Abstract
Initiation of cellular DNA replication is tightly controlled to sustain genomic integrity. In eukaryotes, the heterohexameric origin recognition complex (ORC) is essential for coordinating replication onset. The 3.5 Å resolution crystal structure of Drosophila ORC reveals that the 270 kDa initiator core complex comprises a two-layered notched ring in which a collar of winged-helix domains from the Orc1-5 subunits sits atop a layer of AAA+ ATPase folds. Although canonical inter-AAA+ domain interactions exist between four of the six ORC subunits, unanticipated features are also evident, including highly interdigitated domain-swapping interactions between the winged-helix folds and AAA+ modules of neighboring protomers, and a quasi-spiral arrangement of DNA binding elements that circumnavigate a ~20 Å wide channel in the center of the complex. Comparative analyses indicate that ORC encircles DNA, using its winged-helix domain face to engage the MCM2-7 complex during replicative helicase loading; however, an observed >90° out-of-plane rotation for the Orc1 AAA+ domain disrupts interactions with catalytic amino acids in Orc4, narrowing and sealing off entry into the central channel. Prima facie, our data indicate that Drosophila ORC can switch between active and autoinhibited conformations, suggesting a novel means for cell cycle and/or developmental control of ORC functions.
Introduction
The faithful replication of chromosomes relies on evolutionarily conserved initiator proteins to recruit ring-shaped helicases to DNA in a cell-cycle regulated manner (reviewed in1,2,3). Replication initiators belong to the AAA+ (ATPases associated with a variety of cellular activities) protein superfamily, a large group of multi-subunit nucleotide hydrolases that function as motors or molecular switches in many cellular processes4. AAA+ NTPases assemble into homo- or hetero-oligomeric complexes that actively alter the conformation or position of client macromolecules in response to ATP binding and hydrolysis.
In eukaryotes, replication initiation is promoted by a six-subunit origin recognition complex (ORC)5,6. Five of ORC’s six subunits (Orc1-5) retain AAA+ modules4,7–9, while the sixth (Orc6) is composed of tandem cyclin-box folds similar to transcription factor IIB (TFIIB)10,11. During initiation, ORC binds replication origins, recruiting another AAA+ ATPase, Cdc6, to DNA in a nucleotide-dependent manner (reviewed in3,12). The DNA-bound ORC•Cdc6 complex in turn recruits the MCM2-7 replicative helicase and its associated Cdt1 chaperone to origins, promoting the loading of MCM2-7 complexes onto DNA (reviewed in3,13).
To date, many of the molecular mechanisms by which ORC assembles and operates have remained enigmatic. To better understand ORC function, we therefore determined the crystal structure of the Drosophila complex to 3.5 Å resolution. The structure highlights a domain-swapped organization for ORC and captures the complex in an unanticipated, autoinhibited conformation. Analysis of the structure leads to a revised model for DNA binding and a re-evaluation of proposed ORC•MCM2-7 contacts, which together define a stepwise series of assembly and conformational intermediates that help account for how the complex acts during the early stages of replication initiation.
Crystal structure of the ORC hexamer
Sequence analyses had indicated that the Orc1-5 subunits would share a domain architecture similar to that of archaeal Orc proteins, with AAA+-type ATPase folds fused to at least one C-terminal winged-helix (WH) DNA-binding domain (Fig. 1a)7,8. For its part, the Orc6 C-terminus has been reported to bind to ORC1-5 through a domain insertion in Orc3, leaving its TFIIB-like domain conformationally independent of the ORC core (Fig. 1a)14. For crystallizing Drosophila ORC, we designed a “trimmed” construct lacking the flexible N-terminal extensions of Orc1, Orc2 and Orc314, and the Orc6 TFIIB region (Fig. 1a). Neither modification interfered with ORC assembly, nor did they affect the overall architecture of ORC (Extended Data Fig. 1a–c). This ORC core (referred to as ORC hereafter) crystalized in space group I222 with one Orc1-6 heterohexamer per asymmetric unit. The structure was phased by single-wavelength anomalous dispersion and refined to 3.5 Å with an Rwork/Rfree of 0.22/0.26 (Extended Data Fig. 2a–c and Extended Data Table 1).
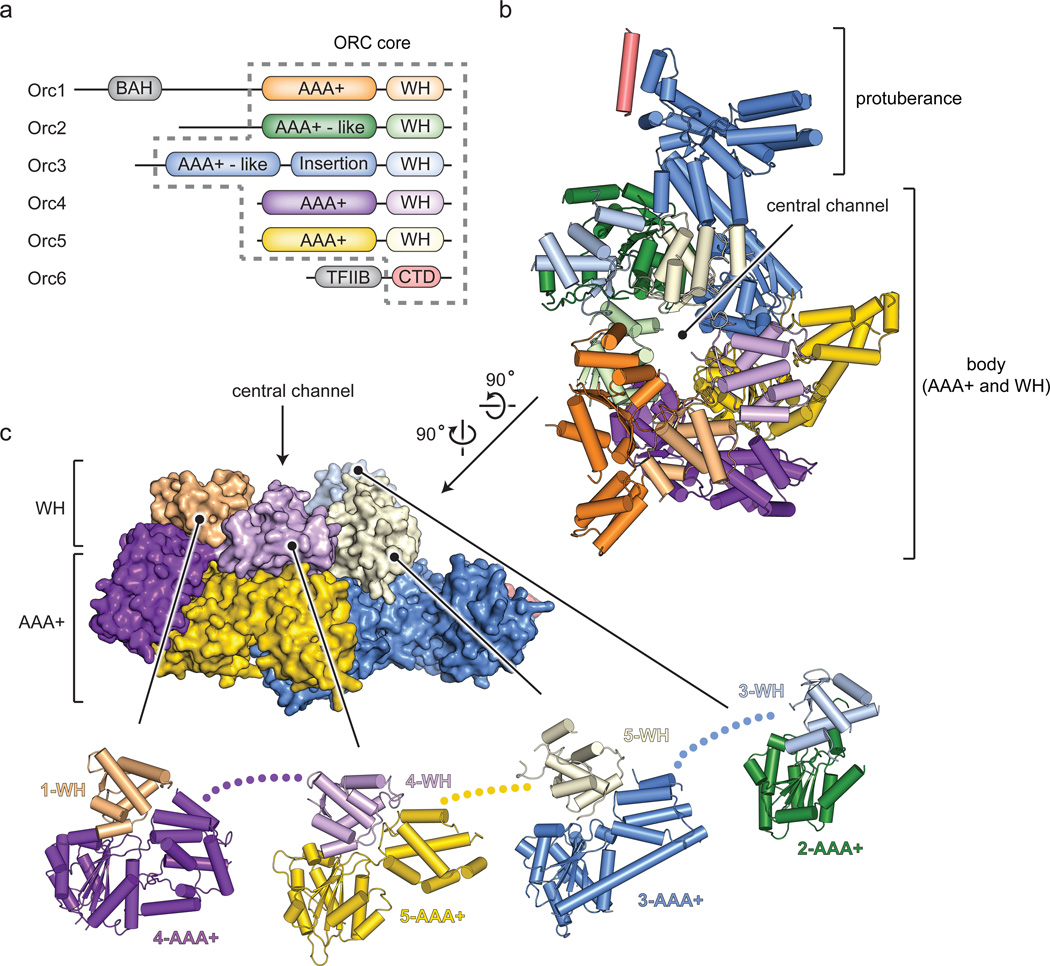
Figure 1.
Structure of Drosophila ORC. a) Domain organization of ORC subunits. Dashed lines demarcate the ORC core used for crystallization (TFIIB and CTD – transcription factor II-like and C-terminal domains in Orc6; BAH – bromo-adjacent homology domain in Orc1). b) Crystal structure of ORC. Domains of individual subunits are colored as in (a). c) Side view of ORC (surface) highlighting the two-tiered, domain-swapped organization of the ORC body (Orc1 AAA+ and Orc2 WH domains, as well as N-terminal Orc2 residues (built as polyalanine) are not shown). Exploded view (cartoon) showing the packing of WH domains against adjacent subunits’ AAA+ regions.
AAA+ and WH domains interlock within the ORC body
The ORC structure shows that the complex forms a lopsided, two-tiered ring with a cashew-shaped protuberance off of one edge (Fig. 1b, c and Supplementary Video 1). Orc1 through Orc5 comprise the ring body, which bears a prominent central channel, while a large domain insertion in Orc3 forms a bi-lobed, α-helical protuberance that engages a short α-helix formed at the Orc6 C-terminus. In contrast to models based on EM reconstructions14,15, the AAA+ subunits are arranged in the order of Orc1-Orc4-Orc5-Orc3-Orc2, thus revising the placement for Orc2 and Orc3 within the pentameric ORC ring (Fig. 1b).
In the structure, Orc1-5 each comprises one AAA+ fold, followed by a single C-terminal WH domain (Extended Data Fig. 3). The AAA+ and WH regions co-associate, but are segregated between the two ring tiers (Fig. 1b, c and Supplementary Video 1). Interestingly, the collar of WH domains is rotationally offset from the AAA+ domains, leading to a domain-swapped organization wherein, apart from Orc2, the WH domain of one subunit packs against the AAA+ domain of its adjoining partner (Fig. 1c). Domain swapping is facilitated by long linkages between the AAA+ and WH modules, a region known to be conformationally flexible in archaeal Orc homologs (Extended Data Fig. 3)16–19. Other deviations from archetypal AAA+ structures include the absence of a small α-helical “lid” subdomain in the AAA+ fold of Orc2, a larger-than normal lid in Orc4, and a complete domain insertion in the lid of Orc3; the latter augmentation forms the protuberance that extends from the principal ORC body to bind Orc6 (Extended Data Figs. 3 and 4; Supplementary Discussion). Globally, the interdigitated interactions between the WH and AAA+ collars, as well as the extensive protein-protein contacts between adjoining WH domains indicate that the WH domains are an important determinant of ORC stability (5,660 Å2 of total surface area are buried between the WH domains, and 15,052 A2 between the WH and AAA+ tiers).
Orientation of DNA binding elements in the WH domain collar
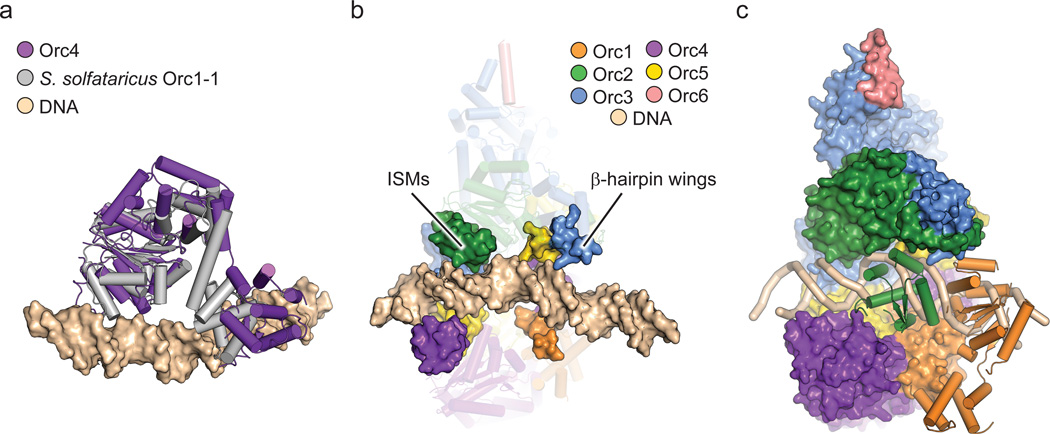
In archaeal Orc homologs, the WH element is responsible for recognizing origin sequences, in which a helix-turn-helix (HTH) motif and a β-hairpin “wing” interact with the adjacent major and minor grooves of double-stranded DNA16,17,20,21 (Fig. 2a). Given the conservation between archaeal Orcs and eukaryotic Orc1-5 proteins, we anticipated that ORC’s WH domains would bind DNA in a similar manner. However, the second α-helix of the HTH in the WH domain (corresponding to the DNA “recognition helix”) is buried against the AAA+ tier in all subunits but Orc2 (Fig. 2b). This arrangement leaves the β-hairpin wings of Orc1, Orc4, Orc5 and Orc3 solvent exposed, which in turn co-localize to form a portion of the interior surface within the central channel in the ORC body (Fig. 2b, c). Given the extensive contacts between the WH and AAA+ tiers, sequestration of the recognition helix seems necessary to maintain ORC integrity. Thus, certain aspects of DNA recognition by the WH domains of ORC likely differ from the approach used by archaeal Orcs.
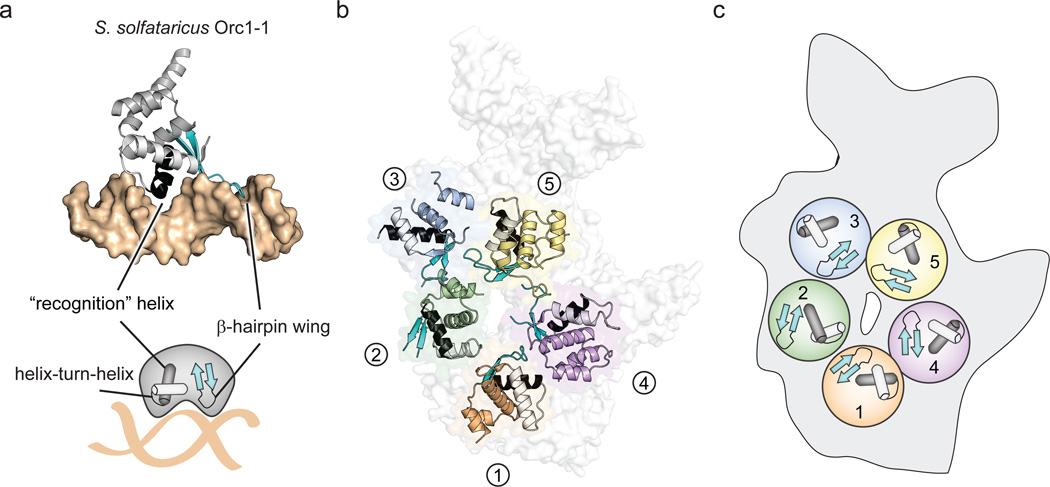
Figure 2.
Eukaryotic ORC and archaeal Orc WH domains. a) WH domain – DNA interactions in archaeal Orc1-1 (PDB code 2qby chain A)16. The WH domain (grey) uses a helix-turn-helix (HTH, black and white) motif and a β-hairpin wing (cyan) motif to engage DNA (tan surface). b) Disposition of WH domains in ORC. The ORC WH domains (excepting Orc2) form a collar in which the recognition helices (black) are buried and the β-hairpin wings form an exposed portion of the central channel (the Orc1 AAA+ domain is not shown). The HTH motif and β-hairpin wings of ORC subunits are colored as in (a). c) Schematic of the HTH and β-hairpin motifs in ORC.
Quasi-canonical AAA+ domain packing within ORC
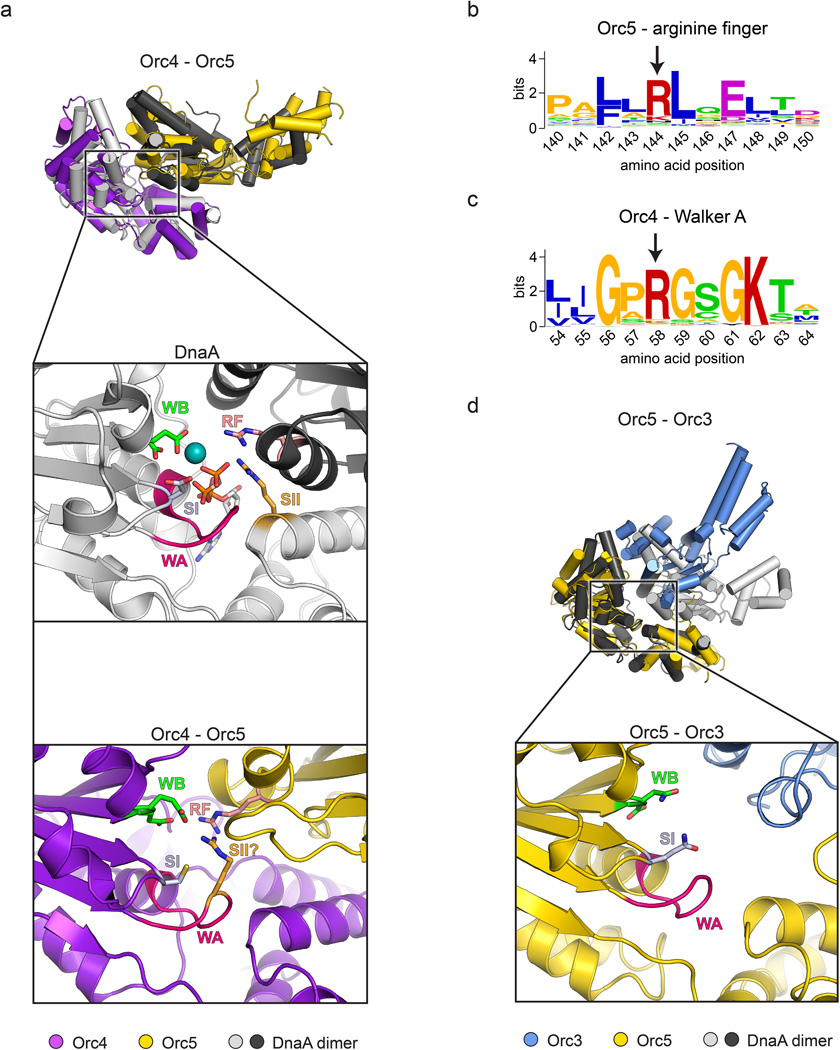
AAA+ ATPases tend to oligomerize into either closed-ring, open-ring, or helical assemblies4,9,22. Based on phylogenetic, biochemical, and EM data, ORC has been proposed to follow a similar trend7,8,14,23,24. The ORC structure shows that, of the various AAA+ domain interactions, the one between Orc4 and Orc5 is most like that seen in typical AAA+ systems, whereby the two subunits associate to form a bipartite nucleotide-binding site at the inter-protomer interface (Fig. 3a and Extended Data Fig. 5a). Metazoan Orc4 manifests conserved Walker A and B ATPase motifs (GKT and D(D/E), respectively), which in related AAA+ ATPases typically contact both β- and γ-phosphates of bound nucleotide and a catalytically important Mg2+ ion. Interestingly, Orc5 donates a well-conserved “Arginine-finger” (Arg144) into the Orc4 active site (Extended Data Fig. 5a, b). Moreover, although Orc4 does not possess a typical “Sensor II” arginine (an amino acid motif in AAA+ ATPases that often aids hydrolysis), its Walker A region does retain a conserved basic amino acid (Arg58)(Extended Data Fig. 5c) that occupies an analogous position (Extended Data Fig. 5a). Overall, the observed structural organization and conservation of catalytically important residues in the Orc4•Orc5 interface raises the possibility that in some organisms, Orc4 might be able to support some level of ATP turnover, conformationally modulate ORC function in response to ATP, or require nucleotide to promote ORC stability (as has been observed for the human complex25,26). Any such activity likely varies between species, however, as S. cerevisiae Orc4 neither contains a conserved Walker A motif nor has been found to bind/hydrolyze ATP27.
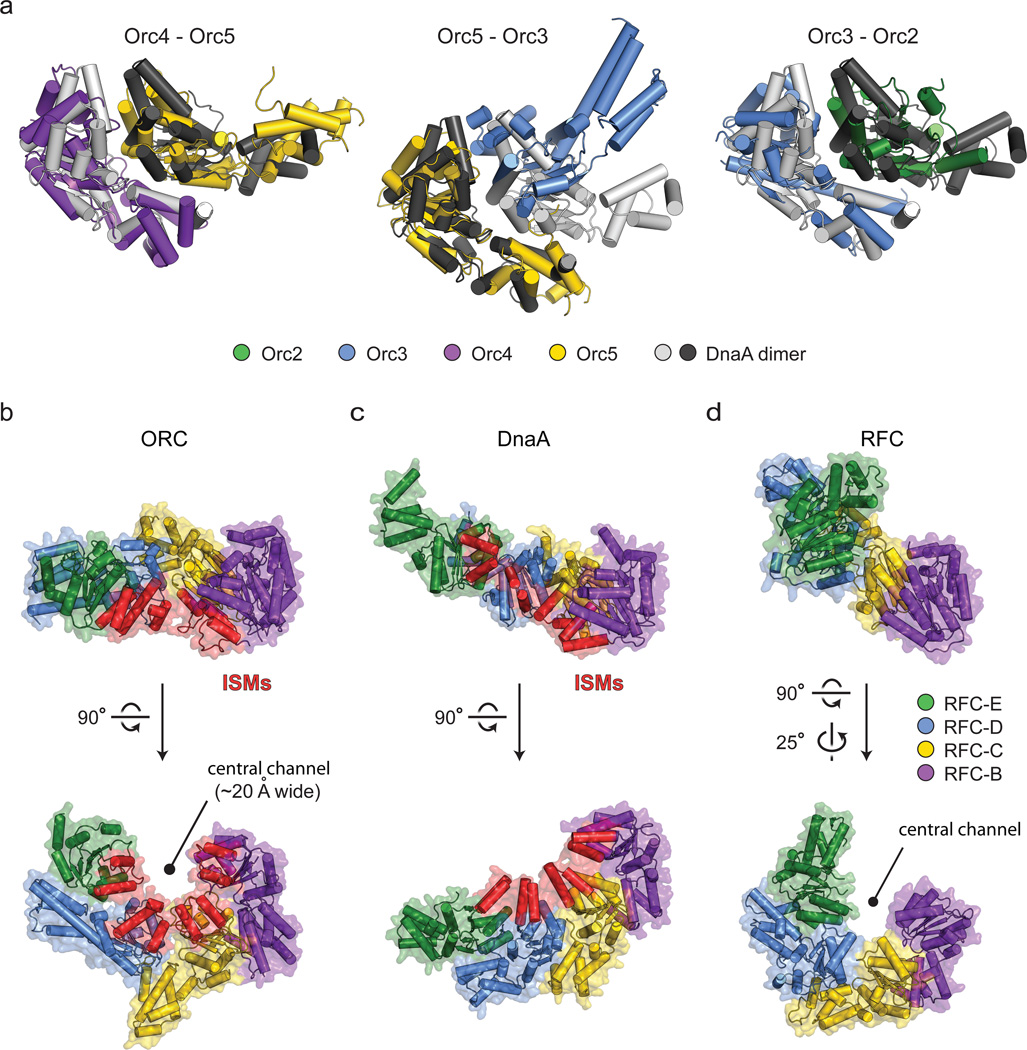
Figure 3.
AAA+/AAA+ domain interactions in ORC. a) Pairwise AAA+ interactions in the Orc2-Orc5 oligomer compared to canonical AAA+ interactions in an ATP-bound DnaA dimer (PDB code 2hcb22). Superpositions were carried out using the AAA+ domain of the left-most protomer of a given ORC subunit pair and DnaA homodimer. b-d) Comparison of AAA+ domain assemblies (to scale) for Orc2-5 (b), ATP-assembled DnaA (PDB code 2hcb22) (c), and Replication Factor C (RFC, PDB code 1sxj31)(d). Upper panels depict side views, with the purple AAA+ domains of DnaA and RFC co-oriented as per the Orc4 AAA+ region. Lower panels show views down the central channel of ORC and RFC, or slightly offset from the helical axis of DnaA. The initiator specific motifs (ISMs, red) of Orc2-5 and DnaA are highlighted (RFC lacks this element). Only the AAA+ domains of ORC, DnaA, and RFC are shown; the RFC-A subunit is omitted. Coloring for ORC is maintained as in panel (a), with sequential subunits in DnaA and RFC colored accordingly.
Other AAA+/AAA+ interactions within ORC deviate to a greater or lesser extent from the canonical packing arrangements exemplified by Orc4•Orc5. For example, the Orc5•Orc3 interface is relatively open, with fewer contacts between AAA+ modules (Fig. 3a and Extended Data Fig. 5d). Although Orc5 possesses a canonical Walker A motif in most eukaryotes, the same is not true for its Walker B motif, nor is there a candidate Orc5 Sensor II residue or an Orc3 arginine finger. These structural features are consistent with biochemical observations showing that Orc5 binds ATP but lacks ATP hydrolysis activity26–30. By comparison, residues for nucleotide binding and hydrolysis are not conserved at the Orc3•Orc2 AAA+/AAA+ interface; hence, the Orc2 and Orc3 AAA+ domains and their interactions appear to play a predominantly structural part in ORC assembly.
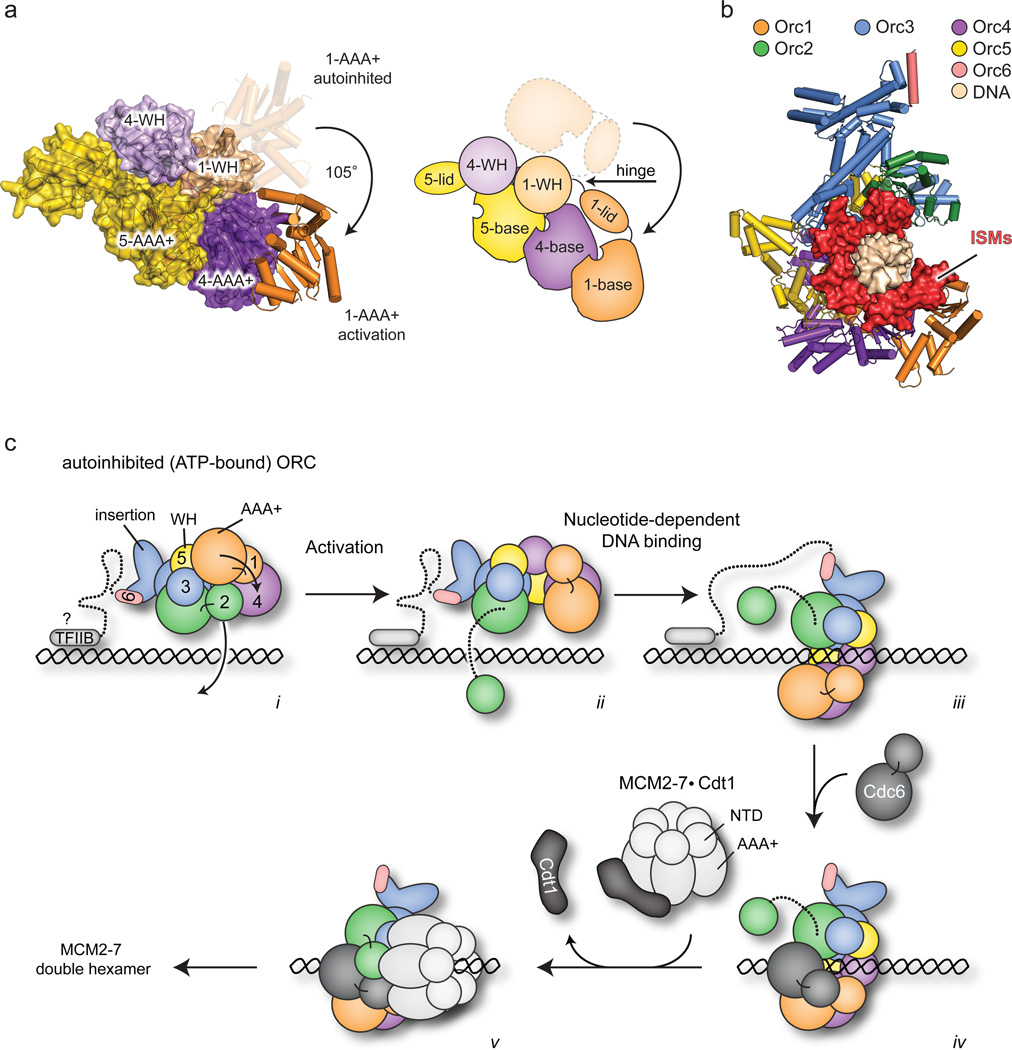
Consistent with its blend of typical and atypical local AAA+/AAA+ interactions, the global organization between ORC’s AAA+ domains is also mixed. Of AAA+ proteins, ORC is most closely related phylogenetically to other replication initiators, including bacterial DnaA, and to DNA polymerase clamp loaders9. In comparing the ORC ATPase assembly to these systems, it can be seen that the Orc4•Orc5 and Orc3•Orc2 AAA+ folds are organized similarly to protomers in ATP-assembled DnaA (Fig. 3a). By contrast, the AAA+/AAA+ organization at the Orc5•Orc3 interface is relatively open. Consequently, the clean helical symmetry observed in ATP-assembled DnaA oligomers22 is broken within the Orc5•Orc3 junction of ORC, a configuration that creates a cracked ring-like architecture reminiscent of clamp loaders31 (Fig. 3b–d). Despite the somewhat more planar arrangement of ATPase folds in ORC, the Orc2-5 initiator specific motifs (or “ISMs”, an α-helical modification that both distinguishes AAA+-family replication initiators and binds DNA directly8,16,17,22,32) nonetheless cluster together, forming a shallow, quasi-spiral shaped set of “threads” that line the interior of the ~20 Å wide central ORC channel (Fig. 3b). Thus, ORC’s structural features are a hybrid of both clamp loader and prokaryotic initiator systems.
An unanticipated Orc1 conformation
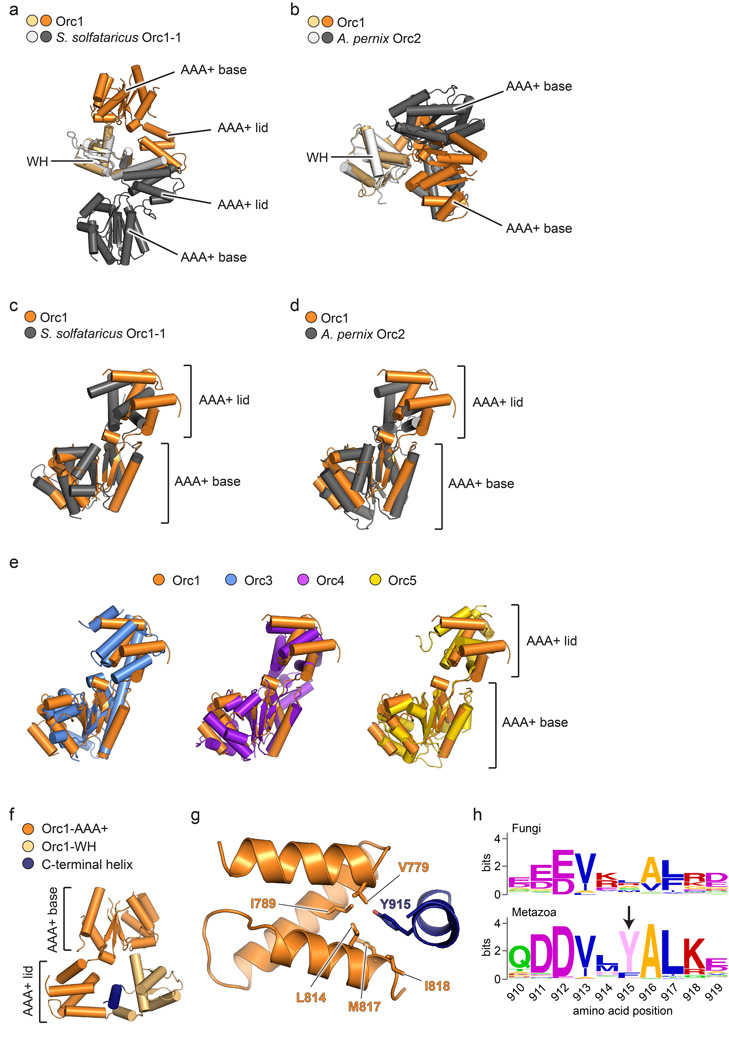
In addition to Orc4 and Orc5, one other ORC subunit also known to bind ATP is Orc127,30. Indeed, Orc1 serves as the major source of ATPase activity in ORC and requires a conserved arginine residue from Orc4 for catalytic activity27–29,33. Given this activity, we expected, as with Orc2-Orc5, to see relatively canonical AAA+ interactions between Orc1 and Orc4 in the structure. Surprisingly, the AAA+ domain of Orc1 is completely disengaged from Orc4, due to buckling of the linker at a single region between the Orc1 ATPase and WH folds (residues 819–821) that gives rise to a ~105° out-of-plane rotation from the Orc2-5 AAA+ domains (Fig. 4a). Although this Orc1 AAA+/WH domain juxtaposition is unique compared to other subunits in the structure (Extended Data Fig. 3), it is similar to one of the conformations adopted by an ADP-bound archaeal Orc homolog, A. pernix Orc218 (Extended Data Fig. 6a, b). Notably, the movement of Orc1 does not significantly affect the relative arrangement of its two AAA+ subdomains, which is maintained as in Orc3-5 and archaeal Orcs (Extended Data Fig. 6c–e). The en bloc reorientation of Orc1 appears stabilized by contacts between the Orc1 AAA+ domain and the WH domains of Orc1, Orc2 and Orc3, together burying a total of 4,256 Å2 at the interface (Fig. 4b, Extended Data Fig. 6f–h).
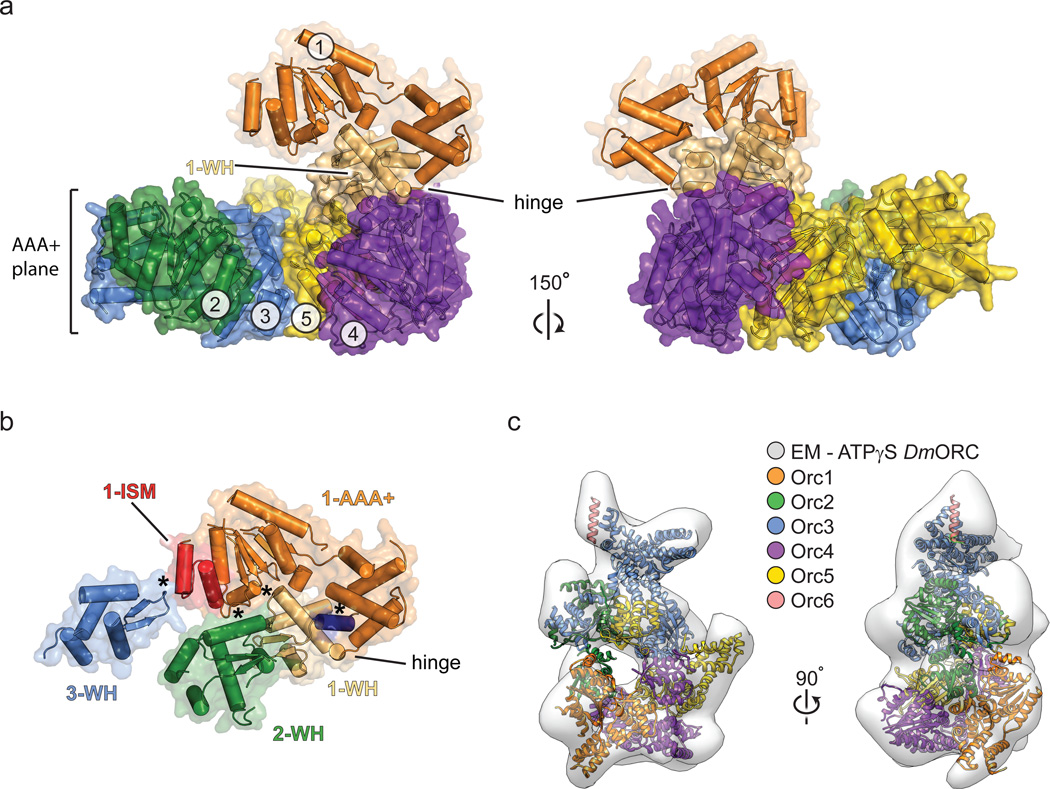
Figure 4.
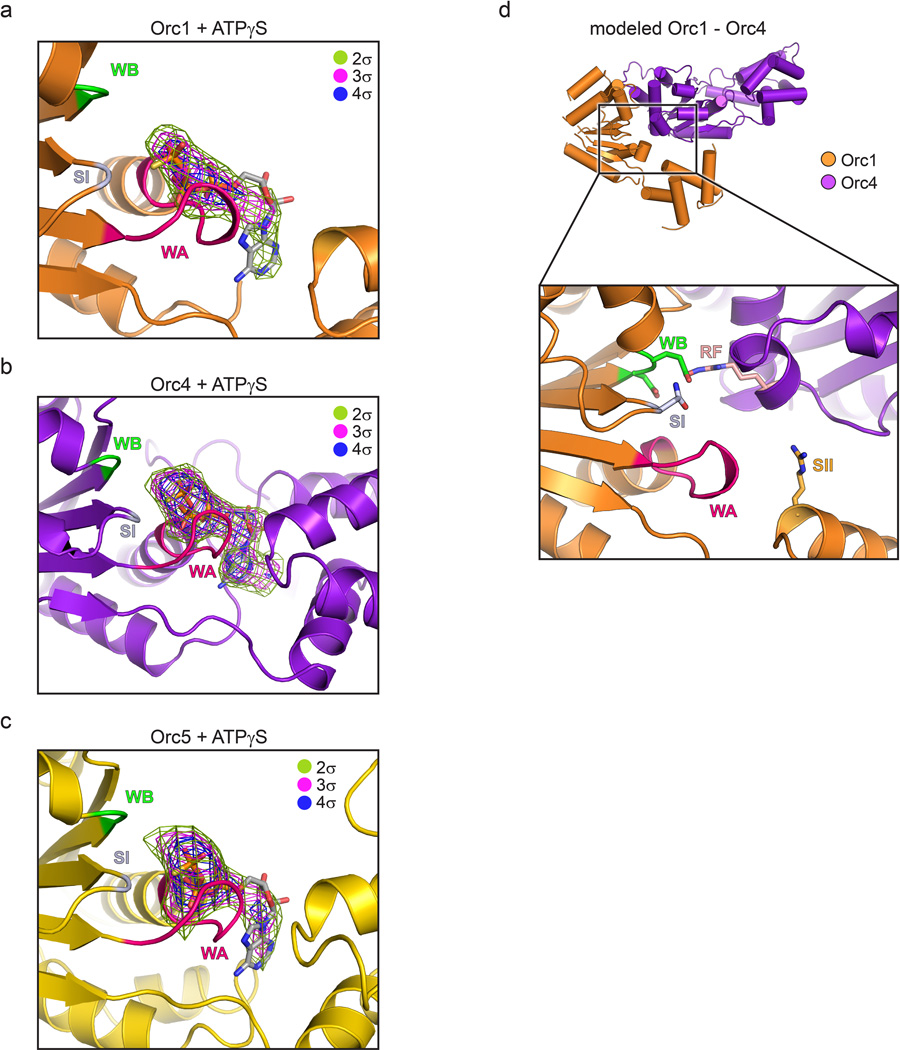
An unanticipated but naturally-occurring Orc1 conformation. a) The Orc1 AAA+ domain is disengaged from Orc4 and sits above the plane of the Orc2-Orc5 AAA+ oligomer. Only the AAA+ domains of Orc1-5 and the WH domain of Orc1 are depicted; the hinge point between the Orc1 AAA+ and WH regions is indicated. b) The Orc1 conformation is stabilized by contacts (asterisks) between its AAA+ domain and the WH domains of Orc1-3 (the C-terminal helix of Orc1 is in dark blue). See also Extended Data Fig. 6f–h. c) Docking study showing that the ORC crystal structure matches the 3D EM volume of Drosophila ORC (EMD-247914). See also Extended Data Fig. 8c and Supplementary Video 2.
A consequence of Orc1’s disposition within the complex is that its nucleotide binding cleft resides ~40 Å away from the arginine finger of Orc4. Importantly, comparison of the crystal structure with a prior 3D EM reconstruction of ATPγS-bound Drosophila ORC14 shows excellent agreement between the two models (Fig. 4c and Supplementary Video 2), indicating that the Orc1 conformation in the crystal corresponds to the predominant state of the complex in solution. Moreover, co-crystallization of ORC with the ATP analog ATPγS, while showing clear density for nucleotide binding to the Orc1, Orc4, and Orc5 AAA+ folds (Extended Data Fig. 7a–c), recapitulates the configuration seen in apo-ORC. Together, these data indicate that Drosophila Orc1 must undergo a large structural change to support ATPase activity, but that ATP-binding is itself insufficient to drive such a rearrangement in a majority of ORC particles.
Implications of the structure for DNA binding by ORC
In the ORC structure, the central channel within the body of the complex is formed by both the Orc2-5 ISMs and the β-hairpin wings of Orc1 and Orc3-5. In archaeal Orcs, these two elements both bind to duplex DNA16,17. To investigate if the ORC central channel could accommodate any of the DNA interactions typified by archaeal Orcs, we superposed the DNA-bound crystal structure of Sulfolobus solfataricus Orc1-116 onto Orc4 (after Orc1, Orc4 is most closely related to archaeal Orcs). Notably, superpositioning of the AAA+ domains of the two proteins (Fig. 5a) resulted in a placement for DNA that aligns the duplex coaxially with the central ORC channel (Fig. 5b). Inspection of the resultant ORC•DNA model not only reveals that the quasi-spiral formed by the Orc2-5 ISMs approximates that of the docked duplex, but that the β-hairpin wings of the Orc1 and Orc3-5 WH domains also reside in a position where they can access the nucleic-acid segment (Fig. 5b).
Figure 5.
The central channel in ORC likely binds DNA. a) Structural alignment of Orc4 onto DNA-bound S. solfataricus Orc1-1 (PDB code 2qby chain A16). b) Reciprocal superposition of DNA-bound archaeal Orc1-1 onto Orc4 in the context of ORC aligns the DNA duplex with ORC’s central channel. The ISMs of Orc2-5 and β-hairpin wings of Orc1 and Orc3-5 (surface) form two semicircular, quasi-spiral collars around the docked DNA (Orc1 AAA+ and Orc2 WH domains are omitted; the remainder of ORC is shown as transparent cartoons). c) The DNA path (tan ribbon) through the central channel is partially blocked by the Orc1 AAA+ (orange cartoon) and the Orc2 WH (green cartoon) domains. The remainder of ORC is shown as surface representation. Coloring per (b).
The superpositioning between DNA-bound archaeal Orc1-1 and the DNA-free Drosophila ORC imaged here has important implications for understanding how the eukaryotic initiator engages origin regions. One is that ORC likely does not bend DNA into a U-turn, as has been suggested23, but instead binds DNA using a mechanism similar to that of sliding clamp loaders, which encircle primer-template junctions34,35. This binding mode is congruent with a recent proposal based on the EM analysis of an ORC•Cdc6•Cdt1•MCM2-7 complex, which posits that ORC helps align the ring of an MCM2-7 complex around DNA24. The apparent sequence-specificity of DNA binding by S. cerevisiae ORC5 (in contrast to metazoan ORC36,37), likely results from specific interactions between amino acid side chains in the ORC channel and nucleotide bases in specific ARS regions.
At present it is unclear from the structure why Drosophila ORC prefers to bind negatively-supercoiled DNA over linear segments36, a binding preference also reported for S. pombe ORC38. Archaeal Orcs are known to underwind DNA upon binding16,17, suggesting that the eukaryotic ORC AAA+ and WH domains may cooperate to do likewise; topological changes in DNA structure induced by S. pombe and human ORC are consistent with such an interpretation38,39. Alternatively, if ORC were to bind B-form DNA, conformational transitions within the ORC body and its associated DNA-binding elements would be required to accommodate the DNA duplex. In this instance, specificity for negatively-supercoiled substrates could arise from the relative positioning of the TFIIB-like DNA-binding elements in Orc610,11,40 and the DNA-recognition elements in the central ORC channel.
Mechanism for ORC activation and implications for helicase loading
Modeling indicates that the DNA binding elements in the central ORC channel can encircle a DNA duplex; however, certain features of the complex would appear to preclude Drosophila ORC from doing so in the state seen both here and by EM. For example, passage of the DNA through the entirety of the central channel is prevented by a constriction formed by both the Orc2 WH domain and the Orc1 AAA+ fold (Fig. 1b). Similarly, although the ATPase region of the ORC ring is cracked open, the Orc2 WH domain and the Orc1 AAA+ fold occlude this crack, thereby preventing the lateral entry of DNA into the central channel from the side of the complex (Figs. 1b and 5c). This observation likely helps explain the weak effect that nucleotide has on DNA binding by Drosophila ORC purified from fly embryos or recombinant sources28,36. It is also interesting to note that by preventing DNA binding, the placement of the Orc1 ATPase region and the Orc2 WH fold also blocks the known interaction site for another critical component of replication initiation, Cdc623,24. Overall, the simplest interpretation of the ORC conformation imaged here is that it corresponds to a naturally autoinhibited form of the complex, and that in some organisms, only a fraction of the total ORC pool that can be obtained from asynchronously dividing cells may be capable of productively altering its interactions with DNA in response to ATP.
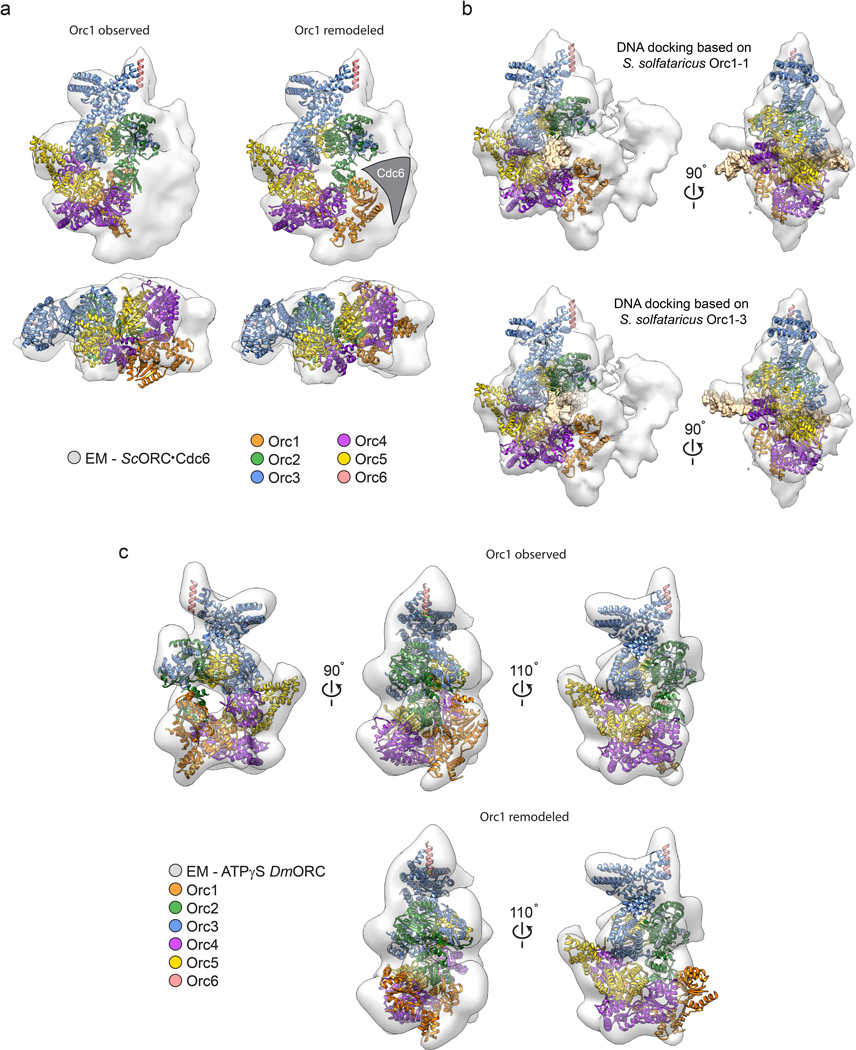
If Drosophila ORC first assembles into an inactive form, then what manner of transition might push the complex into a new state in which its ATPase region is now competent to bind DNA (or Cdc6)? Insights into a simple structural rearrangement that could support such a switch can be gleaned from what is known about archetypal AAA+ ATPase organization. Using the Orc4•Orc5 interaction seen in the crystal structure as a template, we generated a model for the expected arrangement of a functional Orc1•Orc4 AAA+ ATPase center by swiveling the Orc1 AAA+ fold around a single hinge point in the linker region just prior to its WH domain (Supplementary Video 1). The resultant model not only restores expected AAA+ interactions between the Orc1 active site and the Orc4 arginine finger (Fig. 6a and Extended Data Fig. 7d), but also both removes the Orc1-mediated blockage of the putative path for DNA in the central channel and co-aligns the Orc1 ISM with the ISM helix formed by Orc2-Orc5 (Fig. 6b and Supplementary Video 1). Docking of the rearranged model into the cryo-EM densities of S. cerevisiae ORC23,24 shows a reasonable fit for Drosophila ORC containing the repositioned Orc1 AAA+ domain, and further reveals that a region of EM density, which extends from the center of the ORC body, actually corresponds to DNA (Extended Data Fig. 8a, b). In accord with a two-state model, the EM density for S. cerevisiae ORC cannot accommodate the AAA+ domain of Orc1 in the state imaged crystallographically (Extended Data Fig. 8a), nor can the Drosophila ATPγS-ORC EM volume14 account for a remodeled Orc1 conformation for ORC (Extended Data Fig. 8c and Supplementary Video 2).
Figure 6.
Model for ORC activation and its functional consequences. a) A ~105° rotation of the Orc1 ATPase fold about a hinge point within the AAA+/WH linker region juxtaposes the Orc1 active site with the arginine finger surface of Orc4 (only Orc1, Orc4 and the Orc5 AAA+ domain are depicted, both as molecular model and schematic). b) Repositioning of Orc1 extends the circuit of ISMs around the prospective DNA binding site of the central channel. The Orc2 WH domain is omitted; DNA is modeled as per Fig. 5 and shown as surface. c) Model for ORC activation and subsequent Cdc6 and MCM2-7 recruitment/loading. i) Prospective recruitment of autoinhibited Drosophila ORC to DNA by elements such as the TFIIB domain of Orc6. ii) Activation of ORC promotes productive Orc1•Orc4 interactions and Orc2 WH domain repositioning, exposing a gap in the ORC ring. iii) DNA binds to ORC’s central channel through ISM/β-hairpin wing interactions, iv) Cdc6 binds between Orc2 and Orc1, trapping DNA inside the complex. v) ORC•Cdc6 binds to and loads one MCM2-7 hexamer (aided by Cdt1) using the WH domain face of ORC•Cdc6. During loading, two MCM2-7 particles are assembled into a double hexamer47,48
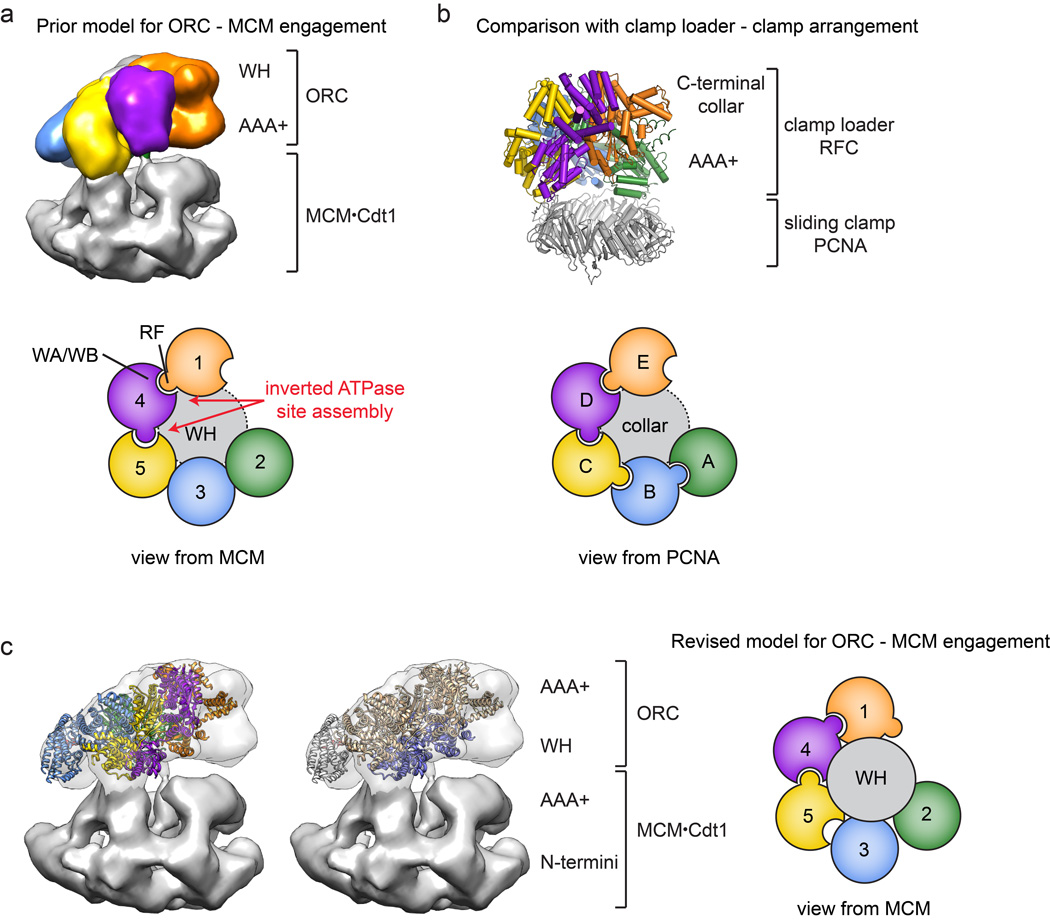
Collectively, the structure and analysis presented here provides a framework for understanding how Drosophila ORC interfaces with its partner proteins and DNA during the initial stages of MCM2-7 loading (Fig. 6c). In this scheme, ORC would start off in an ATP-bound but autoinhibited form – either dissociated from chromatin or in a chromatin-bound state via secondary binding sites/partners – that is restricted in its ability to either bind DNA in its central channel or bind Cdc6 to its ring. Conversion of this state into an activated configuration would involve the en bloc movement of the Orc1 ATPase domain (Supplementary Video 1), allowing Orc1 to engage the arginine finger of Orc4, and unlatching the Orc2 WH domain to open a gap in the Orc1-5 ring. Once open, DNA would bind to the ISM and β-hairpin elements in the central ORC channel, after which Cdc6 would dock into the Orc1/Orc2 gap, trapping DNA within the center of the complex. One prediction of this model is that Cdc6 should bind to ORC using its ATPase center to engage an arginine finger in Orc1 (Drosophila residue Arg734). After formation of a ternary ORC•DNA•Cdc6 complex, the WH domains of Cdc6 and Orc2 would be expected to engage the AAA+ folds of Orc1 and Cdc6, respectively, creating a circuit of WH domains (and their associated β-hairpin elements) that help lock the complex into place. Interestingly, our structural findings and analyses reinterpret a recent 3D EM reconstruction concerning the disposition of ORC and MCM2-7 in a helicase-loading intermediate complex24: rather than using its AAA+ domains to bind MCM2-7, which requires an inverted order of ATPase site-arginine finger interactions around the ORC ring (Extended Data Fig. 9a, b), ORC likely uses its WH domain collar instead (Extended Data Fig. 9c).
A major question still remaining is what event might trigger ORC rearrangement, or why ORC should exist in an autoinhibited state. Based on our EM and co-crystallization data, ATP binding alone is incapable of efficiently driving such a transition for most of the particles in a purified ORC preparation. Interestingly, phosphatase treatment of Drosophila ORC stimulates DNA binding41, suggesting that removal of one or more post-translational marks might help convert ORC to an active form. Moreover, metazoan ORC associates with chromatin in a cell cycle-dependent manner (reviewed in1,42), and the targeting of ORC to chromosomes in metazoans (and in fission yeast) is known to require protein-DNA contacts distinct from those in budding yeast, such as the TFIIB-homology domain in metazoan Orc610,40 or the AT-hook in S. pombe Orc443. The action of these elements, together with the recognition of nucleosomes by the N-terminal BAH domain of Orc144–46, suggest that the formation of nucleotide-dependent contacts between the ATPase region of ORC and DNA may take place after the formation of initial ORC•chromatin encounter complexes. Moreover, metazoans need to stockpile ORC in oocytes, yet keep this pool of ORC from prematurely initiating replication prior to fertilization; hence, ORC may be maternally stored in an inactive form during oogenesis. In these contexts, autoinhibition could provide a novel mechanism for regulating ORC’s productive association with DNA in a cell-cycle dependent and/or developmental stage-specific manner.
Materials and Methods
ORC construct design for crystallization
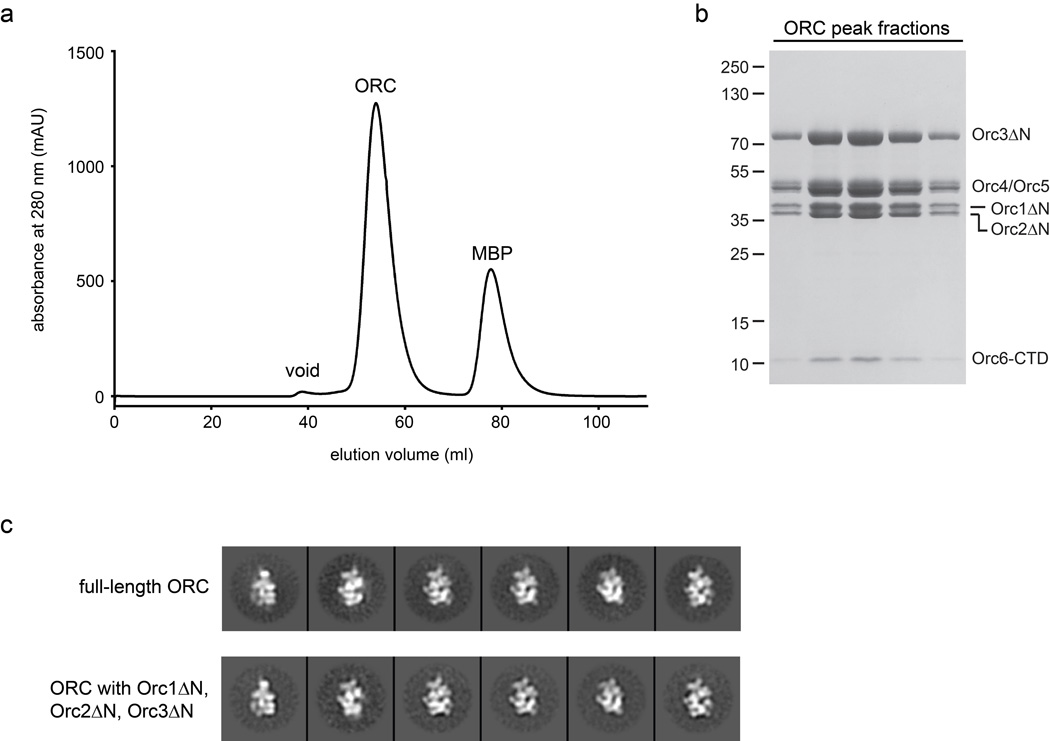
Initial crystallization trials with full-length Drosophila ORC did not yield crystals. To obtain a crystallizable complex, we therefore identified regions in ORC subunits likely to be unconserved, disordered, or flexibly tethered to ORC by multiple sequence alignments and electron microscopy14. Based on these analyses, we designed a Drosophila ORC construct lacking the N-terminal regions that precede the AAA+ domains of Orc1 (amino acids 1–532), Orc2 (amino acids 1–265) and Orc3 (amino acids 1–46). These truncated subunits were found to assemble into a stable hexameric complex and yielded 2D class averages very similar to full-length ORC when analyzed by negative-stain EM, indicating that removal of the termini does not affect overall ORC architecture (Extended Data Fig. 1). In addition, we previously found that the TFIIB-like domain of Orc6 is flexibly tethered to ORC via the Orc6 C-terminal domain14; this region was also removed for ORC crystallization. All biochemical experiments described in the present paper were performed with this “trimmed” ORC construct.
Cloning and baculovirus generation
ORC subunits Orc1 to Orc5 (Orc1: amino acid residues 533–924, Orc2: amino acid residues 266–618, Orc3: amino acid residues 47–721, Orc4: amino acid residues 1–459, Orc5: amino acid residues 1–460) were cloned into a pFastBac-derived polycistronic BioBricks vector (UC Berkeley MacroLab). A hexa-histidine (6xHis) tag was added to the N-terminus of Orc1 and a maltose binding protein (MBP) tag to the N-terminus of Orc4, both followed by a tobacco etch virus (TEV) protease cleavage site. The C-terminus of Orc6 (amino acid residues 187–257) was cloned into a separate pFastBac vector. For Orc6 binding and crosslinking experiments, Orc6 (amino acid residues 187–257) was cloned with an N-terminal 6xHis tag into a ligation-independent-cloning (LIC)-compatible pFastBac vector14. Point mutations (Y225S, A236E, and A236C) were introduced by site-directed mutagenesis and verified by DNA sequencing.
Bacmids were generated in DH10Bac cells and isolated as per the Bac-to-Bac protocol (Invitrogen Life Technologies). Sf9 cells were transfected with bacmid DNA using Cellfectin II (Invritrogen Life Technologies), also according to the manufacturer’s instructions. Baculoviruses were amplified twice in Sf9 cells before infecting large-scale cultures for protein expression.
Expression and purification of ORC for crystallization
For expression, 8 L of High5 cells grown in spinner flasks were co-infected with two baculoviruses: the multibac virus expressing Orc1 through Orc5 and the baculovirus expressing Orc6. 48 h post-infection, cells were harvested by centrifugation and ORC was purified as described in14 with a few modifications. Briefly, harvested cells were resuspended in 200 mL of lysis buffer (50 mM Tris-HCl (pH 7.8), 300 mM KCl, 50 mM imidazole (pH 7.8), 10% glycerol, 200 µM PMSF, 1 µg/L leupeptin) and lysed by sonication. The lysate was clarified by centrifugation, treated with a 20% (NH4)2SO4 precipitation on ice for 30 min, and re-centrifuged. ORC was purified by passing the supernatant solution over a 5 mL HisTrap HP nickel-affinity chromatography column (GE Healthcare), followed by amylose-affinity chromatography using a 20 mL column (New England Biolabs). Purification tags were removed from ORC by incubation with 6xHis-tagged TEV protease49 overnight at 4°C. TEV and any uncleaved His-tagged Orc1-containing material was then removed by an additional nickel-affinity chromatography step in 50 mM Tris-HCl (pH 7.8), 300 mM KCl, 50 mM imidazole (pH 7.8), and 10% glycerol. The flow-through was concentrated in 30K Amicon Ultra-15 concentrators (Millipore) and purified by gel filtration chromatography using a HiPrep 16/60 Sephacryl S-300 HR column (GE Healthcare) equilibrated in 50 mM Tris-HCl (pH 7.8), 300 mM KCl, and 5% glycerol. Peak fractions were pooled and concentrated using 30K Amicon Ultra-15 concentrators (Millipore). Protein was used immediately thereafter for crystallography.
Crystallization
Prior to crystallization, ORC was dialyzed overnight into crystallization buffer (50 mM Tris-HCl (pH 7.8), 200 mM KCl, 5% glycerol, and 0.5 mM TCEP). Crystals grew by sitting-drop vapor diffusion as showers of small plates within a few hours upon combining equal volumes of 10 mg/mL ORC with reservoir solution (50 mM PIPES (pH 7.5), 100 mM ammonium acetate, 10–35 mM MgCl2, and 2.5–5% PEG 20,000) at 19–22°C. These crystals were used as a source for streak seeding to obtain larger crystals, which grew to their maximum size within 2 days. For cryo-protection, the mother liquor was stepwise exchanged (typically 2 steps for 3 h and 15 min, respectively) into 50 mM PIPES (pH 7.5), 25 mM KCl, 10 mM MgCl2, 50 mM ammonium acetate, 4% PEG 20,000, 10% glycerol, 10% ethylene glycol, 20% xylitol, and 0.25 mM TCEP. Crystals were then harvested by looping and flash-frozen in liquid nitrogen (NB – during optimization, a subset of crystals was transferred to 4°C and, after several days of equilibration, cryo-protected and harvested at this temperature; this procedure seemed to slightly increase the number of usable crystals). For phasing, crystals were soaked into cryo-protecting solution containing 1 mM GdCl3 for 3–4 h prior to harvesting. Additional heavy atom-derivatized crystals were obtained by incubating crystals in cryo-protecting solution containing 100 µM ethyl mercuric phosphate for 3 h; data from these soaks were used to identify metal-binding sites for confirming amino acid registers but were not used for phasing.
Crystallization and crystal harvesting for ATPγS-bound ORC was performed as described for apo-ORC but with the following modifications: 1) ORC was dialyzed into crystallization buffer containing MgCl2 (50 mM Tris-HCl (pH 7.8), 200 mM KCl, 5% glycerol, 0.5 mM TCEP, 5 mM MgCl2), 2) prior to crystallization, ATPγS was added to ORC to a final concentration of 1 mM, and 3) for harvesting, 0.5 mM ATPγS was added to the cryo-protectant to prevent dissociation of the nucleotide.
Data collection and structure determination
The diffraction quality of individual crystals varied greatly, necessitating that many hundreds of different crystals be screened to identify acceptable targets for data collection. Crystal screening was performed at beamlines 8.3.1 at the Advanced Light Source (ALS) at Lawrence Berkeley National Lab, X25 at the National Synchrotron Light Source (NSLS) at Brookhaven National Lab, and 23-ID-B at the Advanced Photon Source (APS) at Argonne National Lab. Native diffraction datasets (λ=1.0332 Å) as well as single-wavelength anomalous dispersion (SAD) datasets for gadolinium (λ=1.71083 Å) and sulfur (λ=1.7712 Å, see below) were collected at APS 23-ID-B equipped with a MAR CCD detector. Although complete datasets were typically obtained by exposing multiple regions within a single crystal using the “vector data collection” option to minimize radiation damage, the best native dataset was collected at a single site from a crystal harvested at 4°C. Datasets of ethyl mercuric phosphate-derivatized crystals were collected at NSLS X25 (λ=1.006 Å) on a Pilatus 6M detector.
Diffraction data were processed with XDS50,51. Merging with the program Aimless of the CCP4 software package52 (Extended Data Table 1) indicated that the crystals belonged to space group I222, with unit cell dimension of a=145.5 Å, b=259.0 Å and c=257.0 Å for the best native crystal. The Gd-SAD dataset was obtained by merging data from four different crystals. Despite slight non-isomorphism between crystals, merging data from multiple crystals significantly improved the anomalous signal, phases, and electron density interpretability compared to datasets collected from single crystals.
For initial phasing by SAD, gadolinium sites were identified with SHELXD53. The strongest sites were then used as input into PHASER54 as implemented in PHENIX55 to find additional sites and to obtain initial phases to ~4 Å. Maximum likelihood density modification with RESOLVE56 was used to break the phase ambiguity and to improve electron density maps. At this point, experimental phases were next applied (again using PHENIX) to the native dataset, which was of better quality than the gadolinium derivative. The resulting electron density maps (at 4 Å resolution) allowed identification of all five AAA+ domains and four of the five winged-helix domains, and also revealed density for bulky side chains as expected for this resolution. In parallel to data processing with SHELXD/PHASER/RESOLVE, experimental phases and density maps were also calculated with SHARP and improved by solvent flipping in SOLOMON57,58. Although slightly less featured, the SOLOMON electron density maps were overall very similar to those obtained from RESOLVE but additionally revealed clear protein density in some regions that were poorly defined in RESOLVE density maps. Thus, while model building was performed predominantly into RESOLVE density maps, SOLOMON density maps were used as an additional guide to trace the main chain of the model. Phases were gradually improved by iterative cycles of model building, density modification with phase combination of experimental and model phases in RESOLVE and phase extension to 3.7 Å.
Model building was initiated by automated searches using MOLREP52,59 and by manual docking using UCSF Chimera60,61 to place the AAA+ and winged-helix domains of archaeal Orc1/Cdc6 (PDB codes: 2qby16; 1fnn19) and/or homology models for Drosophila ORC subunits (as generated by Phyre262) into electron density maps. These docked structures were valuable reference points and facilitated tracing of most of the main chain; insights into likely subunit positions (from low-resolution electron microscopy studies of Drosophila ORC14), together with knowledge of the domain architecture of ORC subunits (from sequence predictions), allowed assignment of specific subunits to map density regions. Using COOT63, a nearly complete model of ORC was manually built de novo into phase-combined and B-factor sharpened RESOLVE density maps, guided by the topology of archaeal Orc1/Cdc6 AAA+ and winged-helix domains, as well as by secondary structure prediction and multiple sequence alignments. The initial model was improved by iterative rounds of refinement in PHENIX (real-space, individual xyz, individual ADP), using secondary structure and (in early stages of building) experimental phase restraints, as well as stereochemistry and ADP weight optimization; subsequent rounds of model rebuilding were performed using COOT. During the course of refinement, a slightly higher-resolution and more complete native dataset (to 3.5 Å) was obtained and used for the final rounds of refinement, which also included refinement of TLS parameters. The final model contains the AAA+ and winged-helix domains of Orc1 to Orc5, the Orc3 domain insertion, and the conserved C-terminal helix of Orc6; an N-terminal region of Orc2 (preceding the AAA+ domain) was built as a poly-alanine model, since the amino acid register for this region could not be assigned unambiguously. The final model was validated with MOLPROBITY64 and has excellent geometry (MolProbity score 1.88), with no Ramachandran outliers and only a small fraction (1.9%) of rotamer outliers (Extended Data Table 1).
During the course of model building, several approaches were used to validate the sequence register of the various ORC chains. These included: 1) using Hg-binding sites in ethyl mercuric phosphate-derivatized crystals as fiducials for cysteines and histidines, 2) using sulfur sites in native S-SAD datasets to verify the location of a subset of cysteines and methionines (the weak signal present in these data precluded the use of this information for phasing), and 3) conducting Orc3-Orc6 crosslinking experiments to confirm the register of the Orc6 C-terminal helix (Extended Data Fig. 4h). Hg and S sites were identified from log-likelihood-gradient maps calculated using the MR-SAD option in PHASER54. Additionally, MR-SAD for sulfur sites also revealed the position of two ions with anomalous scattering properties at the wavelength of data collection (λ=1.7712 Å). These ions showed clear density in experimental and 2Fo-Fc maps and were interpreted as a chloride ion in the P-loop of Orc5 and as a potassium ion bound to Orc2.
Once a satisfactory apo-ORC model was obtained, it was used as a search model for molecular replacement (using PHENIX-PHASER) to phase data collected from ORC co-crystallized with ATPγS. The resulting solution (Z-score = 86.7, LLG = 7747) revealed clear (>2-4σ) positive difference density in the nucleotide binding clefts of Orc1, Orc4 and Orc5 that could accommodate ATPγS (Extended data Fig. 7a–c). Since the resolution of these crystals was limited to 4 Å, and since only small structural changes were observed throughout the remainder of ORC, we refrained from building and refining a model against this dataset.
Structure analysis
Structural superpositions and docking into EM maps were performed using UCSF Chimera60,61. Buried surface area at domain/subunit interfaces was calculated with PyMOL (The PyMOL Molecular Graphics System, Version 1.7.0.0 Schrödinger, LLC). Multiple protein sequence alignments were performed with MAFFT65,66 and conservation scores calculated and mapped onto the structure with Consurf67. Sequence logos were generated with WEBLOGO68. Figures were rendered both using PyMOL (The PyMOL Molecular Graphics System, Version 1.7.0.0 Schrödinger, LLC) and UCSF Chimera60,61.
Expression and purification of ORC1-5 and Orc6 for binding and crosslinking studies
ORC containing subunits Orc1 to Orc5 (referred to as ORC1-5) was expressed in High5 cells using a single virus with the multibac approach. Expression and purification were performed as described above for expression of ORC1-6, except that: a) 1 mM β-mercaptoethanol or 1 mM DTT were included in buffers during nickel- and amylose-affinity steps or gel filtration chromatography, respectively, and b) 10% glycerol was maintained in all buffers throughout purification. For crosslinking experiments, 6xHis and MBP tags were removed from ORC1-5, whereas for fluorescence anisotropy and pull-down experiments the affinity tags were left on ORC1-5 by omitting the TEV cleavage and subsequent nickel-affinity steps.
The C-terminus of Orc6 (CTD, residues 187–257) was purified as described previously14. Briefly, 6xHis-tagged Orc6-CTDWT, Orc6-CTDA236E, Orc6-CTDA236C, and Orc6-CTDY225S constructs were expressed in High5 cells and purified by nickel-affinity chromatography in lysis buffer (50 mM Tris-HCl (pH 7.8), 600 mM KCl, 10% glycerol, 50 mM imidazole, 1 mM β-mercaptoethanol, 200 µM PMSF, and 1 µg/L leupeptin). During the second of two wash steps, the salt concentration was decreased to 300 mM KCl. Protein was eluted with 50 mM Tris-HCl (pH 7.8), 300 mM KCl, 10% glycerol, 250 mM imidazole and 1 mM β-mercaptoethanol, concentrated in 3K Amicon Ultra-15 concentrators (Millipore), and further purified on an HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) equilibrated in 50 mM Tris-HCl (pH 7.8), 300 mM KCl, 10% glycerol, and 1 mM DTT. Peak fractions were pooled, concentrated in 3K Amicon Ultra-15 concentrators (Millipore), and stored at −80°C.
Fluorescence anisotropy
Orc6 proteins were N-terminally labeled with Alexa Fluor 488 5-SDP ester (Invitrogen Life Technologies) as described previously14. Binding reactions were performed for 30 min in a total volume of 140 µL containing 30 nM fluorescently labeled wild-type Orc6-CTD, Orc6-CTDY225S, or Orc6-CTDA236E, and different concentrations of ORC1-5 (ranging from 122 pM to 1 µM in two-fold serial dilutions) in a buffer consisting of 50 mM Tris (pH 7.8), 300 mM KCl, 5% glycerol, 1 mM DTT, and 0.1 mg/mL BSA. 40 µL of each reaction were transferred to 384-well plates in triplicates and fluorescence polarization (FP) was measured in a POLARstar Omega plate reader (BMG Labtech) with a 485 nm excitation filter and a 520 nm emission filter. For data analysis, FP measurements were converted into anisotropy units (FA), which ranged from 0.01 to 0.16. The mean FA values obtained from three (for Orc6-CTDY225S and Orc6-CTDA236E) or six (for wild-type Orc6-CTD) independent experiments were fitted to the quadratic binding equation:
where Af and Ab are the measured anisotropy of free and bound, fluorescently labeled Orc6, respectively, [L] is the concentration of Orc6 used in binding assays, [R] is the concentration of ORC1-5, and KD is the apparent binding constant. Due to the limited sensitivity of our plate reader, binding experiments were performed at Orc6 concentrations above the apparent KD for wild-type Orc6 binding to ORC1-5; hence, curve fits are meant to aid visualization rather than to explicitly model the data. Mean and standard deviations of FA values were plotted as a function of ORC1-5 concentration after subtracting Af values from respective mean FA values for visual comparison (Extended Data Fig. 4f).
Crosslinking of Orc6 to ORC1-5
To independently check that the Orc6 C-terminal α-helix was oriented correctly with respect to Orc3, crosslinking studies were performed with either wild-type or an A236C Orc6-CTD mutant and untagged ORC1-5 using the homobifunctional maleimide crosslinker BMOE (bis(maleimido)ethane; Thermo Scientific) under non-reducing conditions. Ala236 of Orc6 was chosen for mutation because inspection of the structure revealed that the insert of Orc3 contained a nearby native cysteine (Cys372), which we reasoned could form a crosslink with an A236C Orc6 mutant if our build register were correct (Extended Data Fig. 4e). Prior to crosslinking, both wild-type Orc6-CTD and Orc6-CTDA236C proteins, as well as untagged ORC1-5, were dialyzed overnight into 50 mM PIPES (pH 7.5), 300 mM KCl, and 10% glycerol to remove reducing agents. Binding of the mutant or wild-type Orc6 protein to ORC1-5 was performed in 50 µL reactions containing 4 µM of each protein in 50 mM PIPES (pH 7.5), 300 mM KCl, and 10% glycerol for 30 min. BMOE was then added to reactions to a final concentration of 0.2 mM for 5 min, after which crosslinking reactions were stopped by adding SDS-PAGE sample-loading dye containing β-mercaptoethanol (100 mM final concentration). Stopped reactions were analyzed by SDS-PAGE and Coomassie staining.
To ensure that Orc6 still bound to ORC1-5 under non-reducing conditions, we performed binding reactions using ORC1-5 containing MBP-tagged Orc4 and either wild-type or A236C Orc6-CTD under non-reducing conditions as described for the crosslinking experiments, but instead subjected the reactions to pull-downs using amylose beads (New England Biolabs). Beads were washed three times with 1 mL of 50 mM PIPES (pH 7.5), 300 mM KCl and 10% glycerol, after which bound proteins were eluted with buffer containing 20 mM maltose and analyzed by SDS-PAGE.
Electron microscopy
4 µL of a 30 nM Drosophila ORC solution containing N-terminally truncated Orc1, Orc2 and Orc3 subunits (in 20 mM Tris (pH 7.8), 125 mM potassium glutamate, 5 mM MgCl2, 1 mM ATPγS) were spotted onto glow-discharged, continuous-carbon film EM grids and stained with four drops of 2% uranyl formate 10 sec each, and blotted. Grids were imaged in a Tecnai T12 BIOTWINN transmission electron microscope operated at 120 keV with a LaB6 cathode as electron source. Data collection, image processing, and 2D classification were performed as described previously14, and 2D class averages were compared to class averages of the full-length ATPγS-ORC dataset reported in14 (Extended Data Fig. 1c).
Extended Data
Extended Data Figure 1.
Deletion of variable N-terminal extensions in Orc1, Orc2 and Orc3 alters neither ORC stability nor overall ORC architecture. a) Gel-filtration chromatography trace of the ORC core used for crystallography, together with b) SDS-PAGE of respective ORC peak fractions from (a), indicate the formation of a stable hexameric complex. c) Full-length ORC and ORC containing N-terminal truncations display similar structural features in 2D EM class averages. Both complexes were imaged by negative-stain EM in the presence of ATPγS. Note that although class averages from ORC with truncated Orc1-3 subunits contain full-length Orc6, Orc6 is not visible due to its flexibile nature14. Class averages for full-length ORC are derived from a dataset used in14.
Extended Data Figure 2.
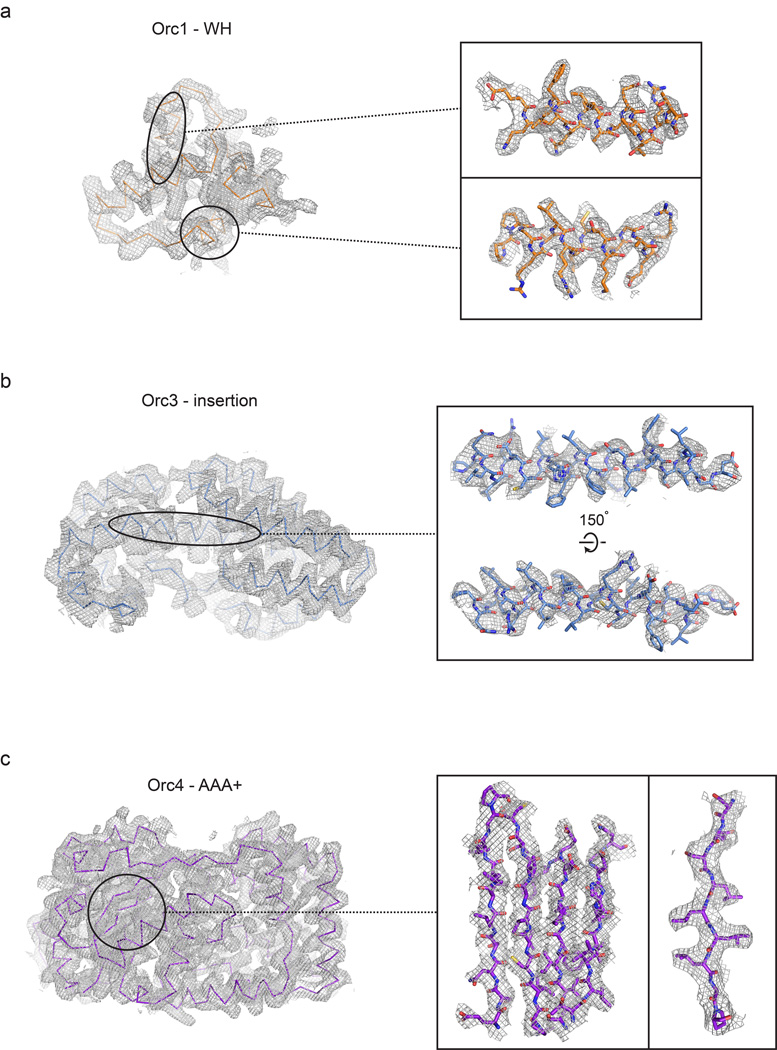
Experimental electron density contoured at 1σ for different regions of the ORC structure. The Orc1 winged-helix (WH) domain is shown in (a), the Orc3 insertion in (b), and the Orc4 AAA+ domain in (c).
Extended Data Figure 3.
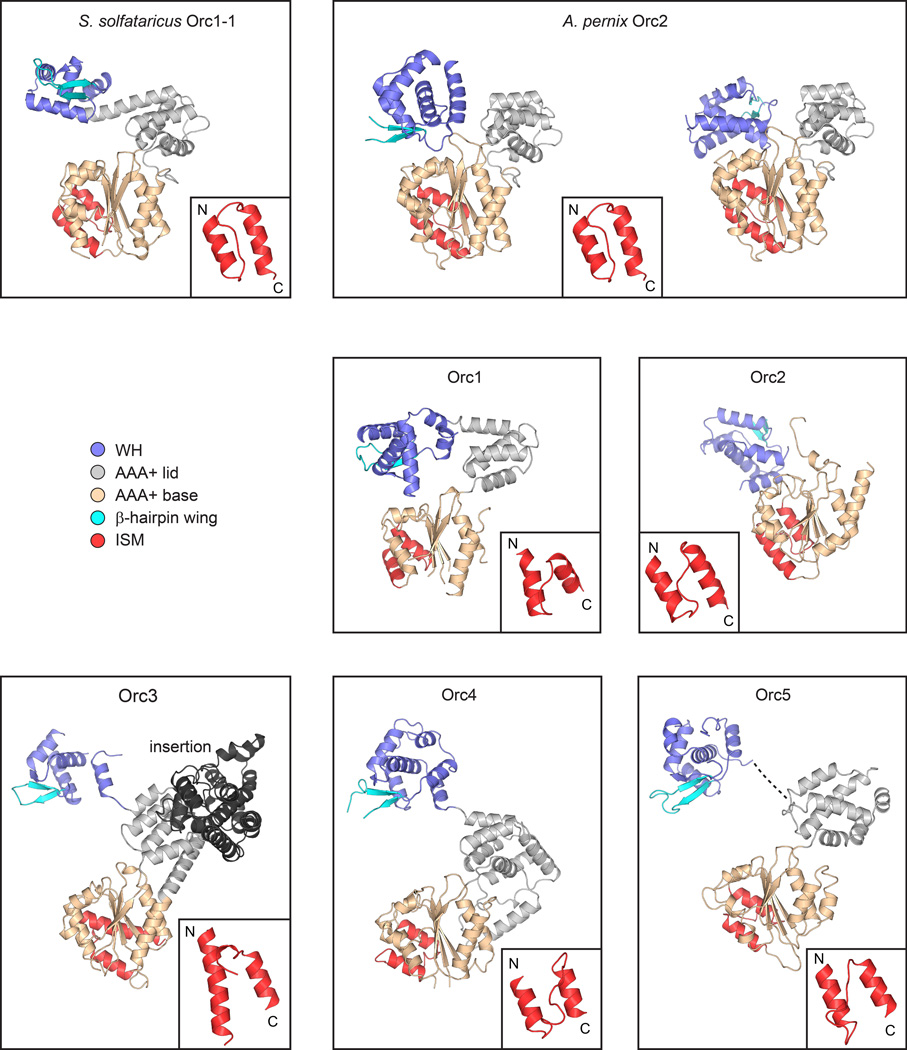
Structure of individual ORC subunits compared to S. solfataricus Orc1-1 (PDB code 2qby chain A16) and A. pernix Orc2 (PDB codes 1w5s chain A (left) and 1w5t chain C (right)18). Different structural elements are colored as indicated. The initiator specific motif (ISM) of the AAA+ ATPase fold is shown in the inset. No electron density was observed for the region linking the AAA+ and WH domains of Orc5 (indicated by a dashed line). The very N-terminal region of Orc2, which could only be built as stretches of polyalanine, is not shown.
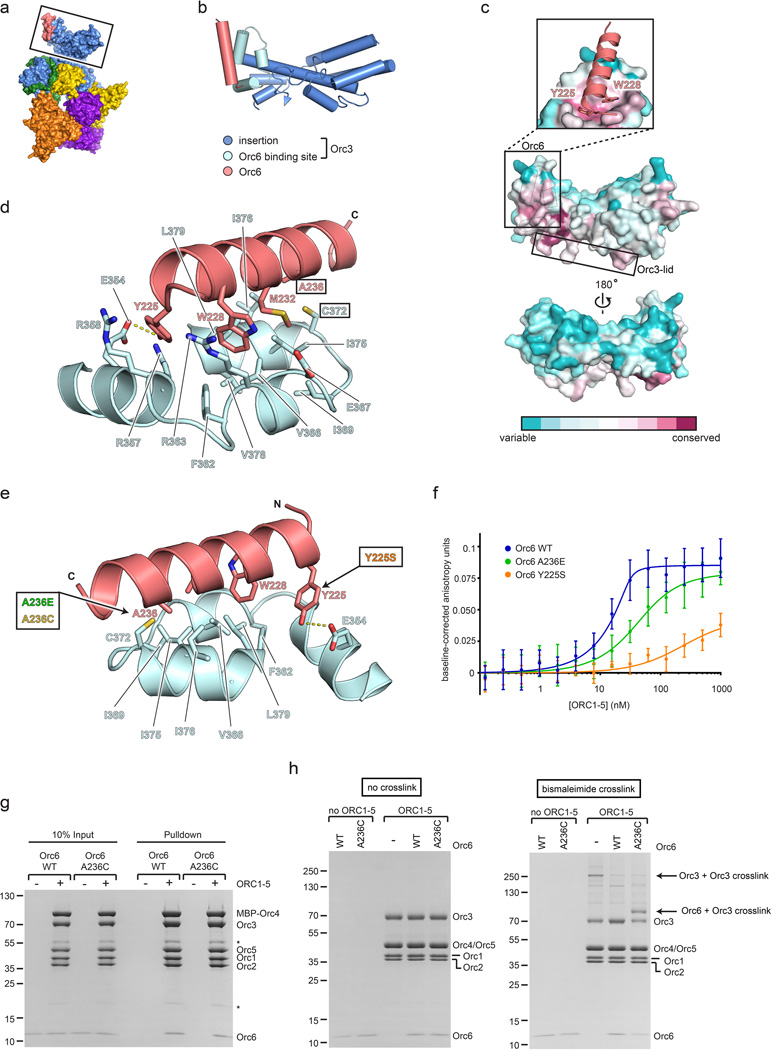
Extended Data Figure 4.
The Orc3 domain insertion forms a conserved, hydrophobic binding platform for Orc6. a) Surface representation of ORC. The Orc3 insertion, which extends from the Orc3 AAA+ lid subdomain and interacts with the C-terminal helix of Orc6, is boxed. b) Secondary structure representation of the boxed region shown in (a). The Orc3 insertion forms a bi-lobed, α-helical fold, three helices of which create a binding site for Orc6. c) Surface conservation of the Orc3 insertion. Conserved Orc3 residues cluster in the region that interacts with the Orc3 lid and in the Orc6 binding pocket. The latter region contacts highly conserved residues in Orc6 (Y225 and W228). d) Close-up view of Orc3•Orc6 interactions, showing a primarily hydrophobic binding site in Orc3 for Orc6 residues (Y225, W228, M232, A236). The Meier-Gorlin syndrome equivalent in Drosophila Orc6, Y225, appears positioned within hydrogen-bonding distance of E354 in Orc3 (dashed line). Colors are as in (b). e to h) Biochemical validation of the binding register for Drosophila Orc6. e) Close-up of the Orc6•Orc3 interface. Orc6-Ala236 faces a hydrophobic surface formed by Orc3 residues and is also in close proximity to a natural cysteine in Orc3 (Cys372). To validate the register of the short C-terminal Orc6 helix and the Orc6•Orc3 interface, we mutated Orc6-Ala236 to either glutamate, which we hypothesized would impede binding to ORC1-5 due to clashes with hydrophobic residues in Orc3, or to cysteine, which we presumed would not affect Orc3 binding but would allow site-specific crosslinking to Orc3-Cys372. f) Orc6A236E has a reduced affinity for the ORC1-5 complex. The C-terminal domains (CTDs) of wild-type (WT) Orc6, Orc6A236E or the Meier-Gorlin syndrome equivalent Orc6Y225S were each N-terminally labeled with Alexa Fluor 488 and tested for ORC1-5 binding using fluorescence anisotropy. As previously shown14, the C-terminal domain of Orc6 binds ORC1-5 with low nanomolar affinity, whereas the Y225S mutation strongly reduces binding. As predicted based on the structure of the Orc6•Orc3 interface, the A236E mutation also reduces the affinity of the Orc6-CTD for ORC1-5. Mean and standard deviations from three (for Orc6Y225S and Orc6A236E) or six (for wild-type Orc6) independent experiments are shown. g) Orc6A236C is able to bind to the ORC1-5 complex. Orc6-CTDWT or Orc6-CTDA236C were incubated with ORC1-5 (containing MBP-tagged Orc4) and subjected to pull-down experiments using amylose resin. Both Orc6-CTDWT and Orc6-CTDA236C co-purified with ORC1-5. The pull-down experiment was performed under non-reducing experimental conditions similar to the crosslinking experiment in panel (h). Asterisks mark two likely proteolytic fragments of Orc3. h) The Orc6-CTDA236C mutant, but not the wild-type Orc6-CTD, specifically crosslinks to Orc3 within the ORC1-5 complex. Orc6-CTDWT or Orc6-CTDA236C, either alone or in the presence of ORC1-5, was incubated with a bifunctional maleimide crosslinker and the proteins subsequently analyzed by SDS-PAGE. In reactions containing ORC1-5 and Orc6-CTDA236C, crosslinking gives rise to a novel band with higher molecular weight than Orc3; the appearance of this band correlates with a decrease in the amount of uncrosslinked Orc3 and Orc6-CTD, and does not appear with reactions containing ORC1-5 and wild-type Orc6-CTD, indicating that this species corresponds to an Orc3-Orc6 crosslink (a moderately strong higher molecular-weight band that appears in the absence of Orc6 likely corresponds to homotypic adducts between exposed cysteines in Orc3). These results are consistent with the structure, which places Orc6-Ala236 in close proximity to Orc3-Cys372. Note that ORC1-5 contained MBP-tagged Orc4 in (g) but that the tag was removed in (h).
Extended Data Figure 5.
ATP-binding site configuration at the Orc4•Orc5 and Orc5•Orc3 interfaces. a) Inter-AAA+ interactions between Orc4 and Orc5 are similar to canonical AAA+ interactions between DnaA protomers (top panel, only Orc4 is used for superpositioning onto the left (light gray) AAA+ domain of an ATP-bound DnaA dimer, PDB code 2hcb22). Close-up views of the nucleotide-binding site are shown for Orc4 (bottom panel) and for DnaA for comparison (middle panel). The resemblance of the Orc4 nucleotide-binding pocket to the active site of functional AAA+ ATPases is somewhat surprising considering that mutations in the active site of Drosophila and human Orc4 have no reported effect on the ATPase activity of ORC as measured in vitro28,29, but may help explain why a Drosophila ORC mutant bearing a Walker A or B substitution in Orc4 exhibits modest DNA replication defects in extracts28. b) The putative arginine finger in Orc5 is well conserved across homologs. A sequence logo of a multiple sequence alignment of the region containing the putative arginine finger (marked with an arrow) in eukaryotic Orc5 protein sequences is shown. Amino acid numberings correspond to the Drosophila Orc5 sequence. c) A potential Sensor-II equivalent arginine (marked with an arrow) in the Orc4 Walker A motif is conserved in eukaryotic Orc4 homologs. A sequence logo of the Walker A motif from a multiple sequence alignment of eukaryotic Orc4 protein sequences is shown. Amino acid positions are numbered as in Drosophila Orc4. d) Inter-AAA+ interactions between Orc5 and Orc3. The top panel shows a superposition derived from placing the AAA+ domain of Orc5 atop the AAA+ domain of the left (dark gray) protomer of an ATP-bound DnaA dimer; the bottom panel shows a close-up view of the nucleotide-binding site at the Orc5•Orc3 interface. Side chains of conserved residues known to be involved in nucleotide binding and hydrolysis in AAA+ ATPases are represented as sticks in both (a) and (d). WA – Walker A, WB – Walker B, SI – Sensor I, SII – Sensor II, RF – arginine finger.
Extended Data Figure 6.
The conformation of Orc1 arises from a reorientation between its AAA+ and WH domains, and not from changes within the AAA+ ATPase domain itself. a) Superpositioning of the WH domains of Orc1 and S. solfataricus Orc1-1 (PDB code 2qby chain A16) reveals different conformations for both proteins, resulting from a large domain rotation of the Orc1 AAA+ domain around a pivot point in the linker preceding its WH domain. b) The Orc1 conformation is most similar to a state seen for A. pernix Orc2 (PDB code 1w5t chain C18). The WH domains of both proteins were superposed as in (a). c to e) Superposing the AAA+ base subdomains of Orc1 and S. solfataricus Orc1-1 (panel (c), PDB code 2qby chain A16), A. pernix Orc2 (panel (d), PDB code 1w5t chain C18), and Orc3, Orc4 or Orc5 (panel (e)) shows that the typical AAA+ configuration between the base and lid subdomains are maintained in Orc1. Only a slight opening of the nucleotide-binding cleft is observed in Orc1, which is likely due to the absence of bound nucleotide. f and g) The most C-terminal α-helix of the Orc1 WH domain mediates interactions with the Orc1 lid subdomain. An overview of the interaction is shown in (f), with a close-up view of contacts between a conserved tyrosine (Tyr915) in the C-terminal Orc1 helix and a hydrophobic pocket of the Orc1 lid depicted in (g). h) The tyrosine in the C-terminal helix of Orc1 is well conserved across metazoan but not fungal Orc1 homologs. Alignments are shown as sequence logos. The numbering of amino acids is based on Drosophila Orc1, and the tyrosine is marked by an arrow.
Extended Data Figure 7.
Nucleotide binding by Orc1, Orc4 and Orc5. For panels (a) to (c), molecular replacement with the apo-ORC model was used to phase diffraction data collected from an ORC-ATPγS co-crystal. Positive Fo-Fc difference density contoured at different sigma levels reveals clear features for nucleotide binding to the AAA+ domains of: a) Orc1, b) Orc4, and c) Orc5. ATPγS is docked into the difference density for reference; due to the moderate (4.0 Å) resolution of the data, this structure was not refined. d) Modeling of canonical AAA+ interactions between Orc1 and Orc4, generated using the Orc4•Orc5 interaction as a reference. Upper panel: structural overview of modeled AAA+ domain positioning between Orc1 and Orc4. Lower panel: Close-up of the modeled Orc1•Orc4 ATPase site. Side chains (taken from their place in the apo-ORC model as a reference) are shown for conserved catalytically important residues. WA – Walker A, WB – Walker B, SI – Sensor I, SII – Sensor II, RF – arginine finger.
Extended Data Figure 8.
Docking of the observed and remodeled ORC structures into the cryo-EM density of S. cerevisiae and Drosophila ORC indicates that the ATPase domain of Orc1 is repositioned into a canonical AAA+/AAA+ interaction with Orc4 when Cdc6 is present, and supports a model where DNA passes through the central channel in ORC. a) The 3D EM volume for S. cerevisiae ORC (as present in a complex with Cdc6, Cdt1, and MCM2-7 and assembled in the presence of DNA – EMD-562524) contains Orc1 in the activated conformation. ORC with Orc1 in the autoinhibited conformation (left panel, as observed in the crystal structure) and remodeled conformation (right panel, remodeled) were docked into the ORC•Cdc6•Cdt1•Mcm2-7 cryo-EM map (only the density for ORC•Cdc6 is shown). The ORC•Cdc6 EM density readily accommodates Orc1 in the activated conformation, but not in its autoinhibited state. The EM density corresponding to Cdc6 is indicated in the right panel. b) DNA passes through the central ORC channel in the DNA•ORC•Cdc6 complex. ORC (with Orc1 in the remodeled conformation) was first docked into the cryo-EM map derived from a DNA•ORC•Cdc6 complex (EMD-538123). The AAA+ domain and its associated DNA from either S. solfataricus Orc1-1 or Orc1-3 (PDB code 2qby16) were then superposed using the AAA+ domain of Orc4 as a guide. Both dockings indicate that a region of density previously assigned to the Orc6 subunit23 actually corresponds to the DNA duplex. Although superpositioning of the AAA+ domain of S. solfataricus Orc1-3 onto Orc4 better positions duplex DNA in the observed EM density (than does the comparable exercise using the Orc1-1•DNA complex), the curvature of the DNA (as present in the Orc1-3•DNA co-crystal structure) results in a greater number of clashes between DNA and ORC subunits. Nevertheless, both docking scenarios are consistent with a DNA binding mode of ORC where DNA runs through the central channel. Note that the handedness of the EM map (EMD-538123) has been corrected in this figure because it has been reported that the original handedness was inverted24. For clarity, the winged-helix domain of Orc2 is omitted from the remodeled ORC structure in (b). c) The autoinhibited ORC conformation observed in the crystal is, unlike the remodeled Orc1 configuration, similar to the ORC conformation observed in Drosophila ORC EM reconstructions. Docking of the ORC crystal structure (top panels) or the remodeled activated ORC structure (bottom panels) into a prior 3D EM reconstruction of Drosophila ORC (EMD-2479)14 reveals excellent agreement between EM and crystal structures, but not between EM and modeled activated ORC structures. The poor fit of the remodeled Orc1 conformation into the EM density suggests that the EM structure represents the autoinhibited state of ORC as seen in the crystal, indicating it is the predominant state in solution. See also Supplementary Video 2.
Extended Data Figure 9.
Docking of the ORC structure into the cryo-EM structure of an S. cerevisiae replication initiation intermediate indicates that ORC recruits the MCM2-7 complex by binding to the ORC winged-helix (WH) domains. A prior model for ORC•MCM2-7 engagement24, proposed from an ORC•Cdc6•Cdt1•MCM2-7 cryo-EM structure generated in the presence of DNA (shown in (a), EMD-562524), used the crystal structure of replication factor C (RFC) bound to the sliding clamp PCNA (shown in (b), PDB code 1SXJ31) to suggest that ORC’s AAA+ domains engage the MCM2-7•Cdt1 complex. However, using the handedness of the EM volume as reported24, this organization of ORC subunits leads to an inverted ATPase site assembly, requiring that the Orc4 arginine finger (which is known to stimulate Orc1 ATP hydrolysis33) points toward the Orc5 nucleotide-binding site rather than the appropriate Orc1 active site. Schematics for the ATP site assemblies of ORC and RFC derived from these structures are shown in the lower panels in (a) and (b). The location of the WH domain collar of ORC and the C-terminal collar of RFC is indicated by a gray circle (WA – Walker A, WB – Walker B, RF – arginine finger). c) Docking of the ORC crystal structure (with Orc1 in its remodeled or “activated” conformation) into the cryo-EM map shown in panel (a) reveals that the WH domains of ORC face an MCM2-7 complex. This switched polarity of WH domains and AAA+ domains in the EM map corrects the ATPase site assembly and is schematized in the right panel.
Extended Data Table 1.
Summary of data collection, phasing and refinement statistics. Highest resolution shell is shown in parenthesis.
| Native | GdCI3 soak | Native - ATPγS | |
|---|---|---|---|
| Data collection: | |||
| Beamline | APS 23ID-B | APS 23ID-B | NSLS X25 |
| Wavelength (A) | 1.0332 | 1.71083 | 1.1 |
| Resolution range (A) | 48.8–3.5 (3.59–3.5) | 50.23–3.96(4.13–3.96) | 58.11–4.0(4.16–4.0) |
| Space group | I222 | I222 | I222 |
| Unit cell dimensions | |||
| a, b, c (Å) | 145.5,259.0,257.0 | 145.3,257.7,256.1 | 144.2,255.3,261.1 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Reflections | |||
| Total | 518,695(38,421) | 2,359,387 (209,066) | 554,356 (62,459) |
| Unique | 61,418(4,491) | 40,955 (3,602) | 41,025(4,568) |
| Completeness (%) | 100.0(100.0) | 97.3 (76.2) | 100.0(100.0) |
| Multiplicity | 8.4 (8.6) | 57.6 (58.0) | 13.5(13.7) |
| Mean 1/σ | 7.7(1.1) | 23.4(1.9) | 16.6(1.3) |
| CCl/2 | 0.995 (0.427) | 0.999 (0.745) | 0.999 (0.678) |
| Rmerge | 0.214(2.181) | 0.355 (3.857) | 0.095(2.174) |
| Rpim | 0.078 (0.788) | 0.047 (0.509) | 0.027 (0.606) |
| Wilson B factor | 118.6 | 165 | 197.8 |
| Phasing: | |||
| FOM before density modification | 0.27 | ||
| FOM after density modification | 0.64 | ||
| Refinement: | |||
| Rwork/Rfree | 0.2243/0.2581 | ||
| Number of atoms | |||
| Protein | 16336 | ||
| Ligands | 2 | ||
| B factors | |||
| Mean | 118.1 | ||
| Macromolecule | 118.1 | ||
| Ligands | 109.2 | ||
| Root mean square deviation | |||
| Bond lengths (Å) | 0.006 | ||
| Bond angles (°) | 0.705 | ||
| Ramachandran plot | |||
| % favored | 92.9 | ||
| % allowed | 7.1 | ||
| % outliers | 0.0 | ||
| Molprobity | |||
| Clashscore | 4.53 |
Supplementary Material
Acknowledgements
We thank the beamline scientists at ALS 8.3.1 (Berkeley Lawrence National Lab), APS 23-ID-B (Argonne National Lab), and NSLS X25 (Brookhaven National Lab) for technical support with X-ray diffraction data collection, and the Nogales lab (University of California, Berkeley) for EM support. We also thank Ann Fisher (University of California, Berkeley) and Yana Li (Johns Hopkins School of Medicine) for assistance with insect cell cultures. This work was supported by the NIH (GM071747 to J.M.B. and CA R37-30490 to M.R.B.) and by a fellowship from the UC Berkeley Miller Institute for Basic Research in Science (to F.B.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
F.B. performed all biochemical and crystallization experiments, collected X-ray diffraction data and determined the structure with guidance from J.M.B. All authors interpreted and discussed results, and wrote the manuscript.
Atomic coordinates and structure factors have been deposited in the RSCB Protein Data Bank under accession code 4XGC.
The authors declare no competing financial interests.
References
- 1.Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell SP, Kaguni JM. Helicase loading at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 5.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 6.Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 7.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarey MG, et al. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 9.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in-a domain distinct from its replication activity. Proc Natl Acad Sci U S A. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, et al. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci U S A. 2011;108:7373–7378. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol. 2014;21:20–25. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
- 14.Bleichert F, et al. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, et al. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2008;105:10326–10331. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 17.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 18.Singleton MR, et al. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J Mol Biol. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, et al. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 20.Dueber EC, et al. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res. 2011;39:3621–3631. doi: 10.1093/nar/gkq1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson NP, et al. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 22.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 23.Sun J, et al. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20:534–544. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci U S A. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 27.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 28.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci U S A. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J. Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc Natl Acad Sci U S A. 2005;102:69–74. doi: 10.1073/pnas.0408690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong D, Coleman TR, DePamphilis ML. Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC. EMBO J. 2003;22:3441–3450. doi: 10.1093/emboj/cdg319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 32.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 34.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonetta KR, et al. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houchens CR, et al. Multiple mechanisms contribute to Schizosaccharomyces pombe origin recognition complex-DNA interactions. J Biol Chem. 2008;283:30216–30224. doi: 10.1074/jbc.M802649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaczynska M, et al. Atomic force microscopic analysis of the binding of the Schizosaccharomyces pombe origin recognition complex and the spOrc4 protein with origin DNA. Proc Natl Acad Sci U S A. 2004;101:17952–17957. doi: 10.1073/pnas.0408369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remus D, Blanchette M, Rio DC, Botchan MR. CDK phosphorylation inhibits the DNA-binding and ATP-hydrolysis activities of the Drosophila origin recognition complex. J Biol Chem. 2005;280:39740–39751. doi: 10.1074/jbc.M508515200. [DOI] [PubMed] [Google Scholar]
- 42.DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- 43.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci U S A. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo AJ, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller P, et al. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 2010;24:1418–1433. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onishi M, et al. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parks TD, et al. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- 50.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 58.Bricogne G, et al. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 59.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 60.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 67.Ashkenazy H, et al. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.