. Previously, GST-His-tagged rPPAD was used to screen for anti-PPAD antibodies.12 The GST tag (25 kDa) adds 9 additional arginine residues to the fusion protein, which may potentially be citrullinated in the process of GST-His-rPPAD autocitrullination. By contrast, rPPAD in our study was designed without GST tag and only contains one additional arginine in the His-tag used (2.5 kDa). Thus, we cannot exclude that the antibodies identified in the previous study may in fact target citrullinated GST in the fusion protein. Such antibodies would not be identified when screening against enzymatically inactive GST-His-rPPAD. Nonetheless, the existence of citrullinated-PPAD-specific antibodies in RA cannot be supported, and appears implausible in the absence of PPAD citrullination in P gingivalis.
While anti-PPAD antibodies are common in patients with RA (and healthy controls), the significance of the antibody response against this bacterial virulence factor for RA-associated PD is not clear. In our study, anti-PPAD antibodies did not correlate with disease activity or anti-CCP levels, reinforcing that anti-PPAD antibodies are not part of the ACPA-response. Antibodies to P gingivalis cell extracts have been reported to correlate with anti-CCP antibodies and disease activity measures in RA,19,43–45 suggesting that anti-PPAD antibodies may represent a unique antibody population. Despite this association, anti-P gingivalis antibodies do not distinguish RA from controls cases,45 and have no predictive value for the development of RA in seropositive arthralgia patients.46 In this context, it is compelling that anti-PPAD levels are significantly lower in RA with PD compared to periodontally healthy controls. Anti-PPAD antibodies may have a protective role for PD in RA, and clearance of PPAD may decrease survival of P gingivalis in the periodontal pocket by limiting ammonia production. The finding that antibodies against major P gingivalis virulence factors confer protection in murine models of periodontal infection and potentially human disease further supports this possibility.18,47–50
Our findings suggest that PPAD autocitrullination is not the underlying mechanism linking P gingivalis-associated PD and RA. Anti-PPAD antibodies may be useful markers to predict the risk of PD progression in patients with RA.
Supplementary Material
Acknowledgments
We wish to thank Lauren DeVine, MS and Robert N. Cole, PhD (Mass Spectrometry and Proteomics Facility, The Johns Hopkins University School of Medicine) for assistance with the citrullination site analyses, Mark A. Reynolds, DDS PhD and Mark E. Shirtliff, PhD (University of Maryland School of Dentistry, Baltimore, MD) for providing bacterial cultures, and Sharon R. Ghazarian, PhD (Biostatistics Core, The Johns Hopkins University School of Medicine) and Janelle M. Montagne (Division of Rheumatology, The Johns Hopkins University School of Medicine) for critical review of the manuscript.
Funding
The study was supported by Grant Number P30AR053503 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAMS or the National Institutes of Health. Additional funding was provided by the Rheumatology Research Foundation (American College of Rheumatology Research and Education Foundation, Within Our Reach), The Donald B. and Dorothy L. Stabler Foundation, the Mackley Fund from Sibley Memorial Hospital, and The Johns Hopkins Arthritis Center Research Fund.
Footnotes
Competing Interests
None declared.
REFERENCES
- 1.Imboden JB. The Immunopathogenesis of Rheumatoid Arthritis. Annu Rev Pathol Mech Dis. 2009;4:417–34. doi: 10.1146/annurev.pathol.4.110807.092254. [DOI] [PubMed] [Google Scholar]
- 2.Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 3.Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat Rev Rheumatol. 2012;8:573–86. doi: 10.1038/nrrheum.2012.134. [DOI] [PubMed] [Google Scholar]
- 4.Schellekens GA, de Jong BA, van den Hoogen FH, et al. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101:273–81. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura K, Sugiyama D, Kogata Y, et al. Meta-analysis: diagnostic accuracy of anti-cyclic citrullinated peptide antibody and rheumatoid factor for rheumatoid arthritis. Ann Intern Med. 2007;146:797–808. doi: 10.7326/0003-4819-146-11-200706050-00008. [DOI] [PubMed] [Google Scholar]
- 6.Van de Stadt LA, de Koning MHMT, van de Stadt RJ, et al. Development of the anti-citrullinated protein antibody repertoire prior to the onset of rheumatoid arthritis. Arthritis Rheum. 2011;63:3226–33. doi: 10.1002/art.30537. [DOI] [PubMed] [Google Scholar]
- 7.Arita K, Hashimoto H, Shimizu T, et al. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–83.. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 8.Shirai H, Mokrab Y, Mizuguchi K. The guanidino-group modifying enzymes: structural basis for their diversity and commonality. Proteins. 2006;64:1010–23. doi: 10.1002/prot.20863. [DOI] [PubMed] [Google Scholar]
- 9.McGraw WT, Potempa J, Farley D, et al. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun. 1999;67:3248–56. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenstein ED, Greenwald RA, Kushner LJ, et al. Hypothesis: The Humoral Immune Response to Oral Bacteria Provides a Stimulus for the Development of Rheumatoid Arthritis. Inflammation. 2004;28:311–8. doi: 10.1007/s10753-004-6641-z. [DOI] [PubMed] [Google Scholar]
- 11.Wegner N, Wait R, Sroka A, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: Implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62:2662–72. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quirke A-M, Lugli EB, Wegner N, et al. Heightened immune response to autocitrullinated Porphyromonas gingivalis peptidylarginine deiminase: a potential mechanism for breaching immunologic tolerance in rheumatoid arthritis. Ann Rheum Dis. 2014;73:263–9. doi: 10.1136/annrheumdis-2012-202726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bingham CO, 3rd, Moni M. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr Opin Rheumatol. 2013;25:345–53. doi: 10.1097/BOR.0b013e32835fb8ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page RC, Eke PI. Case definitions for use in population-based surveillance of periodontitis. J Periodontol. 2007;78:1387–99. doi: 10.1902/jop.2007.060264. [DOI] [PubMed] [Google Scholar]
- 15.De Pablo P, Dietrich T, McAlindon TE. Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. J Rheumatol. 2008;35:70–6. [PubMed] [Google Scholar]
- 16.Pischon N, Pischon T, Kröger J, et al. Association among rheumatoid arthritis, oral hygiene, and periodontitis. J Periodontol. 2008;79:979–86.. doi: 10.1902/jop.2008.070501. [DOI] [PubMed] [Google Scholar]
- 17.Potikuri D, Dannana KC, Kanchinadam S, et al. Periodontal disease is significantly higher in non-smoking treatment-naive rheumatoid arthritis patients: results from a case-control study. Ann Rheum Dis. 2012;71:1541–4. doi: 10.1136/annrheumdis-2011-200380. [DOI] [PubMed] [Google Scholar]
- 18.Scher JU, Ubeda C, Equinda M, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64:3083–94. doi: 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Smit M, Westra J, Vissink A, et al. Periodontitis in established rheumatoid arthritis patients: a cross-sectional clinical, microbiological and serological study. Arthritis Res Ther. 2012;14:R222. doi: 10.1186/ar4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44.. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 21.Griffen AL, Lyons SR, Becker MR, et al. Porphyromonas gingivalis Strain Variability and Periodontitis. J Clin Microbiol. 1999;37:4028–33. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquis RE, Bender GR, Murray DR, et al. Arginine deiminase system and bacterial adaptation to acid environments. Appl Environ Microbiol. 1987;53:198–200. doi: 10.1128/aem.53.1.198-200.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casiano-Colón A, Marquis RE. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 1988;54:1318–24. doi: 10.1128/aem.54.6.1318-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederman R, Brunkhorst B, Smith S, et al. Ammonia as a potential mediator of adult human periodontal infection: inhibition of neutrophil function. Arch Oral Biol. 1990;35(Suppl):205S–209S. doi: 10.1016/0003-9969(90)90159-8. [DOI] [PubMed] [Google Scholar]
- 26.Shawcross DL, Wright GAK, Stadlbauer V, et al. Ammonia impairs neutrophil phagocytic function in liver disease. Hepatol Baltim Md. 2008;48:1202–12. doi: 10.1002/hep.22474. [DOI] [PubMed] [Google Scholar]
- 27.Sato K, Yukitake H, Narita Y, et al. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett. 2013;338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 28.Slakeski N, Seers CA, Ng K, et al. C-Terminal Domain Residues Important for Secretion and Attachment of RgpB in Porphyromonas gingivalis. J Bacteriol. 2011;193:132–42.. doi: 10.1128/JB.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoji M, Sato K, Yukitake H, et al. Por Secretion System-Dependent Secretion and Glycosylation of Porphyromonas gingivalis Hemin-Binding Protein 35. PLoS ONE. 2011;6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez SB, Stitt BL, Ash DE. Expression of peptidylarginine deiminase from Porphyromonas gingivalis in Escherichia coli: Enzyme purification and characterization. Arch Biochem Biophys. 2009;488:14–22. doi: 10.1016/j.abb.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundberg K, Kinloch A, Fisher BA, et al. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008;58:3009–19. doi: 10.1002/art.23936. [DOI] [PubMed] [Google Scholar]
- 32.Lundberg K, Wegner N, Yucel-Lindberg T, et al. Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6:727–30. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 33.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 34.Eke PI, Page RC, Wei L, et al. Update of the case definitions for population-based surveillance of periodontitis. J Periodontol. 2012;83:1449–54. doi: 10.1902/jop.2012.110664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senshu T, Sato T, Inoue T, et al. Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal Biochem. 1992;203:94–100.. doi: 10.1016/0003-2697(92)90047-b. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez SB, Stitt BL, Ash DE. Cysteine 351 is an essential nucleophile in catalysis by Porphyromonas gingivalis peptidylarginine deiminase. Arch Biochem Biophys. 2010;504:190–6. doi: 10.1016/j.abb.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrade F, Darrah E, Gucek M, et al. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62:1630–40. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beausoleil SA, Villén J, Gerber SA, et al. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 39.Glew MD, Veith PD, Peng B, et al. PG0026 Is the C-terminal Signal Peptidase of a Novel Secretion System of Porphyromonas gingivalis. J Biol Chem. 2012;287:24605–17. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veith P, Talbo G, Slakeski N, et al. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem J. 2002;363:105–15. doi: 10.1042/0264-6021:3630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis MA, Thickett A, Slaney JM, et al. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect Immun. 1999;67:3816–23. doi: 10.1128/iai.67.8.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veith PD, Talbo GH, Slakeski N, et al. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem J. 2002;363:105–15. doi: 10.1042/0264-6021:3630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikuls TR, Payne JB, Reinhardt RA, et al. Antibody responses to Porphyromonas gingivalis (P. gingivalis) in subjects with rheumatoid arthritis and periodontitis. Int Immunopharmacol. 2009;9:38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arvikar SL, Collier DS, Fisher MC, et al. Clinical correlations with Porphyromonas gingivalis antibody responses in patients with early rheumatoid arthritis. Arthritis Res Ther. 2013;15:R109. doi: 10.1186/ar4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikuls TR, Payne JB, Yu F, et al. Periodontitis and Porphyromonas gingivalis in Patients with Rheumatoid Arthritis. Arthritis Rheum Published Online First. 2014 Jan 8; doi: 10.1002/art.38348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Smit M, van de Stadt LA, Janssen KMJ, et al. Antibodies against Porphyromonas gingivalis in seropositive arthralgia patients do not predict development of rheumatoid arthritis. Ann Rheum Dis. 2014:annrheumdis–2013–204594. doi: 10.1136/annrheumdis-2013-204594. [DOI] [PubMed] [Google Scholar]
- 47.Genco CA, Odusanya BM, Potempa J, et al. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect Immun. 1998;66:4108–14. doi: 10.1128/iai.66.9.4108-4114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuboniwa M, Amano A, Shizukuishi S, et al. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect Immun. 2001;69:2972–9. doi: 10.1128/IAI.69.5.2972-2979.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson FC, Genco CA. Prevention of Porphyromonas gingivalis-Induced Oral Bone Loss following Immunization with Gingipain R1. Infect Immun. 2001;69:7959–63. doi: 10.1128/IAI.69.12.7959-7963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sweier DG, Shelburne PS, Giannobile WV, et al. Immunoglobulin G (IgG) Class, but Not IgA or IgM, Antibodies to Peptides of the Porphyromonas gingivalis Chaperone HtpG Predict Health in Subjects with Periodontitis by a Fluorescence Enzyme-Linked Immunosorbent Assay. Clin Vaccine Immunol CVI. 2009;16:1766–73. doi: 10.1128/CVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.