Abstract
Chondroitin/dermatan sulfate (CS/DS) proteoglycans consist of unbranched sulfated polysaccharide chains of repeating GalNAc-GlcA/IdoA disaccharide units, attached to serine residues on specific proteins. The CS/DS proteoglycans are abundant in the extracellular matrix where they have essential functions in tissue development and homeostasis. In this report a phylogenetic analysis of vertebrate genes coding for the enzymes that modify CS/DS is presented. We identify single orthologous genes in the zebrafish genome for the sulfotransferases chst7, chst11, chst13, chst14, chst15 and ust and the epimerase dse. In contrast, two copies were found for mammalian sulfotransferases CHST3 and CHST12 and the epimerase DSEL, named chst3a and chst3b, chst12a and chst12b, dsela and dselb, respectively. Expression of CS/DS modification enzymes is spatially and temporally regulated with a large variation between different genes. We found that CS/DS 4-O-sulfotransferases and 6-O-sulfotransferases as well as CS/DS epimerases show a strong and partly overlapping expression, whereas the expression is restricted for enzymes with ability to synthesize di-sulfated disaccharides. A structural analysis further showed that CS/DS sulfation increases during embryonic development mainly due to synthesis of 4-O-sulfated GalNAc while the proportion of 6-O-sulfated GalNAc increases in later developmental stages. Di-sulfated GalNAc synthesized by Chst15 and 2-O-sulfated GlcA/IdoA synthesized by Ust are rare, in accordance with the restricted expression of these enzymes. We also compared CS/DS composition with that of heparan sulfate (HS). Notably, CS/DS biosynthesis in early zebrafish development is more dynamic than HS biosynthesis. Furthermore, HS contains disaccharides with more than one sulfate group, which are virtually absent in CS/DS.
Introduction
Chondroitin/dermatan sulfate (CS/DS) proteoglycans consist of core proteins with attached linear sulfated polysaccharides of repeating glucuronic acid and N-acetylgalacotsamine (GlcA-GalNAc) in CS or iduronic acid and N-acetylgalactosamine (IdoA-GalNAc) in DS (Fig. 1). CS/DS is produced by virtually all vertebrate cells and has multiple functions during animal development, including fundamental processes as cell division and cytokinesis [1–3]. CS/DS is deposited extensively in cartilage ECM, where the high overall charge provides electrostatic forces to promote high water content, enabling joints to withstand mechanical compression. CS/DS biosynthesis produces chains varying in length, charge density and sulfation pattern, which affect physical docking to ECM molecules, such as growth factors, thus functioning as an extra level of regulation to dictate the function of CS/DS binding proteins [4, 5].
Fig 1. Modification reactions in CS/DS biosynthesis.
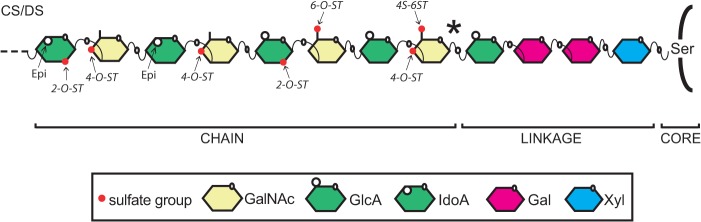
The CS/DS chain is modified by sulfotransferases and epimerases. A GlcNAc added to the linkage region (asterisk) commits the nascent chain to HS synthesis instead of CS/DS synthesis. Epi: GlcA-C5-epimerase, Gal: galactose, GalNAc: N-Acetylgalcotamine, GlcA: glucuronic acid, GlcNAc: N-Acetylglucosamine, IdoA: iduronic acid, Ser: Serine, Xyl: xylose, 2-O-ST: 2-O-sulfotransferase, 4-O-ST: 4-O-sulfotranferases, 4S-6ST: N-Acetylgalactosamine-4-sulfate-6-O-sulfotransferase, 6-O-ST: 6-O-sulfotransferases.
The biosynthesis of CS/DS is initiated with the synthesis of a GlcA–Gal–Gal–Xyl–O-Ser tetrasaccharide structure attached to one of more than 30 known CS/DS core proteins [5, 6]. This structure constitutes the linkage region for CS/DS as well as for heparan sulfate (HS). In HS biosynthesis, a GlcNAc is added to the linkage region as the fifth sugar followed by repeating additions of GlcNAc and GlcA. In biosynthesis of CS/DS a GalNAc is added to the linkage region (Fig. 1). Sometimes there may be a competition for link structures by CS/DS and HS specific enzymes [7]. However, if HS or CS will be synthesized also depends on other factors such as amino acid properties in close vicinity to the attachment serine residue [8] and sulfation of the link structure [9]. Elongation of CS/DS by repeating additions of GalNAc and GlcA is the result of a concerted action of CS/DS glycosyltransferases [5, 6, 10].
The common sulfate donor, 3′-phosphoadenosine 5′-phosphosulfate (PAPS) is used by all sulfotransferases during CS/DS modification. CS modifications are initiated by the addition of a sulfate group to the GalNAc C-4 or C-6 position by chondroitin 4-O- or 6-O-sulfotransferases, respectively (Fig. 1). A portion of GlcA residues are converted by DS epimerases to IdoA, and IdoA-GalNAc disaccharides may be sulfated on the C-4 position of the GalNAc residue by a dermatan 4-O-sulfotransferase. Subsequently, a subset of 4-O-sulfated or 6-O-sulfated disaccharides may be disulfated by the sulfation of the C-2 position on GlcA/IdoA by a 2-O-sulfotransferase or sulfated at the C-6 position of a previously C-4 sulfated GalNAc residue by a GalNAc4S-6-O-sulfotransferase. Typically, the resulting molecule consists of both GlcA and IdoA residues and is hence a hybrid between CS and DS.
Eight CS/DS sulfotransferases were identified in mammals: two chondroitin 6-O-sulfotransferases CHST3 (C6ST-1) [11] and CHST7 (C6ST-2) [12], three chondroitin 4-O-sulfotransferases CHST11 (C4ST-1) [13], CHST12 (C4ST-2) [13], CHST13 (C4ST-3) [14], one dermatan 4-O-sulfotransferase CHST14 (D4ST-1) [15], one N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase CHST15 (GALNAC4S-6ST) [16] and one uronyl-2-sulfotransferase (UST) [17]. Conversion of GlcA residues into the stereoisomer IdoA is mediated by two dermatan sulfate epimerases, DSE (DS-epi1) [18] and DSEL (DS-epi2) [19].
A number of genetic alterations in genes involved in the CS/DS biosynthesis have been investigated in human patients [20] transgenic mice [21]. Human patients with mutations in the chondroitin 6-O-sulfotransferase CHST3 gene develop chondrodysplasia and are diagnosed with Larsen syndrome [22, 23], although targeted mutations in the Chst3 gene in mice did not result in any apparent phenotype [24]. Different homozygous mutations in human CHST14 and DSE have been linked to Ehlers-Danlos syndrome (EDS), where patients display, among other symptoms, distinctive craniofacial dysmorphism [25, 26]. Mice lacking a functional Chst14 or Dse gene have a reduced body size and show increased skin fragility, corresponding to human symptoms [25, 27]. A lower survival frequency of these mice has also been seen. Interestingly, a genome-wide association study suggested CHST11 as a plausible candidate gene for increased osteoarthritis susceptibility [28]. Mice lacking functional Chst11 (C4ST-1) develop chondrodysplasia [5, 29] while mice deficient of functional Chst15 (GalNac4S-6ST) lack GalNAc 4,6 disulfate residues but develop no apparent abnormalities [30].
In the developing mouse embryo chst11 is expressed in the branchial arches and the limb buds as well as in the notochord [31]. Chst12 (C4ST-2) is expressed in the stomach and gut as well as in the skeleton in a 14,5 day old mouse embryo [32]. Chst13 (C4ST-3) is expressed in the liver and kidney, in the bones of the upper and lower jaws and in the eye socket [33]. No expression was found in the developing brain, in contrast to adult brain Chst13 expression. Chst15 is expressed during mouse embryonic development at E8.5 in the neuroepithelium of the forebrain region, and is particularly strong in the dorsal aspects of the neural folds [22]. Furthermore, expression was detected in the pharyngeal region and the foregut as well as in the somites [22].
Little is known of the spatio-temporal regulation of CS/DS biosynthesis during zebrafish development. Hayes and coworkers have used antibodies recognizing CS/DS sulfation motifs to study zebrafish skeletogenesis, and describe non-sulfated disaccharides and disaccharides with sulfate groups at C-4 and C-6 positions (Fig. 1), but also more subtle changes in sulfation pattern indicating dynamic control of CS/DS biosynthesis during development [34]. Expression of CS/DS modification enzymes during zebrafish embryonic development has not been systematically studied but a study of zebrafish chst11 (C4ST-1) shows persistent expression in somites and brain, and transient expression in the pectoral fins [35].
We have previously characterized CS/DS polymerization during zebrafish development [10] and in this study we systematically investigate the phylogeny and expression of all known CS/DS modification enzymes as well as the temporal variation of CS/DS structures. We conclude that CS/DS biosynthesis is a highly dynamic regulated process during vertebrate development.
Experimental Procedure
Ethic statement
All animal experiments in this project were approved by Uppsala Djurförsöksetiska nämnd, Uppsala, Sweden (Permit number C262/11).
Animals
Wild-type zebrafish (AB, WIK) embryos were obtained in natural crosses and cultured in system water. 0.003% 1-phenyl-2-thiourea (Sigma) was added to the water to inhibit pigmentation. Embryos were anesthetized in tricaine methanesulfonate (Sigma-Aldrich) prior to fixation and staged using hours post fertilization (hpf) [36].
Phylogenetic analysis
Gene and protein sequences for CHST3, CHST7, CHST11–15, UST, DSE, and DSEL from human, mouse, zebrafish and stickleback were collected from the Ensembl database [37]. Ensembl accession numbers are displayed in Table 1. Amino acid sequences were aligned with Clustal Omega [38]. The default alignment parameters were applied. The sequences were realigned and bootstrapped 1000 times using SEQBOOT from Phylip 3.695v [39]. Protein distances were calculated using PROTDIST. The Jones–Taylor–Thornton matrix was used for the calculation. The neighbor-joining trees were calculated from the 1000 different distance matrixes, previously generated with PROTDIST, using NEIGHBOR from Phylip 3.695. Majority rule consensus tree was constructed with CONSENSE from the same package. The trees were plotted using TreeView 1.6.6 [40].
Table 1. Accession numbers of CS/DS modification enzymes.
| Human/Mouse | Alternative name | Ensembl Nr ENSG00000 | Chr. Pos. | Ensembl Nr ENSMUSG000000 | Chr. Pos. | Zebrafish | aa % | Ensembl Nr ENSDARG000000 | Chr. Pos. | Stickleback | Ensembl Nr ENSGACG000000 | Chr. Pos. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHST3 | Chondroitin 6-O-sulfotransferase 1 | 122863 | 10:73.72m | 57337 | 10: 60.18m | chst3a | 58 | 61070 | 13: 29.74m | chst3a | 02438 | grV:1.01m |
| chst3b | 53 | 77844 | 12: 50.62m | chst3b | 08386 | grVI:9.61m | ||||||
| CHST7 | Chondroitin 6-sulfotransferase 2 | 147119 | X:46.43m | 37347 | X: 20.06m | chst7 | 53 | 44341 | 6: 37.57m | chst7 | 13833 | grVIII:17.98m |
| CHST11 | Chondroitin 4-O-sulfotransferase 1 | 171310 | 12:104.85m | 34612 | 10: 82.99m | chst11 | 79 | 34375 | 4: 8.24m | chst11 | 19340 | grIV:24.26m |
| CHST12 | Chondroitin 4-O-sulfotransferase 2 | 136213 | 7:2.44m | 36599 | 5: 140.51m | chst12a | 61 | 28786 | 3: 41.72m | chst12a | 07992 | grXI:4.98m |
| chst12b | 55 | 77198 | 1: 10.92m | chst12b | 16733 | grIX: 5.13m | ||||||
| si:dkey-26i13.4 | 94981 | 1: 10.82m | ||||||||||
| si:dkey-26i13.3 | 95482 | 1: 10.81m | ||||||||||
| si:dkey-26i13.5 | 93636 | 1: 10.80m | ||||||||||
| si:dkey-26i13.6 | 92200 | 1: 10.79m | ||||||||||
| si:dkey-26i13.7 | 92242 | 1: 10.78m | ||||||||||
| CHST13 | Chondroitin 4-O-sulfotransferase 3 | 180767 | 3:126.24m | 56643 | 6: 90.31m | chst13 | 52 | 04018 | 23: 34.99m | chst13a | 10830 | grXVII:11.00m |
| chst13b | 04801 | grXII:5.08m | ||||||||||
| CHST14 | Dermatan 4-sulfotransferase 1 | 169105 | 15:40.76m | 74916 | 2:118.93m | chst14 | 59 | 43011 | 20: 25.78m | chst14 | 14327 | grVIII:18.79m |
| CHST15 | N-acetylgalactosamine 4-S 6-O-sulfotransferase | 182022 | 10:125.77m | 30930 | 7:132.24m | chst15 | 56 | 74527 | 17: 21.34m | chst15a | 02542 | grV:1.18m |
| chst15b | 09700 | grVI:11.74m | ||||||||||
| UST | DS/CS 2-O ulfotransferase | 111962 | 6:149.07m | 47712 | 10:8.20m | ust | 67 | 06044 | 20:31.45m | ust | 11589 | grXVIII:11.86m |
| DSE | Chondroitin-glucuronate 5-epimerase | 111817 | 6:116.58m | 39497 | 10:34.15m | dse | 66 | 17988 | 20:0.55m | dse | 06171 | grXVIII:3.37m |
| DSEL | 171451 | 18:65.17m | 38702 | 1:111.86m | dsela | 62 | 74504 | 24:0.05m | dsel | 04870 | grXXI:10.44m | |
| dselb | 58 | 61461 | 2:27.46m |
CS/DS modification enzymes and their alternative names in human, mouse, zebrafish and stickleback. Amino acid (aa) sequence identity (%) to human orthologues is displayed for predicted zebrafish proteins. Ensembl accession numbers and chromosomal positions are also presented.
cDNA cloning
All probes were amplified by PCR from zebrafish cDNA template, transcribed using T7 polymerase and DIG-labeling kit (Roche). Nucleotide removal was performed with the RNeasy kit (Qiagen) and the riboprobes were checked on an agarose gel. Primers for the respective genes are listed in Table 2.
Table 2. Primers used to make RNA probes for the in situ hybridization of CS/DS modification enzymes.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| chst3a | TGTGGGCGAGTTCTTTAAC | AAGTTTCATGGTCGGTCCAC |
| chst3b | TGTGGCTTTGGTCATCATAGA | GATGAAGCGCATGTGCAG |
| chst7 | ATGAAGAGAAGGCTTCAGAAAAAATACATCATCTTA | TTAGGTCTTCTCCATGTTCCTCTTCTGATAATCCAG |
| chst11 | TGGAGATCCCTTCCAATGAG | CAGGGCTTCAGACCTCAGTC |
| chst12a | TCCTGGAGACCCTCACAATC | TTTGTCTCCAGTGCTCGTTG |
| chst12b | GCAGCTGCAGTTTTTCCTTC | ATGCTCATTGAACGGCTTCT |
| chst13 | AAACAGAAGCGGTTTGTTCTCCG | TCATCTCAACTTCAAGTACGCAGGCAT |
| chst14 | TTTCAAGAAAGCGCAGAAATCAGAGGTT | TAGTGCCTGCAGTATTCAGTGGTTGTGT |
| chst15 | CCACAAGTATGGCCTCCTAAGCT | CACAGAAAAGCAGGGTCCTTTAA |
| dse | TCAGCGTACATGGAGACTCG | GCTGTTAGCATTGTGCCTGA |
| dsela | TGTTTGACTTTGGGGTCACA | CAGGAAGGGCAGCTTTAGTG |
| dselb | AGCTGCCGAATTCAAAAAGA | CTCTGGACCACTCACAAGCA |
Whole mount in situ hybridization
In situ hybridization was performed essentially as previously described [41]. Zebrafish embryos and larvae of different stages were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS), dehydrated in methanol and stored at-20°C. Upon using, embryos were rehydrated and permeabilized with Proteinase K (5μg/ml). Hybridization was performed at 70°C and riboprobes were detected with BM purple AP Substrate precipitating solution (Roche). Embryos and larvae were cleared in glycerol and photographed with a Nikon SMZ1500 Stereomicroscope. Images and figures were adapted in Adobe Photoshop and Adobe Illustrator.
Structural Analysis of CS/DS and HS by Reverse Phase Ion Pair (RPIP)-HPLC
CS/DS and HS were isolated from zebrafish embryos and larvae at different stages in duplicate or triplicate samples with 250 embryos at 30 hpf and 100 embryos at 2 days post fertilization (dpf), 3 dpf and 4 dpf. CS/DS was degraded into disaccharides by enzymatic cleavage, and detected in a HPLC-based system essentially following the protocol as described earlier [42], with the modification of washing the DEAE column with 0.4M NaCl to remove hyaluronic acid. In short, GAGs were isolated by proteolytic cleavage, nuclease treatment, and DEAE ion exchange chromatography. The purified GAGs were cleaved with chondroitinase ABC and 10% of the sample was used to identify CS/DS disaccharide components. HS was purified from cleaved CS/DS disaccharides by a second DEAE purifications step followed by cleavage with heparin lysase I-III. The two generated samples of CS/DS and HS disaccharides were subjected to RPIP-HPLC analysis followed by post-column derivatization with cyanoacetamide and detection in a fluorescence detector, essentially as described [42]. The identity and the amount of the disaccharides were established by comparing the samples with CS/DS and HS disaccharide standards, respectively.
Results
Most CS/DS modification enzymes are present as single orthologues in zebrafish
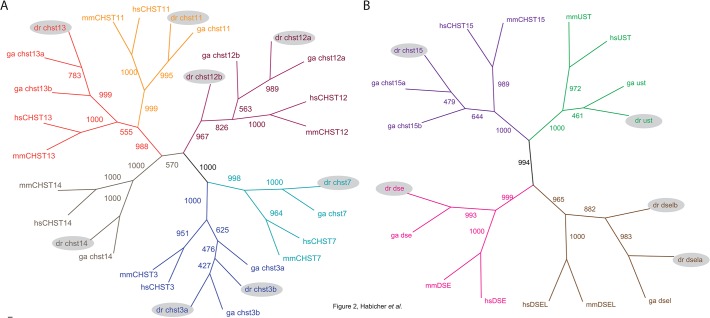
BLAST searches using previously reported amino acid sequences of human and mouse CS/DS modification enzymes [5, 43] as templates were performed to identify orthologous genes in zebrafish (D. rerio). Amino acid sequence alignments of identified zebrafish enzymes and human orthologues can be found in S1 Fig. We included another teleost fish, stickleback (G. aculeatus), into the analysis in order to better understand the divergence of genes in different teleost lineages after the teleost specific genome duplication. One orthologue of CS/DS 6-O-sulfotransferase CHST7 and two orthologues of CS/DS 6-O-sulfotransferases CHST3, named chst3a and chst3b, were identified in both teleost species, sharing 53–58% identity with human proteins (Fig. 2A). Three genes in the group of CS/DS 4-O-sulfotransferases CHST11, CHST13 and CHST14 displayed single orthology to zebrafish genes chst11, chst13 and chst14, respectively. However, in stickleback two co-orthologues of CHST13, chst13a and chst13b were identified. Zebrafish 4-O-sulfotransferases share 52–79% amino acid identity with corresponding human proteins.
Fig 2. Phylogenetic analyses of CS/DS 4-O- and 6-O-sulfotransferases (A) and other CS/DS modification enzymes (B).
The consensus neighbor-joining trees were calculated with 1000 bootsrap replicates. Zebrafish orthologues are marked with grey shadow. Species names are abbreviated as hs, human; mm, mouse; dr, zebrafish, and ga, stickleback. The accession numbers are listed in the Table 1.
Notably, seven sequences show orthology to the human CS/DS 4-O-sulfotransferase CHST12. Zebrafish chst12a displays 61% identity in protein sequence and is located on chromosome 3, while chst12b shares 55% identity with human protein and is located on chromosome 1. The additional five genes are located as tandem copies in close proximity to chst12b on chromosome 1 (Table 1). There is evidence for expression of chst12a and chst12b in zebrafish in the form of pooled RNASEQ data, displayed in the Ensembl database, but no conclusive evidence that the tandem duplicates of chst12, named chst12 (3-, 4-, 5-, 6- and 7), are functional. In stickleback we find two copies of chst12, where one is a clear orthologue of zebrafish chst12a. With the help of additional analysis of chst12 genes in other teleosts, we could see a variation in the number of chst12 copies between different teleost species (data not shown). There is only one single chst12 gene in gar, a group of fish that diverged prior to the teleost specific genome duplication, indicating that the rest of the genes in teleosts are lineage specific duplicates.
The human 4-sulfate 6-O-sulfotransferase CHST15 does not belong to the same family as CS/DS 6-O-sulfotransferases. There is one orthologue of human 4-sulfate 6-O-sulfotransferase CHST15 in zebrafish, named chst15 sharing 56% identical amino acid residues. Other teleosts like stickleback (Fig. 2B), cod, fugu, medaka and platyfish have two co-orthologues of this sulfotransferase gene, named chst15a and chst15b (data not shown). A single orthologue was identified for CS/DS 2-O-sulfotransferase (ust) in teleosts, where zebrafish ust is 67% identical to the human protein sequence. There are two dermatan sulfate epimerase genes in mammals, DSE and DSEL. Human DSE shares 66% amino acid identity with the zebrafish orthologue dse. In the zebrafish genome dsel seems to be duplicated. dsela displays higher identity of the predicted amino acid sequence to the human DSEL (62%) and stickleback dsel (73%) compared to dselb (58% and 63%) (Table 1).
CS/DS modification enzymes are spatially and temporally regulated in early zebrafish development
Chondroitin and Dermatan Sulfate 4-O-sulfotransferases
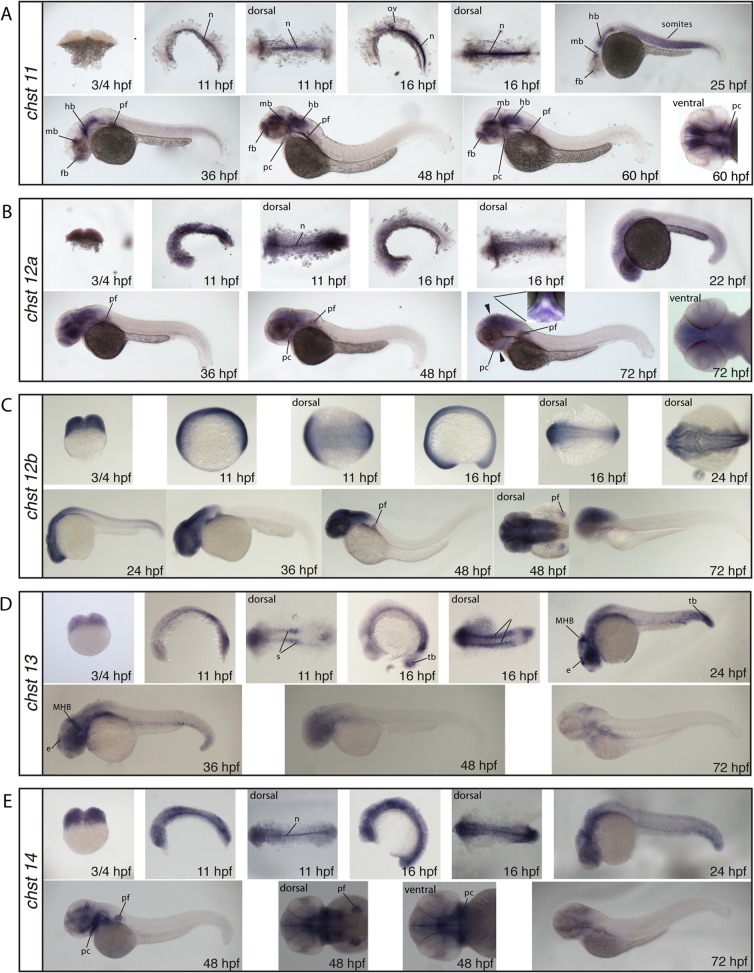
With the exception of chst11, all 4-O-sulfotransferases show expression at the 2-cell stage (¾ hours post fertilization (hpf)) indicating maternal contribution (Fig. 3). We find that chst11, chst12a and chst14 are expressed in the notochord at the somite stages (11–16 hpf) (Fig. 3), in accordance with previously published findings for chst11 [35]. Staining of chst11 is shifted from the notochord to the somites, the hindbrain, the midbrain and weak staining is seen in the forebrain at 24 hpf (Fig. 3A). By 36 hpf, the somite expression fades; the forebrain, some cells of the midbrain and the pectoral fin bud show staining, and the hindbrain has a very strong expression (Fig. 3A). At 48 and 60 hpf, the for-, mid- and hindbrain as well as the pectoral fins remain stained and in addition, forming pharyngeal cartilages weakly express chst11 (Fig. 3A). chst12a shows a weak overall expression with stronger expression in the anterior regions at 24 hpf. At 36 hpf, the head and pectoral fins are prominently stained (Fig. 3B). From 48 hpf, chst12a expression is localized to the head, the pectoral fins and the pharyngeal cartilage (Fig. 3B). chst12b shows at the 5- and 15- somite stages (11–16 hpf) weak overall expression (Fig. 3C). At 24 hpf, expression becomes stronger in the central nervous system, and at 36 and 48 hpf, the central nervous system and pectoral fins are prominently stained. At 72 hpf, this staining is slightly weaker (Fig. 3C). chst13 displays clear labeling in the somites and the tail bud at 5- and 15-somite stages (11–16 hpf) (Fig. 3D). At 24 hpf, expression in the tail bud, the midbrain-hindbrain boundary (MHB) and a region overlapping with the epiphysis stains (Fig. 3D). By 36 hpf, staining of chst13 is still prominent in the midbrain-hindbrain boundary and the epiphysis, and at 48 and 72 hpf, staining is weak and restricted mainly to the head (Fig. 3D). At 24 hpf, chst14 displays strong labeling in the head region and is prominent in the MHB (Fig. 3E). At 48 hpf, strong staining is detected in the pharyngeal cartilage and in the pectoral fins (Fig. 3E).
Fig 3. Whole mount in situ hybridization of the CS/DS 4-O-sulfotransferases chst11 (A), chst12a (B), chst12b (C), chst13 (D) and chst14 (E).
All images, if not otherwise stated in the figure, are shown in lateral views. Arrowheads indicate the position of the transverse section shown in B at 72 hpf. e: epiphysis, hb, hindbrain, mb: midbrain, MHB: midbrain-hindbrain boundary, n: notochord, ov: otic vesicle, pc: pharyngeal cartilage, pf: pectoral fin, s: somites, tb: tailbud
Chondroitin Sulfate 6-O-sulfotransferases
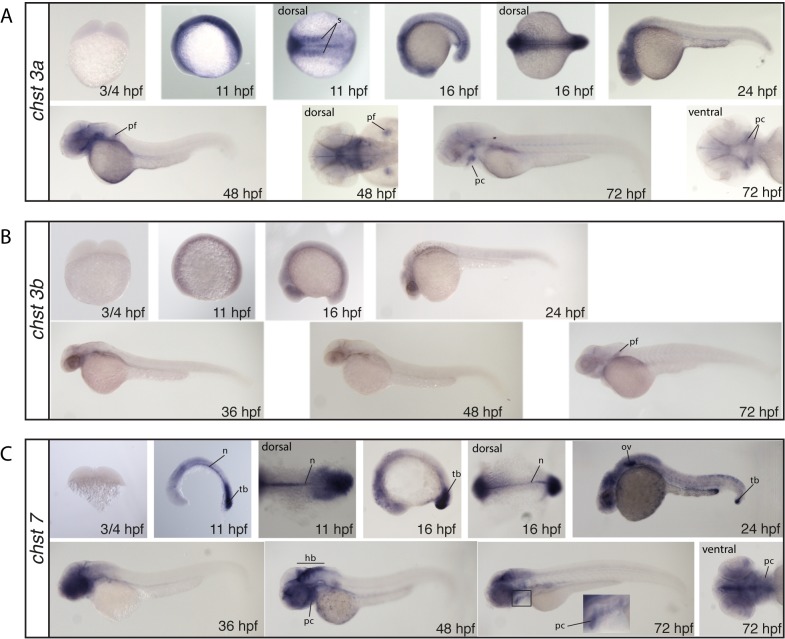
chst3a, chst3b and chst7 lack staining at the 2-cell stage (¾ hpf) indicating no maternally deposited mRNA (Fig. 4). The chst3a probe shows weak overall staining at the 5- and 15-somite stages (11–16 hpf), as well as strong somite staining (Fig. 4A). At 24 and 48 hpf staining is limited to the head and the pectoral fins (at 48 hpf). At 72 hpf, staining is restricted to the pharyngeal cartilages and the pectoral fins (Fig. 4A). chst3b displays only very weak staining from the 5 somite stage (11 hpf) to 48 hpf (Fig. 4B). At 72 hpf, the pectoral fins show increased staining (Fig. 4B). At the 5- and 15-somite stage (11–16 hpf), the chst7 probe strongly stains the notochord and tail bud (Fig. 4C). At 24 hpf, staining is still detected at the tail bud but starts shifting to the head region where it gets prominent from 36 hpf (Fig. 4C). At 48 and 72 hpf the hindbrain stains strongly as well as the pharyngeal cartilages. (Fig. 4C).
Fig 4. Whole mount in situ hybridization of the CS/DS 6-O-sulfotransferases chst3a (A), chst3b (B) and chst7 (C).
All images show lateral views, if not otherwise stated in the figure. hb: hindbrain, n: notochord, ov: otic vesicle, pc: pharyngeal cartilage, pf: pectoral fin, tb: tailbud, s: somites
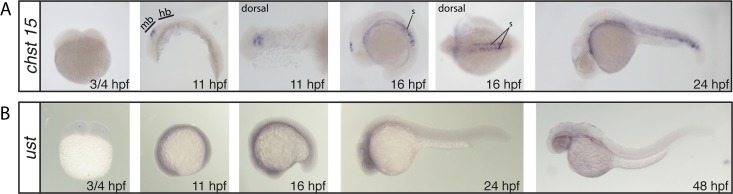
N-Acetylgalactosamine 4-sulfate 6-O-sulfotransferase and uronyl-2-sulfotransferase chst15 and ust show no staining at the 2-cell stage (¾ hpf) indicating no maternal contribution (Fig. 5). At the 5-somite stage (11 hpf) chst15 displays a very distinct staining in a subset of cells in the dorsal midbrain (Fig. 5A). This staining persists at the 15-somite stage (16 hpf), where in addition somites stain strongly (Fig. 5A). At 24 hpf, weak somite staining is still present but fades after this stage (Fig. 5A). The probe for ust shows weak overall staining at 5- and 15-somite stages (Fig. 5B). At 24 hpf, weak staining in the head region is observed, while at 48 hpf almost no staining is detected (Fig. 5B).
Fig 5. Whole mount in situ hybridization of N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase, chst15 (A) and CS/DS 2-O-sulfotransferase, ust (B).
All images show lateral views, if not otherwise stated in the figure. mb: midbrain, hb: hindbrain, s: somites
Dermatan sulfate epimerases
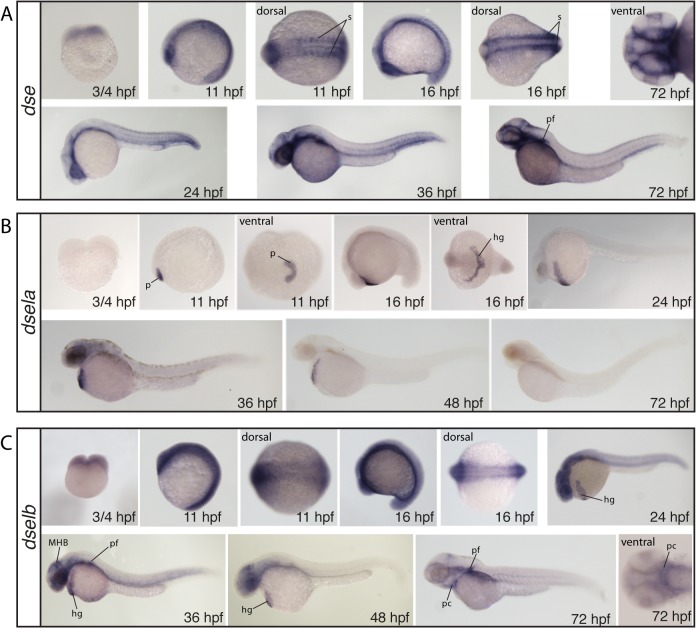
Both probes for dse and dselb stain at the 2-cell stage (¾ hpf), indicating maternal deposition of mRNA (Fig. 6A,C) while no staining was seen at this stage for dsela (Fig. 6B). A general weak staining of dse is detected at the 5- and 15-somite stage (11–16 hpf) while somites and the eyes stain strongly (Fig. 6A). At 24 and 36 hpf, only the posterior end of the somites remain stained and staining appears strong in the most ventral regions of the head (Fig. 6A). At 72 hpf staining is still strong and general in the most ventral regions of the head and pectoral fins (Fig. 6A). dsela stains very strong in the polster at the 5-somite stage and shows clear staining in cells of the hatching gland at the 15-somite stage (Fig. 6B). The expression in the hatching gland remains persistent at 24, 36 and 48 hpf, while no staining is detected at 72 hpf (Fig. 6B). dselb shows a general expression at the somite stages, and at 24 hpf, somites as well as the head region and the hatching gland are stained (Fig. 6C). At 36 and 48 hpf, in addition to the hatching gland, also the midbrain-hindbrain boundary and the pectoral fins are strongly labeled (Fig. 6C). By 72 hpf, staining is weak and restricted to the pectoral fins and the cartilage elements (Fig. 6C).
Fig 6. Whole mount in situ hybridization of the DS epimerases dse (A), dsela (B) and dselb (C).
All images show lateral views, if not otherwise stated in the figure. hg: hatching gland, MBH: midbrain-hindbrain boundary, p: polster, pc: pharyngeal cartilage, pf: pectoral fin, s: somites
Disulfated disaccharides are rare in zebrafish CS/DS
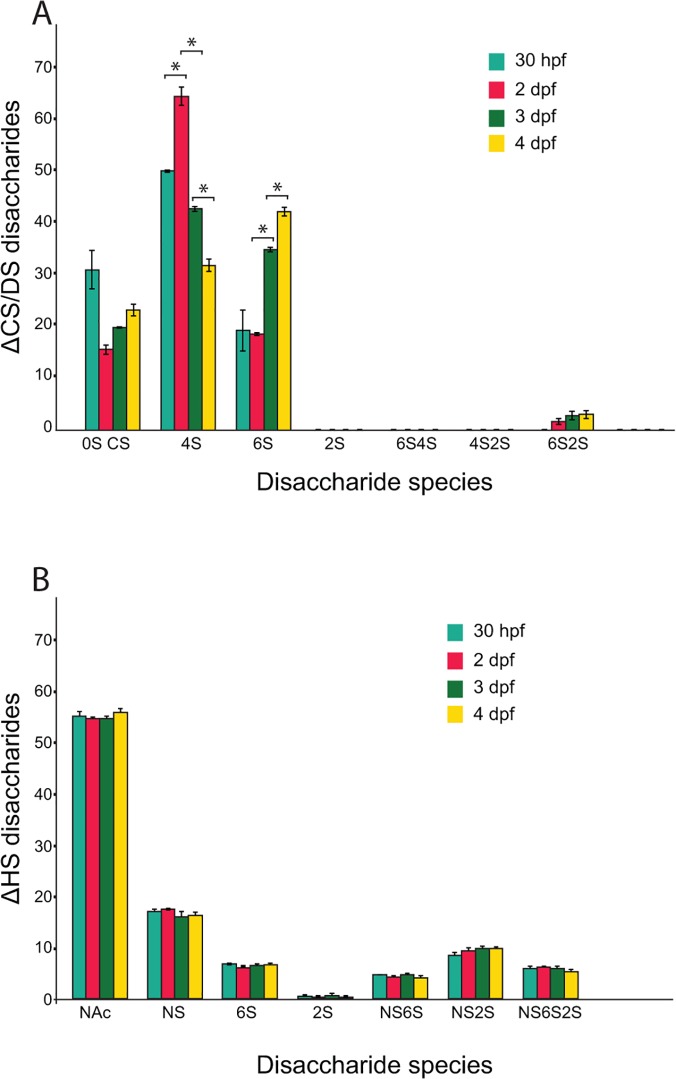
We next investigated the relative sulfation of CS/DS during early zebrafish development. CS/DS was isolated from zebrafish embryos at different developmental stages, cleaved into disaccharides and analyzed by reverse-phase ion-pairing chromatography as previously described [42]. We found that CS/DS sulfation increases from 70 sulfate groups per 100 disaccharides at 30 hpf to 80 sulfate groups 2–4 days post fertilization (dpf) (data not shown). 4-O-sulfate groups are the most common modification (Fig. 7A), in accordance with the strong expression of CS/DS 4-O-sulfotransferases (Fig. 3). The proportion of 6-O-sulfated disaccharides increases from 2dpf (Fig. 7A) correlating with a massive increase in CS/DS synthesis in pharyngeal cartilage [10], suggesting that a high proportion of 6-O-sulfation marks CS/DS synthesis in the cartilage during early zebrafish development. Notably, only low amounts of 2-O-sulfation and disulfated disaccharides could be detected (Fig. 7A), in agreement with the limited spatial expression of the ust and chst15 enzymes (Fig. 5).
Fig 7. Disaccharide composition of CS/DS (A) and HS (B) as measured by RPIP-HPLC analysis.
Significant differences (p-value <0.01) in the proportion of specific CS/DS disaccharide species are indicated (star) (A). In contrast, the changes in proportion of specific HS disaccharide species during development between 30 hpf to 4 dpf are not significant (p-value >0.01). (B). Disaccharide species for CS/DS are indicated as 0S CS (ΔHexA- GalNAc/ΔHexA-GlcNAc), 4S (ΔHexA-GalNAc4S), 6S (ΔHexA-GalNAc6S), 2S (ΔHexA2S-GalNAc), 6S4S (ΔHexA-GlcNAc4S6S), 4S2S (ΔHexA2S-GalNAc4S), and 6S2S (ΔHexA2S-GalNAc6S). Disaccharide species for HS are indicated as NAc (ΔHexA-GlcNAc), NS (ΔHexA- GlcNS), 6S (ΔHexA-GlcNAc6S), 2S (ΔHexA2S-GlcNAc), NS6S (ΔHexA-GlcNS6S), NS2S (ΔHexA2S-GlcNS), and NS6S2S (ΔHexA2S-GlcNS6S).
We next compared the changes in CS/DS modifications to HS. Interestingly, compared to the changes in CS/DS modifications (Fig. 7A), HS modifications remains stable during zebrafish development (Fig. 7B).
We conclude that CS/DS sulfation during early zebrafish development is a dynamic process dominated by variable proportions of 6-O- and 4-O-sulfated disaccharides with a marginal contributions of 2-O-sulfated and disulfated disaccharides.
Discussion
In this study we present a characterization of what, to the current understanding of CS/DS biosynthesis [2, 4, 5, 43], is the complete set of zebrafish enzymes that modify CS/DS.
The phylogenetic analysis
The zebrafish and stickleback lineages diverged early in teleost evolution but after the teleost-specific genome duplication. Subfunctionalization or neofunctionalization of duplicated genes, or gene loss of one of the duplicates, has occurred both before and after the separation of the two lineages [44]. Our phylogenetic analysis of CS/DS modification enzymes identified orthologous genes of all mammalian enzymes in the teleosts zebrafish and stickleback (Fig. 2).
We have analyzed the orthologous relationship between human, mouse, stickleback, and zebrafish genes and found that three mammalian genes, CHST3, CHST12, and DSEL are present as duplicates in zebrafish. In comparison, in the stickleback genome, dsel is present as a single orthologue and the zebrafish single orthologues chst13 and chst15 are duplicated. Our phylogenetic analysis furthermore indicates that the two families of 4-O- and 6-O-sulfotransferases are more similar to each other than to the family of 4-sulfate 6-O-sulfotransferase. This family is represented by a single gene, CHST15, in most vertebrates, including zebrafish, whereas some other teleost fishes such as stickleback, medaka and pufferfishes, retain both copies of the gene.
CS/DS biosynthesis is dynamic during embryo development
Our analysis of GAG structures, isolated from whole zebrafish embryos, reveals that CS/DS biosynthesis is dynamic during early zebrafish development compared to an apparent overall stability of HS biosynthesis (Fig. 7). This does not exclude changes in HS structures in certain tissues. The variation of modifications in CS/DS structure during early zebrafish development is characterized by an increase in total sulfation with changes in the proportion of 4-O-sulfated respective 6-O-sulfated disaccharides (Fig. 7A). The shift from 4-O-sulfation to 6-O-sulfation and the relative absence of di-sulfated disaccharides during zebrafish embryogenesis and early larval development are reminiscent of Xenopus laevis development [45]. Evidence has accumulated for interactions between CS/DS and a large number of proteins (reviewed in [2, 4, 5]) and it is an intriguing possibility that CS/DS structure, regulated by combinatorial expression of CS/DS modification enzymes (Fig. 3–6), mediates evolutionary conserved instructive functions during animal development. The ratio 4-O-sulfated / 6-O-sulfated disaccharides has also been shown to be crucial during mouse neural development [46]. However, it should be noted that HS disaccharides with multiple sulfate groups, believed to be most important for HS-protein interaction [47], are common during zebrafish development (Fig. 7B). In contrast, CS/DS disaccharides with multiple sulfate groups are rare (Fig. 7A). Also the restricted expression of Chst15 and Ust (Fig. 5), known to be necessary for synthesis of di-sulfated CS/DS disaccharides (reviewed in [5]), suggests that CS/DS disaccharides during zebrafish development are usually decorated with single sulfate groups.
Composition of CS/DS in zebrafish cartilage and bone
CS/DS is accumulated in zebrafish larvae cartilage structures in large amounts [7] and both CS/DS glycosyltransferases [10], 4-O-sulfotransferases (Fig. 3) and 6-O-sulfotransferases (Fig. 4) are distinctly expressed in cartilage structures during zebrafish development. This suggests cartilage CS/DS to be dominated by non-sulfated, 4-O-sulfated and 6-O-sulfated CS/DS disaccharides, also indicated by the whole embryo composition analysis (Fig. 7). These components can further be detected in most cartilage and bone structures by antibodies recognizing CS/DS motifs in zebrafish larvae at 4 dpf and 8 dpf [34] and similar proportions of different CS/DS disaccharides is found in vertebral CS/DS during zebrafish ageing [48].
Cooperation between biosynthesis enzymes
CS/DS biosynthesis enzymes produce tissue specific structures during zebrafish development [34] presumably as a result of differential expression of CS/DS modification enzymes (Figs. 3–6). However, we have previously shown that it is difficult to predict the in vivo effect of varying amounts of biosynthesis enzymes, indicating a complex regulation of CS/DS biosynthesis [7, 49]. Interestingly, evidence has recently accumulated suggesting that cooperation between CS/DS glycosyltransferases and CS/DS modification enzymes strongly affects CS/DS biosynthesis. Expression of CHST11 (C4ST-1) has been shown to be critical for synthesis of long CS/DS chains in mouse fibroblasts [50] and it was further shown that chain elongation by the CS/DS glycosyltransferase CSGALNACT2 depends on CHST11 (C4ST-1) expression [51]. During zebrafish development, such cooperation would be most likely to occur in the notochord and in brain tissue with strong expression of both Csgalnact2 [10] and Chst11 (Fig. 3). In contrast, cooperation between CSGALNACT1 and CHST12 affects CS/DS biosynthesis differently by increasing the number of chains attached to a core protein rather than affecting chain length [52, 53]. While chst12a and chst12b are widely expressed (Fig. 3), the restricted expression of csgalnact1 [10] would suggest that such cooperation would be manifested primarily in pharyngeal cartilage structures. Furthermore, the proportion of GlcA to IdoA in CS/DS chains appears to be controlled by functional cooperation between CHST14 (D4ST-1) and DSE (DS-epi1) [54, 55]. chst14 expression is widespread in zebrafish embryos (Fig. 3) while the strong expression of dse in pharyngeal cartilage and the notochord suggests that functional collaboration resulting in an elevated proportion of IdoA between these enzymes may occur in these structures (Fig. 6). As only a few studies so far have investigated the role of enzymatic collaboration during CS/DS modification, more examples might well be revealed in future studies. It should also be noted that the role of CS/DS enzymes with the ability to catalyze the same enzymatic reaction is still poorly investigated in organisms. For example, a confirmation of the role for CHST7 in CS/DS biosynthesis during animal development is still lacking.
Conclusions and future directions
In this study we present a detailed phylogenetic analysis of CS/DS modification enzymes and we show that CS/DS modification enzymes are differentially expressed while CS/DS structure varies significantly during zebrafish development. This study together with our previous characterization of zebrafish CS/DS glycosyltransferases [10] thus lay the foundation to design experiments in zebrafish strains with genetically manipulated CS/DS biosynthesis to further understand the regulation of CS/DS biosynthesis, the function of CS/DS modification enzymes and the subsequent effects on embryonic development.
Supporting Information
A, alignment of CS/DS 4-O sulfotransferases. B, CS/DS 6-O sulfotransferases. C, CS/DS 4-sulfate 6-O sulfotransferases. D, CS/DS 2-O-sulfotransferases and E, CS/DS epimerases. Identical amino acids are marked with red letters on yellow background, conserved residues are indicated by blue letters on light blue background and blocks of similar residues marked with black letters on green background.
(PDF)
Acknowledgments
We thank professor Lena Kjellén for comments on an earlier version of the manuscript. Zebrafish embryos were provided by the Science for Life Laboratory Zebrafish facility in Uppsala.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Knut and Alice Wallenberg Foundation (https://www.wallenberg.com/kaw/en), and the Swedish Research Council VR (http://www.vr.se/inenglish.4.12fff4451215cbd83e4800015152.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Esko JD, Kimata K, Lindahl U. Proteoglycans and Sulfated Glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor (NY)2009. [PubMed]
- 2. Prabhakar V, Sasisekharan R. The biosynthesis and catabolism of galactosaminoglycans. Adv Pharmacol. 2006;53:69–115. Epub 2007/01/24. [pii] 10.1016/S1054-3589(05)53005-9 . [DOI] [PubMed] [Google Scholar]
- 3. Maeda N, Ishii M, Nishimura K, Kamimura K. Functions of chondroitin sulfate and heparan sulfate in the developing brain. Neurochem Res. 2010;36(7):1228–40. Epub 2010/11/27. 10.1007/s11064-010-0324-y . [DOI] [PubMed] [Google Scholar]
- 4. Nandini CD, Sugahara K. Role of the sulfation pattern of chondroitin sulfate in its biological activities and in the binding of growth factors. Adv Pharmacol. 2006;53:253–79. Epub 2007/01/24. [pii] 10.1016/S1054-3589(05)53012-6 . [DOI] [PubMed] [Google Scholar]
- 5. Mikami T, Kitagawa H. Biosynthesis and function of chondroitin sulfate. Biochimica et biophysica acta. 2013;1830(10):4719–33. Epub 2013/06/19. 10.1016/j.bbagen.2013.06.006 . [DOI] [PubMed] [Google Scholar]
- 6. Thelin MA, Bartolini B, Axelsson J, Gustafsson R, Tykesson E, Pera E, et al. Biological functions of iduronic acid in chondroitin/dermatan sulfate. The FEBS journal. 2013;280(10):2431–46. Epub 2013/02/28. 10.1111/febs.12214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holmborn K, Habicher J, Kasza Z, Eriksson AS, Filipek-Gorniok B, Gopal S, et al. On the roles and regulation of chondroitin sulfate and heparan sulfate in zebrafish pharyngeal cartilage morphogenesis. J Biol Chem. 2012;287(40):33905–16. Epub 2012/08/08. 10.1074/jbc.M112.401646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esko JD, Zhang L. Influence of core protein sequence on glycosaminoglycan assembly. Curr Opin Struct Biol. 1996;6(5):663–70. Epub 1996/10/01. doi: S0959-440X(96)80034-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 9. Sugahara K, Kitagawa H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr Opin Struct Biol. 2000;10(5):518–27. Epub 2000/10/24. doi: S0959-440X(00)00125-1 [pii]. . [DOI] [PubMed] [Google Scholar]
- 10. Filipek-Gorniok B, Holmborn K, Haitina T, Habicher J, Oliveira MB, Hellgren C, et al. Expression of chondroitin/dermatan sulfate glycosyltransferases during early zebrafish development. Dev Dyn. 2013;242(8):964–75. Epub 2013/05/25. 10.1002/dvdy.23981 . [DOI] [PubMed] [Google Scholar]
- 11. Fukuta M, Kobayashi Y, Uchimura K, Kimata K, Habuchi O. Molecular cloning and expression of human chondroitin 6-sulfotransferase. Biochimica et biophysica acta. 1998;1399(1):57–61. Epub 1998/08/26. . [DOI] [PubMed] [Google Scholar]
- 12. Kitagawa H, Fujita M, Ito N, Sugahara K. Molecular cloning and expression of a novel chondroitin 6-O-sulfotransferase. The Journal of biological chemistry. 2000;275(28):21075–80. Epub 2000/04/27. 10.1074/jbc.M002101200 . [DOI] [PubMed] [Google Scholar]
- 13. Hiraoka N, Nakagawa H, Ong E, Akama TO, Fukuda MN, Fukuda M. Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. The Journal of biological chemistry. 2000;275(26):20188–96. Epub 2000/04/27. 10.1074/jbc.M002443200 . [DOI] [PubMed] [Google Scholar]
- 14. Kang HG, Evers MR, Xia G, Baenziger JU, Schachner M. Molecular cloning and characterization of chondroitin-4-O-sulfotransferase-3. A novel member of the HNK-1 family of sulfotransferases. The Journal of biological chemistry. 2002;277(38):34766–72. Epub 2002/06/25. 10.1074/jbc.M204907200 . [DOI] [PubMed] [Google Scholar]
- 15. Evers MR, Xia G, Kang HG, Schachner M, Baenziger JU. Molecular cloning and characterization of a dermatan-specific N-acetylgalactosamine 4-O-sulfotransferase. The Journal of biological chemistry. 2001;276(39):36344–53. Epub 2001/07/27. 10.1074/jbc.M105848200 . [DOI] [PubMed] [Google Scholar]
- 16. Ohtake S, Ito Y, Fukuta M, Habuchi O. Human N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase cDNA is related to human B cell recombination activating gene-associated gene. The Journal of biological chemistry. 2001;276(47):43894–900. Epub 2001/09/27. 10.1074/jbc.M104922200 . [DOI] [PubMed] [Google Scholar]
- 17. Kobayashi M, Sugumaran G, Liu J, Shworak NW, Silbert JE, Rosenberg RD. Molecular cloning and characterization of a human uronyl 2-sulfotransferase that sulfates iduronyl and glucuronyl residues in dermatan/chondroitin sulfate. The Journal of biological chemistry. 1999;274(15):10474–80. . [DOI] [PubMed] [Google Scholar]
- 18. Maccarana M, Olander B, Malmstrom J, Tiedemann K, Aebersold R, Lindahl U, et al. Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. The Journal of biological chemistry. 2006;281(17):11560–8. Epub 2006/03/01. 10.1074/jbc.M513373200 . [DOI] [PubMed] [Google Scholar]
- 19. Pacheco B, Malmstrom A, Maccarana M. Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate. The Journal of biological chemistry. 2009;284(15):9788–95. Epub 2009/02/04. 10.1074/jbc.M809339200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mizumoto S, Ikegawa S, Sugahara K. Human genetic disorders caused by mutations in genes encoding biosynthetic enzymes for sulfated glycosaminoglycans. The Journal of biological chemistry. 2013;288(16):10953–61. 10.1074/jbc.R112.437038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mizumoto S, Yamada S, Sugahara K. Human genetic disorders and knockout mice deficient in glycosaminoglycan. BioMed research international. 2014;2014:495764 10.1155/2014/495764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thiele H, Sakano M, Kitagawa H, Sugahara K, Rajab A, Hohne W, et al. Loss of chondroitin 6-O-sulfotransferase-1 function results in severe human chondrodysplasia with progressive spinal involvement. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(27):10155–60. Epub 2004/06/25. 10.1073/pnas.0400334101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Unger S, Lausch E, Rossi A, Megarbane A, Sillence D, Alcausin M, et al. Phenotypic features of carbohydrate sulfotransferase 3 (CHST3) deficiency in 24 patients: congenital dislocations and vertebral changes as principal diagnostic features. American journal of medical genetics Part A. 2010;152A(10):2543–9. Epub 2010/09/11. 10.1002/ajmg.a.33641 . [DOI] [PubMed] [Google Scholar]
- 24. Bartolini B, Thelin MA, Rauch U, Feinstein R, Oldberg A, Malmstrom A, et al. Mouse development is not obviously affected by the absence of dermatan sulfate epimerase 2 in spite of a modified brain dermatan sulfate composition. Glycobiology. 2012;22(7):1007–16. 10.1093/glycob/cws065 . [DOI] [PubMed] [Google Scholar]
- 25. Malfait F, Syx D, Vlummens P, Symoens S, Nampoothiri S, Hermanns-Le T, et al. Musculocontractural Ehlers-Danlos Syndrome (former EDS type VIB) and adducted thumb clubfoot syndrome (ATCS) represent a single clinical entity caused by mutations in the dermatan-4-sulfotransferase 1 encoding CHST14 gene. Human mutation. 2010;31(11):1233–9. Epub 2010/09/16. 10.1002/humu.21355 . [DOI] [PubMed] [Google Scholar]
- 26. Muller T, Mizumoto S, Suresh I, Komatsu Y, Vodopiutz J, Dundar M, et al. Loss of dermatan sulfate epimerase (DSE) function results in musculocontractural Ehlers-Danlos syndrome. Human molecular genetics. 2013;22(18):3761–72. Epub 2013/05/25. 10.1093/hmg/ddt227 . [DOI] [PubMed] [Google Scholar]
- 27. Maccarana M, Kalamajski S, Kongsgaard M, Magnusson SP, Oldberg A, Malmstrom A. Dermatan sulfate epimerase 1-deficient mice have reduced content and changed distribution of iduronic acids in dermatan sulfate and an altered collagen structure in skin. Molecular and cellular biology. 2009;29(20):5517–28. Epub 2009/08/19. 10.1128/MCB.00430-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynard LN, Loughlin J. The genetics and functional analysis of primary osteoarthritis susceptibility. Expert reviews in molecular medicine. 2013;15:e2 10.1017/erm.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kluppel M, Wight TN, Chan C, Hinek A, Wrana JL. Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development. 2005;132(17):3989–4003. Epub 2005/08/05. 10.1242/dev.01948 . [DOI] [PubMed] [Google Scholar]
- 30. Ohtake-Niimi S, Kondo S, Ito T, Kakehi S, Ohta T, Habuchi H, et al. Mice deficient in N-acetylgalactosamine 4-sulfate 6-o-sulfotransferase are unable to synthesize chondroitin/dermatan sulfate containing N-acetylgalactosamine 4,6-bissulfate residues and exhibit decreased protease activity in bone marrow-derived mast cells. The Journal of biological chemistry. 2010;285(27):20793–805. Epub 2010/05/05. 10.1074/jbc.M109.084749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kluppel M, Vallis KA, Wrana JL. A high-throughput induction gene trap approach defines C4ST as a target of BMP signaling. Mechanisms of development. 2002;118(1–2):77–89. Epub 2002/09/28. . [DOI] [PubMed] [Google Scholar]
- 32. Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS biology. 2011;9(1):e1000582 Epub 2011/01/27. 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Zwaag B, Burbach JP, Scharfe C, Oefner PJ, Brunner HG, Padberg GW, et al. Identifying new candidate genes for hereditary facial paresis on chromosome 3q21-q22 by RNA in situ hybridization in mouse. Genomics. 2005;86(1):55–67. Epub 2005/06/15. 10.1016/j.ygeno.2005.03.007 . [DOI] [PubMed] [Google Scholar]
- 34. Hayes AJ, Mitchell RE, Bashford A, Reynolds S, Caterson B, Hammond CL. Expression of glycosaminoglycan epitopes during zebrafish skeletogenesis. Developmental dynamics: an official publication of the American Association of Anatomists. 2013;242(6):778–89. 10.1002/dvdy.23970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mizumoto S, Mikami T, Yasunaga D, Kobayashi N, Yamauchi H, Miyake A, et al. Chondroitin 4-O-sulfotransferase-1 is required for somitic muscle development and motor axon guidance in zebrafish. Biochem J. 2009;419(2):387–99. Epub 2009/01/08. doi: BJ20081639 [pii] 10.1042/BJ20081639 . [DOI] [PubMed] [Google Scholar]
- 36. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 1995;203(3):253–310. Epub 1995/07/01. 10.1002/aja.1002030302 . [DOI] [PubMed] [Google Scholar]
- 37. Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2013. Nucleic acids research. 2013;41(Database issue):D48–55. Epub 2012/12/04. 10.1093/nar/gks1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539 Epub 2011/10/13. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Felsenstein J. PHYLIP—Phylogeny Interference Package (Version 3.2). Cladistics. 1989;5:164–6. [Google Scholar]
- 40. Page RD. TreeView: an application to display phylogenetic trees on personal computers. Computer applications in the biosciences: CABIOS. 1996;12(4):357–8. Epub 1996/08/01. . [DOI] [PubMed] [Google Scholar]
- 41. Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. Epub 2008/01/15. doi: nprot.2007.514 [pii] 10.1038/nprot.2007.514 . [DOI] [PubMed] [Google Scholar]
- 42. Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, et al. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem. 2004;279(41):42732–41. . [DOI] [PubMed] [Google Scholar]
- 43. Kusche-Gullberg M, Kjellen L. Sulfotransferases in glycosaminoglycan biosynthesis. Current opinion in structural biology. 2003;13(5):605–11. Epub 2003/10/22. . [DOI] [PubMed] [Google Scholar]
- 44. Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. Journal of molecular evolution. 2004;59(2):190–203. Epub 2004/10/16. 10.1007/s00239-004-2613-z . [DOI] [PubMed] [Google Scholar]
- 45. Yamada S, Onishi M, Fujinawa R, Tadokoro Y, Okabayashi K, Asashima M, et al. Structural and functional changes of sulfated glycosaminoglycans in Xenopus laevis during embryogenesis. Glycobiology. 2009;19(5):488–98. Epub 2009/02/05. doi: cwp005 [pii] 10.1093/glycob/cwp005 . [DOI] [PubMed] [Google Scholar]
- 46. Miyata S, Komatsu Y, Yoshimura Y, Taya C, Kitagawa H. Persistent cortical plasticity by upregulation of chondroitin 6-sulfation. Nat Neurosci. 2012;15(3):414–22, S1–2. 10.1038/nn.3023 . [DOI] [PubMed] [Google Scholar]
- 47. Kreuger J, Spillmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174(3):323–7. Epub 2006/08/02. doi: jcb.200604035 [pii] 10.1083/jcb.200604035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayes AJ, Reynolds S, Nowell MA, Meakin LB, Habicher J, Ledin J, et al. Spinal Deformity in Aged Zebrafish Is Accompanied by Degenerative Changes to Their Vertebrae that Resemble Osteoarthritis. PloS one. 2013;8(9):e75787 Epub 2013/10/03. 10.1371/journal.pone.0075787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ledin J, Ringvall M, Thuveson M, Eriksson I, Wilen M, Kusche-Gullberg M, et al. Enzymatically active N-deacetylase/N-sulfotransferase-2 is present in liver but does not contribute to heparan sulfate N-sulfation. J Biol Chem. 2006;281(47):35727–34. Epub 2006/09/21. 10.1074/jbc.M604113200 . [DOI] [PubMed] [Google Scholar]
- 50. Uyama T, Ishida M, Izumikawa T, Trybala E, Tufaro F, Bergstrom T, et al. Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J Biol Chem. 2006;281(50):38668–74. Epub 2006/10/17. doi: M609320200 [pii] 10.1074/jbc.M609320200 . [DOI] [PubMed] [Google Scholar]
- 51. Izumikawa T, Okuura Y, Koike T, Sakoda N, Kitagawa H. Chondroitin 4-O-sulfotransferase-1 regulates the chain length of chondroitin sulfate in co-operation with chondroitin N-acetylgalactosaminyltransferase-2. The Biochemical journal. 2011;434(2):321–31. Epub 2010/12/09. 10.1042/BJ20101456 . [DOI] [PubMed] [Google Scholar]
- 52. Izumikawa T, Koike T, Kitagawa H. Chondroitin 4-O-sulfotransferase-2 regulates the number of chondroitin sulfate chains initiated by chondroitin N-acetylgalactosaminyltransferase-1. The Biochemical journal. 2012;441(2):697–705. Epub 2011/09/29. 10.1042/BJ20111472 . [DOI] [PubMed] [Google Scholar]
- 53. Sakai K, Kimata K, Sato T, Gotoh M, Narimatsu H, Shinomiya K, et al. Chondroitin sulfate N-acetylgalactosaminyltransferase-1 plays a critical role in chondroitin sulfate synthesis in cartilage. The Journal of biological chemistry. 2007;282(6):4152–61. Epub 2006/12/06. 10.1074/jbc.M606870200 . [DOI] [PubMed] [Google Scholar]
- 54. Malmstrom A, Fransson LA. Biosynthesis of dermatan sulfate. I. Formation of L-iduronic acid residues. J Biol Chem. 1975;250(9):3419–25. . [PubMed] [Google Scholar]
- 55. Pacheco B, Maccarana M, Malmstrom A. Dermatan 4-O-sulfotransferase 1 is pivotal in the formation of iduronic acid blocks in dermatan sulfate. Glycobiology. 2009;19(11):1197–203. Epub 2009/08/08. 10.1093/glycob/cwp110 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, alignment of CS/DS 4-O sulfotransferases. B, CS/DS 6-O sulfotransferases. C, CS/DS 4-sulfate 6-O sulfotransferases. D, CS/DS 2-O-sulfotransferases and E, CS/DS epimerases. Identical amino acids are marked with red letters on yellow background, conserved residues are indicated by blue letters on light blue background and blocks of similar residues marked with black letters on green background.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.