Abstract
Cellular RNA-dependent RNA polymerases (RDRs) are fundamental components of RNA silencing in plants and many other eukaryotes. In Arabidopsis thaliana genetic studies have demonstrated that RDR2 and RDR6 are involved in the synthesis of double stranded RNA (dsRNA) from single stranded RNA (ssRNA) targeted by RNA silencing. The dsRNA is subsequently cleaved by the ribonuclease DICER-like into secondary small interfering RNAs (siRNAs) that reinforce and/or maintain the silenced state of the target RNA. Models of RNA silencing propose that RDRs could use primer-independent and primer-dependent initiation to generate dsRNA from a transcript targeted by primary siRNA or microRNA (miRNA). However, the biochemical activities of RDR proteins are still partly understood. Here, we obtained active recombinant RDR2 and RDR6 in a purified form. We demonstrate that RDR2 and RDR6 have primer-independent and primer-dependent RNA polymerase activities with different efficiencies. We further show that RDR2 and RDR6 can initiate dsRNA synthesis either by elongation of 21- to 24- nucleotides RNAs hybridized to complementary RNA template or by elongation of self-primed RNA template. These findings provide new insights into our understanding of the molecular mechanisms of RNA silencing in plants.
Introduction
RNA-dependent RNA polymerases (RDRs) are required in RNA silencing, a conserved RNA-based mechanism involved in stability, protection, inheritance and expression of eukaryotic genomes at transcriptional or post-transcriptional levels [1–6]. RDRs produce long double-stranded RNAs (dsRNAs) that are subsequently cleaved by Dicer-like (DCL) nucleases into small interfering RNAs (siRNAs) ranging from 21 to 24 nucleotides (nt) [1–5]. siRNAs are then loaded onto ARGONAUTE (AGO) proteins that are part of the RNA silencing complex. One strand of the siRNA duplex then guides the ARGONAUTE complex to transcripts with complementary sequences, triggering transcript cleavage or the inhibition of translation [2–4,6]. ARGONAUTE complexes can also target genomic loci in a process known as RNA directed DNA methylation (RdDM) that promotes transcriptional gene silencing (TGS) [2–6].
In Arabidopsis thaliana, six RDRs have been identified. RDR1, RDR2 and RDR6 are functional in several siRNAs pathways whereas the functions of RDR3a, RDR3b and RDR3c, which belong to a distinct phylogenetic clade, have yet to be identified [1,4,5]. RDR2 synthesizes dsRNA from single stranded RNAs (ssRNAs) produced by the plant-specific RNA polymerase IV. RDR2-dependent dsRNA are then cleaved by DCL3 to produce the abundant 24-nt siRNAs that target heterochromatic and repetitive regions of the genome for direct sequence-specific DNA methylation and histone modification that leads to TGS [3–5,7–9]. RDR6 has the broadest range of substrates and is involved in the biogenesis of viral siRNAs [10], siRNAs from aberrant RNA transgenes during post-transcriptional gene silencing (PTGS) [11], tasiRNAs [12–15] and natsiRNA [16]. RDR6-dependent dsRNA are then cleaved by DCL4 or DCL2 to produce 21- or 22-nt long siRNA respectively [1–3,5]. RDR6 and RDR2 are also involved in PTGS transitivity and TGS transitivity, respectively. Transitivity is a phenomenon that leads to the production of secondary siRNAs upstream and downstream of the primary siRNA or miRNA binding site on a target transcript [1,17–23]. The production of secondary siRNAs allows the amplification of the RNA silencing effects [1,17–23]. However, it is not well understood how RDR6 and RDR2 are recruited to their RNA templates during transitivity. The cleavage of a transcript targeted by an miRNA produced from a miRNA:miRNA* duplex that contains asymmetrically positioned bulged bases is considered sufficient for the initiation of RDR6-dependent transitivity [17,23]. It is also unclear how RDR6 and RDR2 initiate complementary RNA synthesis during the amplification of RNA silencing. Initiation of RNA synthesis could occur as a primer-independent (de novo) process or a primer-dependent process using primary siRNA or miRNA as the primers for elongation [1,4,5]. Most known RNA polymerases rely on a primer-independent initiation mechanism, with only a few exceptions where short oligonucleotides or proteins are used as primers [4]. For Arabidopsis RDR6 there is in vivo evidence suggesting that both modes of initiation are used for complementary RNA synthesis [20,24,25]. RDR2 mediated production of nuclear secondary siRNAs is essential for RdDM during TGS in Arabidopsis and requires primary siRNAs [1,19]. However, the precise mechanism of RNA synthesis initiation was not determined.
In vitro studies on purified RDRs from tomato leaves and on wheat germ extract have detected both primer-dependent and primer-independent activities [26,27]. Tomato RDR shares significant homology with all plant RDRs [4,28] therefore suggesting that primer-dependent RNA polymerase activity might be an intrinsic property of plant RDRs. However, while in vitro primer-independent (de novo) RNA polymerase activity has been observed for RDR6, no in vitro priming activity was detected [29,30]. RDR2 activity was studied in combination with PolIV on immunoprecipitates from Arabidopsis thaliana [31]. This shows that in absence of PolIV, RDR2 has no polymerase activity. In combination with active site mutated PolIV, RDR2 can synthesize short RNA in the absence of a primer but did not synthetize dsRNA by elongation of smRNAs hybridized to complementary RNA template [32].
Here we report on the in vitro enzymatic activities of purified recombinant RDR2 and RDR6. We demonstrate that RDR2 and RDR6 display primer-independent (de novo) RNA polymerase activity. We show that RDR2 and RDR6 can both elongate self-primed RNA templates. In addition, we have found that RDR2 and RDR6 can synthesize dsRNA by elongation of smRNAs hybridized to complementary RNA template. Our results demonstrate that the primed initiation mode is an intrinsic property of Arabidopsis RDR2 and RDR6 and that RDR6 priming activity is more sensitive to the in vitro experimental conditions and/or the absence of cellular co-factors than RDR2. These findings provide important new insights into our understanding of the molecular mechanisms of RNA silencing in Arabidopsis.
Materials and Methods
Plant material and transformation
Arabidopsis thaliana (Columbia ecotype) plants and N. benthamiana plants were grown on soil at 22°C in day/night cycle (12-h light/12-h dark) in growth rooms. Arabidopsis transgenic lines were obtained by Agrobacterium-mediated transformation using the floral dip procedure [32] as modified by Clough and Bent [33]. Agrobacteria containing the expression vectors pEarley100.RDR2-HA and pEarley100.RDR6-HA were used to complement rdr2–2 and rdr6–11 mutant plants, respectively. Transformants (named RDR-HAcomp) were selected on soil sprayed with a 150mg/L Basta (Glufosinate ammonium) solution. For phenotypic analysis, T2 plants were selected on soil and treated with Basta. Young flowers were harvested from six individual transgenic plants for pEarley100.RDR2-HA lines and pEarley100.RDR6-HA lines, quickly frozen in liquid nitrogen, and stored at −80°C for Northern blot analysis.
Constructions of plasmids
The full-length cDNA of RDR2 and RDR6 were amplified from the ATG to the stop codon by PCR using primers AttB1-RDR2/AttB2-RDR2, and AttB1-RDR6/AttB2-RDR6 (S1 Table). The resulting amplicons were cloned into pDONR207 (Invitrogen) to give pDONR207-RDR2 and pDONR207-RDR6 plasmids. Amplification of RDR2-HA was realized on pDONR207-RDR2 with primers Age1-RDR2 and SmaI-RDR2-2xHA. Amplification of RDR6-HA was realized on pDONR207-RDR6 with primers Age1-RDR6 and XhoI-RDR6-2xHA. The RDR6-HA amplicon was then digested by AgeI/XhoI and cloned into pEAQ-HT plasmid [34] to obtain pEAQ-RDR6-HA. The RDR2-HA amplicon was then digested by AgeI/SmaI and cloned into pEAQ-HT plasmid to obtain pEAQ-RDR2-HA. Cloned RDR sequences were verified by DNA sequencing. RDR mutants were obtained by linear amplification of pDONR207-RDR2 and pDONR207-RDR6 plasmids with primers RDR2mutFW and RDR6mutFW respectively, to substitute by an Ala residue the putative catalytic Asp residues located at amino acid 834 of RDR2 and 867 of RDR6. Amplification products were then digested with DpnI to eliminate the plasmid templates and introduced into E. coli. Mutations were checked by DNA sequencing after plasmid purification of pDONR207-RDR2m and pDONR207-RDR6m. Amplification of RDR2m-HA was realized on pDONR207-RDR2m with primers Age1-RDR2 and SmaI-RDR2–2xHA and cloned into pEAQ-HT plasmid to obtain pEAQ-RDR2m-HA. Amplification of RDR6m-HA was realized on pDONR207-RDR6m with primers Age1-RDR6 and XhoI-RDR6-2xHA and cloned into pEAQ-HT plasmid to obtain pEAQ-RDR6m-HA. The full-length cDNA of RDR2 and RDR6 with 2xHA tag in 3’ end were amplified by PCR using primers AttB1-RDR2/AttB2-2xHA and AttB1-RDR6/ AttB2-2xHA with pEAQ-RDR2-HA and pEAQ-RDR6-HA templates, respectively. The resulting amplicons were cloned into pDONR207 (Invitrogen) to give pDONR207-RDR2-HA and pDONR207-RDR6-HA plasmids. A LR recombinant reaction was carried out between the expression vector pEARLEY100 [35] and either pDONR207-RDR2-HA or pDONR207-RDR6-HA to obtain pEarley100.RDR2-HA and pEarley100.RDR6-HA plasmids used for rdr mutant complementation.
Expression, purification of recombinant RDR proteins and immuno blotting
Transient expression of recombinant RDRs using agrobacterium containing either pEAQ-RDR2-HA, pEAQ-RDR6-HA, pEAQ-RDR2m-HA or pEAQ-RDR6m-HA plasmids has been realized in N. benthamiana according to the protocol established by Voinnet et al. [36]. Infected leaves were collected after 60h of incubation, frozen in nitrogen and conserved at −80°C. Protein extraction and purification were done according to Curaba and Chen [29] with some modifications. Typically, 5g of harvested infiltrated tissues were ground in a mortar. One volume of tissue powder was mixed with two volumes of extraction buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 0.1% (v/v) triton, 10% (v/v) glycerol, 1mM phenylmethylsulfonyl fluoride, 0.05% (v/v) 2-mercaptoethanol, 0.5% (v/v) Polyvinylpyrrolidone (PVP)) and one protease inhibitor mixture tablet per 20 ml (Roche), and incubated for 20 min at 4°C. Lysate was centrifuged 10 min at 1,500 g at 4°C, then for 30 min at 20,000 g at 4°C and the supernatant was collected (when necessary, additional centrifugation steps were performed to eliminate dark pigments). 200 μl of anti-HA affinity matrix (Roche) were stacked in a chromatography column and washed 3 times with 5ml of extraction buffer. The supernatant was passed through the column 2 times. The beads were washed 3 to 4 times with 20 volumes of the washing buffer (20 mM Tris-HCl, pH 7.6, 100 mM NaCl, 0.1 mM EDTA, 0.05% (v/v) Tween 20, 0.05% (v/v) 2-mercaptoethanol and one protease inhibitor tablet/20 ml) and finally resuspended in 3 volumes of storage buffer (20 mM HEPES- KOH, pH 7.6, 20 mM NaCl, 0.2 mM EDTA, 20% (v/v) glycerol, 0.05% (v/v) 2-mercaptoethanol and one protease inhibitor tablet/20 ml), and stored in aliquots at −80°C. RDR-HA-bound on anti-HA-agarose beads were analyzed on a 4–20% polyacrylamide gradient-SDS gel and transferred to Protran BA 85 Nitrocellulose membrane (Whatman). Monoclonal anti-HA antibody (Sigma) were used as primary antibody, and Sheep Anti-Mouse IgG ECL Antibody, HRP Conjugated (GE Healthcare) as secondary antibody. ECL Plus Western Blotting Detection Reagents (GE Healthcare) and Fluorescent Image analyser FLA-5000 (Fuji) were used for detection and image acquisition.
Preparation of RNA duplexes
Chemically synthesized and PAGE purified heteropolymeric single-stranded RNA (ssRNA) templates were obtained from Dharmacon (S2 Table). In a 20μL reaction assay containing 500 mM of NaCl, 150 pmol of RNA37 was mixed with either 100 pmol of 5’[32P]-labelled RNA21, RNA22 or RNA24. The mixture was heated at 95°C for 2 min and slowly cooled for 3h at room temperature. Water was added to the mixture up to 50 μl before purification on G25 Sephadex columns (Health care) to remove NaCl and free [γ-32P] ATP.
Polymerase activity assays
RDR activity assays were performed according to Makeyev and Bamford [37] with some modifications. Assays were conducted in 20μl reaction mixtures assembled on ice, containing 50 mM HEPES-KOH (pH 7.6), 10 mM MgCl2, 0.1 mM EDTA, 100 μM each of cold ATP, CTP, GTP and UTP, 100 nM of single-stranded or partially double-stranded RNA. In some experiments RNA templates were labeled by 5’ phosphorylation using [γ-32P]ATP (3000 Ci/mmol) and T4 kinase (Promega). In experiments with unlabeled templates, 150 nM of [α-32P]GTP (3000 Ci/mmol) were added to the reaction mixtures. To start the reaction, about 2.2 pmol of RDR bound on anti-HA-agarose beads were added and tubes were incubated at 25°C for 4h in a thermomixer set for a gently 30s-long agitation (300 rpm) every 10 min. After incubation, reactions were placed on ice and 4 μl of polymerisation products were either directly mixed with a formamide/EDTA gel-loading buffer or treated with RNase T1 before addition of loading buffer (see Nuclease Treatments). Polymerisation products were then heated 5 min at 65°C and loaded on a 14% acrylamide-bisacrylamide [19:1], 7 M urea gels, using TTE buffer (89 mM Tris/HCl (pH 8.0), 28 mM Taurine and 0.5 mM EDTA). The gels were exposed and analysed using photo-stimulated plates and a Fluorescent Image Analyzer (Fluorescent Image Analyser FLA3000, Fuji).
Nuclease treatments
Four microliters of the polymerase reaction mixture supplemented with 5 units of RNaseT1 (Invitrogen) were incubated 30 min at 30°C and the reaction was placed on ice. Then, 1μl of 200 mM Dithiothreitol in addition to the formamide/EDTA loading buffer was added to the mixture and samples were heated 5 min at 65°C before loading on polyacrylamide/Urea gels.
Results
Production of functional RDR2 and RDR6 with a double C-terminal HA-tag
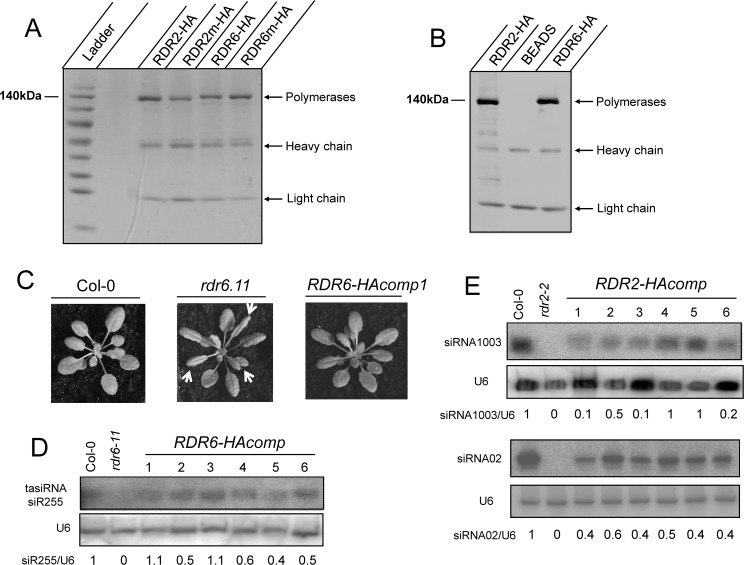
To produce RDR2 and RDR6 we chose a transient expression system in N. benthamiana. Full-length RDR2 and RDR6 coding sequences with a double HA tag at the 3’ end were cloned into the pEAQ-HT plasmid. pEAQ-HT contains a gene for the expression of the P19 inhibitor of RNA silencing in planta and the introduced coding sequences are flanked by the 5’-untranslated region (UTR) and the 3’-UTR from Cowpea mosaic virus (CPMV) RNA-2. These features allow for an extremely high-level production of proteins [34]. Tagged RDR2 and RDR6 (RDR2-HA and RDR6-HA) expressed in N. benthamiana leaves were immunopurified with HA-conjugated beads and the purity of the enzymes was checked on polyacrylamide gel stained with coomassie blue and by western blot analysis showing that in each case a single polypeptide was purified (Fig. 1A and 1B). To ensure that RDR activities observed in polymerase assays were not due to contaminating polymerases, we also produced mutated versions of RDR2-HA (RDR2m-HA) and RDR6-HA (RDR6m-HA) where a catalytic aspartate residue critical for RDR activity is substituted by alanine [29, 37, 38]. We obtained about 100 ng of purified RDR per 1μl of HA-conjugated beads (Fig. 1A). The functionality of the HA-tagged RDR6 and RDR2 proteins was confirmed by stable expression under the control of the cauliflower mosaic virus (CaMV) 35S promoter in A. thaliana rdr6.11 and rdr2.2 mutants respectively. The rdr6.11 mutant has distinctive downward curling of the rosette leaves. Expression of RDR6-HA fully restored the wild-type phenotype and the production of an RDR6-dependent tasiRNA (siR255) (Fig. 1C and 1D). rdr2.2 mutant does not show an obvious visible phenotype, but is impaired in the production of some endogenous 24nt siRNAs. Northern blot analysis showed that expression of RDR2-HA restored the production of these siRNAs (siRNA1003, siRNA02) (Fig. 1E).
Fig 1. Purification of RDR-HA and RDRm-HA and complementation of rdr2–2 and rdr6–11 mutations by RDR2-HA and RDR6-HA.
(A) Coomassie blue stained protein gel of immunopurified RDRs. After immunoprecipitation, 2.5μl of HA-conjugated beads, corresponding to 250ng of proteins, were loaded on a 4–20% polyacrylamide gradient-SDS gel. Heavy and light chains from HA antibody are visible. (B) Western blot performed with anti-HA antibodies. After immunoprecipitation, 1μl of HA-conjugated beads were loaded on a 4–20% polyacrylamide gradient-SDS gel. The secondary antibody cross reacts with the heavy and light chains of the HA antibody. (C) 24-day-old Columbia wild type (Col-0) plant, rdr6–11 mutant and one rdr6–11 transgenic line containing the 35S::RDR6-HA transgene (RDR6-HAcomp1). The downward curling rosette leaf phenotype of rdr6–11 mutants (arrows) was rescued by RDR6-HA expression. (D) RNA blot analysis of the accumulation of the TAS1-derived tasiRNA siR255. The analysis was performed on rdr6–11 mutant and six independent 35S::RDR6-HA rdr6–11 transgenic lines. (E) RNA blot analysis of the accumulation of the heterochromatic siRNA1003 and siRNA02. The analysis was performed on rdr2–2 mutant and six independent 35S::RDR2-HA rdr2–2 transgenic lines (RDR2-HAcomp). U6 snRNA hybridization served as control for total RNA loading and normalized values with Col-0 control set to 1 are indicated.
RDR2-HA and RDR6-HA initiate in vitro RNA synthesis using a primer-independent (de novo) mechanism
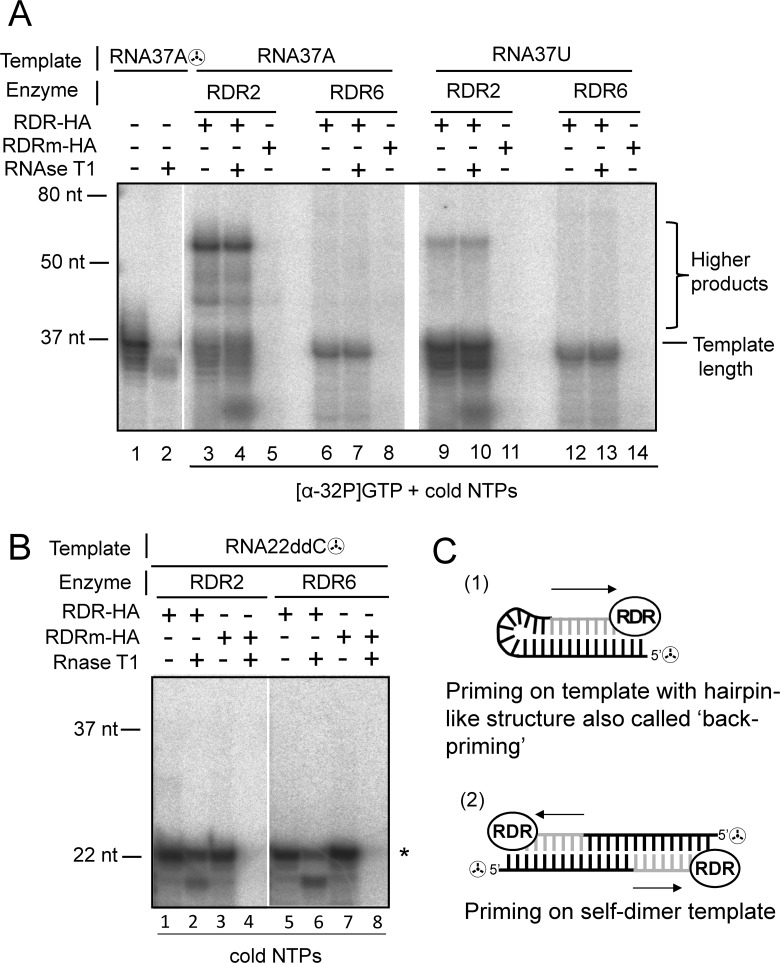
RNA polymerase activity assays for RDR2-HA and RDR6-HA were performed using two short chemically synthesized and PAGE purified heteropolymeric ssRNA templates with a 37 nt random sequence containing a monophosphate at the 5’ terminus, a feature of small RNAs generated by DICER-like enzymes (S2 Table). The recombinant RDRs and cold ssRNA templates (RNA37A and RNA37U) were incubated in the presence of all four cold ribonucleoside triphosphates (NTPs) and trace amounts of [α-32P] GTP (Fig. 2A). For RDR2-HA and RDR6-HA analysis of reaction products on a denaturing polyacrylamide gel showed the production of labeled RNA of various sizes (Fig. 2A lanes 3,6,9,12). No labeling was detected using the mutated RDRs (Fig. 2A lanes 5,8,11,14). The band-product pattern is strikingly different between RDR2-HA to RDR6-HA: the former generates labeled products longer than the template and a major product nearly up to twice the size of the template (referred thereafter as “higher products”) (Fig. 2A lanes 3 and 9), whereas for RDR6-HA the major product is of apparent template size (Fig. 2A lanes 6 and 12). To show that the RDR2-HA and RDR6-HA exhibit authentic polymerase activity leading to synthesis of RNA complementary to the ssRNA template and resulting in production of dsRNA, polymerase products were treated with RNAse T1. RNAse T1 digests ssRNA, but not dsRNA, by cleaving between guanosine 3'-phosphate residues and the 5'-OH residues of adjacent nucleotides. In this experiment 5'[32P]-labeled RNA37A was used as a size marker (Fig. 2A lane1) and 5'[32P]-labeled RNA37A treated with RNAse T1 was used as a control for RNAseT1 digestion (Fig. 2A lane 2). Without prior polymerase action the 5’[32P] labeled RNA37A ssRNA template is degraded by RNAse T1 (Fig. 2A compare lane 1 and 2) whereas polymerase products are resistant to RNase T1 (Fig. 2A lanes 4,7,10,13) thus confirming their double-stranded nature. Similar results are obtained when RNA polymerase assays with RDR2-HA and RDR6-HA are performed using 5’[32P]-labeled ssRNA37 template and cold NTPs (S1 Fig.). RDR2-HA is able to synthesize “higher products”, whereas RDR6-HA is not. However, we note that RNaseT1 treatment of RDR6-HA generated products only partially removes the radiolabeled input template, indicating that the RDR6 products are dsRNA (S2 Fig. lane 5). To establish the nature of the “higher products” we performed polymerase assays using a 5’[32P]-labeled RNA template blocked at its 3′ end by a dideoxycytidine (RNA22ddC) (S2 Table). In this case, both polymerases produced the same banding pattern with the longer product at template size and without the “higher products” (Fig. 2B). The absence of template elongation by the 3' ddC nucleotide shows that the "higher products" produced by RDR2-HA are the result of RNA synthesis starting at the 3’ end by either self-priming or intermolecular priming mechanism (Fig. 2C). Furthermore, this experiment demonstrates that the two polymerases are able to initiate RNA synthesis de novo in the absence of self-priming. For both enzymes, we could also observe RNAse T1 resistant products that were smaller than the template length (Fig. 2B lanes 2 and 6) indicating that de novo synthesis may start internally on the provided template. Altogether, these results show that both RDR2-HA and RDR6-HA are active RNA polymerases that are able to initiate RNA synthesis using a primer-independent (de novo) mechanism. In addition, RDR2-HA is able to initiate RNA synthesis using a self-priming mechanism.
Fig 2. RDR2-HA and RDR6-HA initiate in vitro RNA synthesis using a de novo (primer-independent) mechanism.
(A) RNA polymerase assays of purified RDR2-HA and RDR6-HA on 37nt long ssRNA templates (RNA37A and RNA37U) in presence of cold NTPs and trace amount of [α32-GTP]. lane1: 5'[32P]-labeled RNA37A (RNA37A☢) is used as a size marker. lane2 RNA37A☢ treated with RNAse T1 as a control for RNAseT1 digestion in conditions that almost completely degrade the ssRNA template (B) Purified RDR2-HA and RDR6-HA were incubated with cold NTPs and a 22-nt 5’[32P]-labeled ssRNA template blocked at its 3′ end by dideoxycytidine (RNA22ddC☢). (*) A single star indicates the position of the 5’[32P]-labeled template. (C) Schematic representation of mechanisms that allow RNA polymerases to generate products higher than the RNA template size (“higher products”). Arrows indicate the direction of RNA polymerization. Template RNA in black, synthesized RNA by RDR in grey.
RDR2-HA and RDR6-HA initiate in vitro RNA synthesis using a primer-dependent initiation mechanism
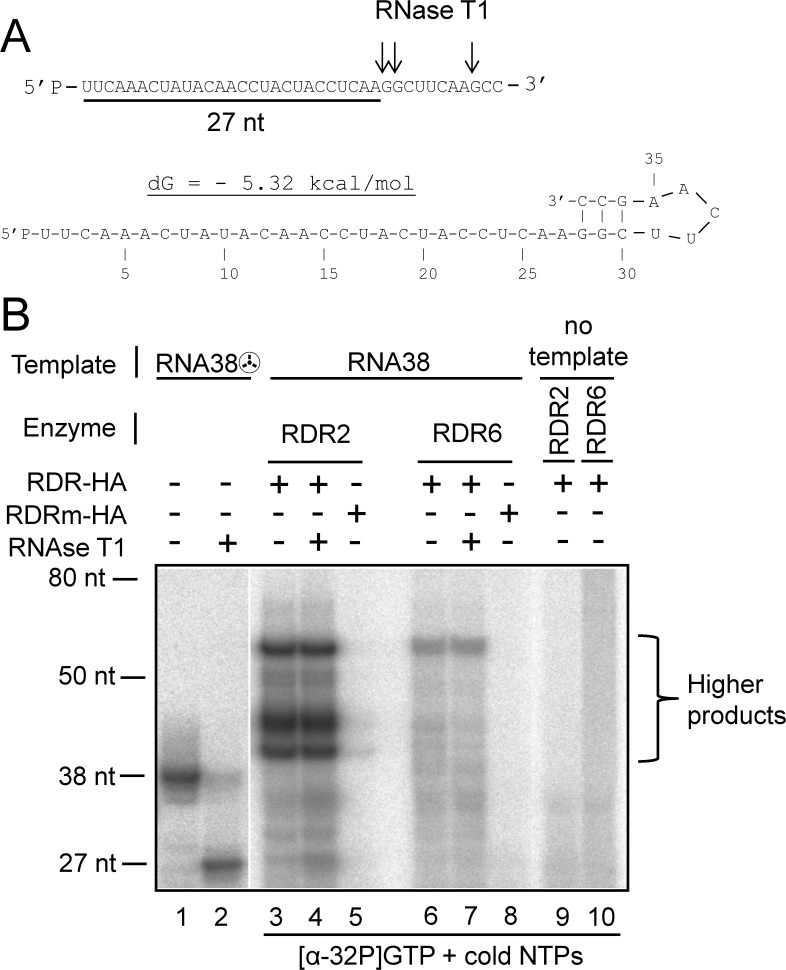
In polymerase assays using non labeled ssRNA template, cold NTPs and trace amount of [α-32P]GTP, RDR2-HA was able to synthesize “higher product” (Fig. 2A). Similar results were obtained when RDR2-HA was incubated with 5’[32P]-labeled ssRNA37 templates and cold NTPs. In these conditions detection of “higher products” is only possible if there are generated by elongation of the radiolabeled input template (S1 Fig. lanes 1 and 2). Therefore the “higher products” generated by RDR2-HA are likely due to a priming activity that corresponds to self-primed elongation of the ssRNA template from its 3'-terminal hydroxyl by intramolecular (Fig. 2C.1) or intermolecular template interactions (Fig. 2C.2). This hypothesis is also supported by our assays with a ddC blocked template (Fig. 2B). RDR6-HA did not perform self-primed elongation of RNA37A and RNA37U templates (Fig. 2A and S1 Fig.). As RDR6 is a very close paralog of RDR2 [4] we hypothesized that they share catalytic properties and that experimental conditions or template features could account for the difference in the priming efficiency between RDR2-HA and RDR6-HA. As 3’ terminal secondary structures can promote primer-dependent initiation [39] we designed an ssRNA template that can adopt a stable hairpin-like conformation in the 3' terminus region (named RNA38, Fig. 3A) and performed RNA polymerase assays using this template, cold NTPs and trace amounts of [α-32P]GTP. Under these conditions both RDR2-HA and RDR6-HA produce “higher products” resistant to RNAase T1 (Fig. 3B, lanes 3,4,6,7). To confirm that the “higher products” observed were due to a self-primed elongation of the RNA38 template, we incubated each RDR with 5’[32P]-labeled RNA38 template and cold NTPs. In these conditions, we observe a product pattern similar to that observed with labeled RNA38 and [α-32P]GTP (Fig. 3B and S2 Fig.). The “higher products” were double-stranded, as shown by the RNase T1 resistance assay (Fig. 3B lanes 4 and 7, S2 Fig. lanes 2 and 5). Unlike RDR6-HA, RDR2-HA generates from RNA37 and RNA38 templates, clearly detectable primer-dependent products of various sizes (Fig. 2A lanes 3,4,9,10, 3B lanes 3 and 4, S1 Fig. lanes 1 and 2, S2 Fig. lanes 1 and 2).
Fig 3. Polymerisation activity of RDR2-HA and RDR6-HA on an ssRNA template designed to adopt a stable hairpin-like conformation in the 3' terminus region.
(A) Sequence and RNAse T1 cleavage sites of the 38-nt-long ssRNA38 template and schematic representation of the energetically most-favorable predicted secondary structure of ssRNA38 template using OligoAnalyser 3.1 (IDT). (B) Purified RDR2-HA and RDR6-HA were incubated with RNA38 template, cold NTPs and trace amounts of [α32-GTP]. 5'[32P]-labeled ssRNA38 is used as a size marker (RNA38A☢) (lane1) and as a control for RNAseT1 digestion (lane2).
This size heterogeneity observed for RDR2-HA products could arise from a more efficient transcription initiation of RNA templates with 3’ terminal nucleotide primed at different positions. Another nonexclusive hypothesis could be that RDR2-HA start transcription initiation of RNA templates with 3’ terminal nucleotide primed at a unique position but has reduced elongation processivity which gives rise to abortive products.
We conclude that both RDR2-HA and RDR6-HA are able to perform elongation of self-primed ssRNA templates. However, contrary to RDR2-HA, RDR6-HA has elongated only one of the three RNA templates used, RNA38. This could be due to different nucleotide sequences or to the nature of the 3’ terminal nucleotide of the templates (S2 Table). In addition, RNA38 molecules can be self-primed with more stable secondary structures than RNA37A and RNA37U, this could allow RDR6-HA to perform self-primed elongation of RNA38.
RDR2-HA and RDR6-HA initiate in vitro dsRNA synthesis by elongation of 21-, 22- and 24- nt small RNAs hybridized to a complementary RNA template
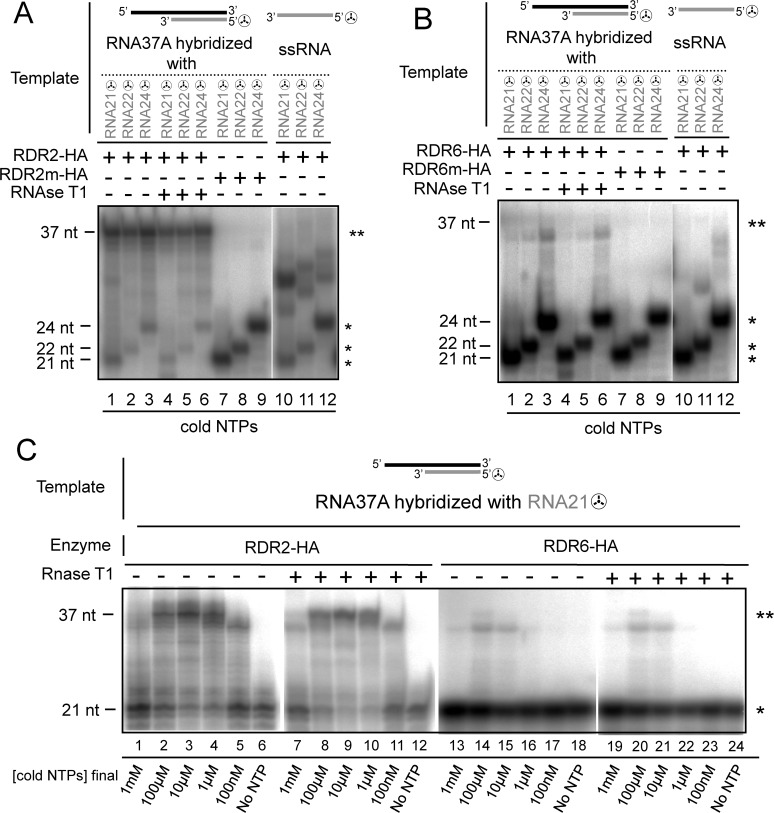
RNA silencing models propose that secondary siRNAs are produced by RDR2- and RDR6-mediated elongation of small RNAs (smRNA) i.e. primary siRNA or miRNA hybridized to target ssRNA [1,19]. In vivo studies have shown that AtRDR6 has primer elongation activity [20], this activity has not been demonstrated in vitro with RDR6 alone [29] or with SGS3, a dsRNA-binding protein that physically interacts with RDR6 [40] and that stabilizes RDR6 templates in vivo [15]. Therefore, our next challenge was to determine whether purified RDR6-HA and RDR2-HA were able to elongate smRNA hybridized to a complementary ssRNA template and to determine if this priming activity is dependent on smRNA size. 5’[32P]-labeled smRNAs of 21-, 22- and 24-nt corresponding in size to smRNAs involved in RNA silencing were used as primers by hybridization to the 3’ complementary region of a 37-nt ssRNA template (S3 Fig.). These partial dsRNA mimic the in vivo hybridization of an siRNA or miRNA to a target RNA. Each RDR was incubated with the 5’[32P]-labeled partial dsRNA templates and cold NTPs (Fig. 4). These experimental conditions only allow detection of products resulting in elongation of 5’[32P]-labeled smRNAs. For RDR2-HA, we obtained a strongly labeled RNase T1-resistant product of 37-nt size, which corresponds to synthesis of the full-length dsRNA by elongation from the 5’[32P]-labeled smRNAs (Fig. 4A lanes 1–6, 4C lanes 2–4 and 8–10, S4 Fig. lanes 3 and 4). The intensity of the 37-nt signal was similar, irrespective of the hybridized smRNA size (Fig. 4A lanes 1–6). No polymerisation was observed with mutated RDR2-HA (Fig. 4A lanes 7–9, S4 Fig. lane 5). To exclude the possibility that the 37-nt signal was produced by self-priming activity on non hybridized 5’[32P]-labeled smRNAs, reactions containing only the 5’[32P]-labeled smRNA were performed (Fig. 4A lanes 10–12). In this case, “higher products” corresponding to self-primed elongation of smRNAs were observed but no 37-nt product was detected. For RDR6-HA, we observed signals corresponding to dsRNA produced by smRNA elongation but the intensity of the signals was much weaker than that observed for RDR2-HA (Fig. 4B lanes 1–6, 4C lanes 13–15 and 19–21, S4 Fig. lanes 6 and 7). In addition, the main band was a few nucleotides below the template size indicating that RDR6-HA performed an incomplete synthesis of complementary strand. When each mutated RDR was incubated with partial dsRNA templates (i.e. no polymerase activity), a strong RNAse T1 resistant signal corresponding to the 5’[32P]-labeled smRNAs showed that in our experimental conditions, hybridization of the smRNAs to complementary RNA37 template was effective (Fig. 4A lanes 7–9, 4B lanes 7–9). Altogether, these results show that RDR2-HA and RDR6-HA are able to initiate in vitro dsRNA synthesis by elongation of smRNAs hybridized to complementary RNA template.
Fig 4. Polymerisation activity of RDR2-HA and RDR6-HA on partial dsRNA templates.
(A) Purified RDR2-HA and RDR2m-HA were incubated with partial dsRNAs (S3 Fig.) formed with RNA37 hybridized with either 5’[32P]-labeled 21-, 22- or 24-nt ssRNA (RNA21☢, RNA22☢, RNA24☢) (lanes 1–9). Purified RDR2-HA was incubated with either RNA21☢, RNA22☢, RNA24☢ oligonucleotides (lanes 10–12). (B) Purified RDR6-HA and RDR6m-HA were incubated with partial dsRNAs formed with RNA37 hybridized with either RNA21☢, RNA22☢ or RNA24☢ oligonucleotides (lanes 1–9). Purified RDR6-HA was incubated with RNA21☢, RNA22☢, RNA24☢ oligonucleotides (lanes 10–12). (C) Purified RDR2-HA and RDR6-HA were incubated with partial dsRNAs formed with RNA37A hybridized with 5’[32P]-labeled RNA21☢ oligonucleotide and with various concentrations of all four cold NTPs. (*) A single star indicates the position of the 5’[32P]-labeled ssRNA. (**) A double star indicates the position of the full-length elongated RNA template.
In vitro primer-dependent RNA polymerisation by RDR6-HA and RDR2-HA is dependent on the concentration of NTPs
Two studies have reported in vitro primer-independent RNA polymerisation of recombinant RDR6 but no primer-dependent RNA polymerisation was detected [29,30]. Although weak, we showed a primer-dependent activity for RDR6-HA (Fig. 4B and 4C, S4 Fig.). One hypothesis is that in vitro primer-dependent activity of recombinant RDR6 could have been prevented by the N-terminal localisation of the HA tag in contrast to the C-terminally HA-tagged RDR6 used in our study. Another hypothesis is that primer-dependent RNA polymerisation is sensitive to differences in the polymerase reaction mixtures, such as NTP concentration that is known to modulate the ratio between priming and de novo activity of viral RDR [41]. To test this latter hypothesis we used RDR6-HA and RDR2-HA to perform 21-nt smRNA elongation assays on partial dsRNA template with different NTPs concentrations (Fig. 4C). No priming activity was detected for RDR6-HA at 1μM and 100nM cold NTPs. At 100μM and 10μM cold NTPs we observed a weak signal a few nucleotides below the 37 nt full length product (Fig. 4C lanes 14 and 15) indicating that RDR6-HA performed an incomplete primer elongation as observed above (Fig. 4B). At 1mM cold NTPs, the concentration used in previous studies on recombinant RDR6, the signal is at the limit of detection (Fig. 4C lane 13). Therefore NTP concentration strongly influences the efficiency of RDR6-HA primer-dependent activity and could explain why this activity was not detected by others [29,30]. For RDR2-HA, a signal corresponding to the synthesis of full-length dsRNA by elongation of the 5’[32P]-labeled smRNAs was detected at 100 μM, 10μM and 1 μM cold NTPs (Fig. 4C, lanes 2–4) and a signal corresponding to an incomplete smRNA elongation was detected at 1mM and 100nM cold NTPs (Fig. 4C lanes 1 and 5). As observed for all smRNAs tested at 100 μM (Fig. 4A and 4B), the efficiency of primer-dependent activity of RDR2-HA was much higher than for RDR6-HA at each NTP concentration tested (Fig. 4C). These results indicate that under in vitro conditions, priming activity is dependent on NTP concentration and that RDR2-HA is more efficient than RDR6-HA at elongating smRNA hybridized to a complementary RNA template. RDR6-HA might exhibit stronger primer-dependent activity in the presence of cellular factors or under other yet-to-be defined experimental conditions. Altogether, our results show that RDR2-HA and RDR6-HA are both potent primer-dependent and primer-independent RNA polymerases.
Discussion
Our findings demonstrate that in vitro purified recombinant RDR2-HA, like RDR6-HA has an RNA dependent RNA polymerase activity. This is consistent with the involvement of RDR2 in the biogenesis of dsRNA that give rise to heterochromatic and repeat-associated siRNAs, the most abundant endogenous siRNAs in plants. [5,7–9]. RDR2-HA and RDR6-HA can initiate dsRNA synthesis in vitro by a primer-independent (de novo) mechanism (Fig. 2). These results are in agreement with recent in vivo studies on the role of RDR6 in the tasiRNA-generating pathway. It was shown that for the TAS1, TAS2 and TAS3 ssRNA templates, RDR6-mediated synthesis of complementary RNA is initiated internally at the second, third or fourth nucleotide from the 3’ end, suggesting a de novo initiation mechanism for RDR6 polymerase activity in vivo [24,25].
We showed that RDR2-HA and RDR6-HA synthesized RNA products that were longer than the RNA template size and nearly twice the size of the template. These products are dsRNA and result from a self-primed elongation of the ssRNA template from its 3'-terminal hydroxyl. RDR6-HA could not perform in vitro self-primed elongation of all ssRNA templates tested but was active on an RNA template designed to adopt a stable hairpin-like conformation at the 3' terminus region. In contrast, RDR2-HA was able to efficiently extend from a RNA 3’-hydroxyl end at a variety of positions on an RNA template (Fig. 2 and Fig. 3). RDR2 and RDR6 share almost identical active site architecture with similar amino acids residues but have a different additional large N-terminal domain with uncharacterized function that could explain the difference in priming efficiency [4]. This difference in priming efficiency may reflect a requirement for cellular co-factors in RDR6 priming or other yet-to-be defined experimental conditions.
Does elongation of self-primed RNA template by RDRs have a functional significance in RNA silencing? Improperly terminated and unpolyadenylated mRNA (known as aberrant RNAs) from sense transgenes are targeted by RDR6-mediated RNA silencing in Arabidopsis [42]. In such cases mRNA with defective 3’-ends lack the full complement of RNA-binding proteins. The naked 3’ ends can then lead to the spontaneous formation of 3’ hairpin-like structures, allowing priming of dsRNA synthesis by RDR6. In support of this view, enhanced RDR6-dependent RNA silencing phenotypes have been observed in Arabidopsis mutants that are deficient for mRNA 3’-end formation [43]. RDR2 is involved in TGS and synthesizes dsRNA from RNAs produced by the plant-specific DNA-dependent RNA polymerase IV [6,31,44]. We observed efficient self-primed elongation of ssRNA template by RDR2 (Fig. 2 and 3). A recent study address the activity of RDR2 in combination with the plant-specific DNA-dependent RNA polymerase IV with which it interacts [31]. In this work, it is shown that RDR2 alone has no polymerase activity, while RDR2, in combination with inactive PolIV, can efficiently synthesize short RNA in the absence of a primer. In addition, this study did not show efficient elongation of a primed RNA hybridized to complementary RNA template by RDR2 in combination with inactive PolIV, while in this condition efficient primer independent synthesis from short RNA template is found. Several causes may explain the differences with our results. We purified RDR2 from overexpressing N. Benthamiana as a single polypeptide while in the study of Haag et al., it was immunopurified from Arabidopsis in combination with PolIV, therefore some Arabidopsis inhibitors of RDR2 polymerase activity may have copurified. For exemple, PolIV even inactive may inhibit RDR2 polymerase activity. However our results are still compatible with a model were selfed-primed PolIV transcripts are converted to double-stranded RNA by RDR2 activity to serve as DCL3 substrates. Double-stranded RNA synthesis from self-primed PolIV transcripts by RDR2 would however suppose that the 3’ end of nascent RNA be previously released from the PolIV transcribing complex.
In vivo, RDR2 priming-dependent activity could be involved in the production of dsRNA from unpolyadenylated RNAs that adopt 3’ hairpin-like structures, such as those produced by PolIV transcription of transposons, retroelements and other repetitive DNA elements [3,7–9,44]. In addition, the occurrence of hairpin-like structures at unpolyadenylated 3’ ends could be a feature that promotes the recruitment of RDR2 and RDR6 to their RNA substrates. In such cases, RDR recruitment and initiation of complementary RNA synthesis would be independent of primary siRNA or miRNA targeting. This hypothesis is supported by a recent study showing the RDR6-dependent production of phased and unphased siRNAs from RNAs that do not appear to be targeted by siRNAs or miRNAs [45].
In Arabidopsis, RDR2 and RDR6 are involved in the transcription of RNA targeted by primary siRNA or miRNA leading to production of secondary siRNAs [1–5,19,20]. One possible mechanism for the synthesis of the complementary strand 5’ of the smRNA binding site is the elongation of smRNA hybridized to the target RNA [1,19,20,46,47]. Indeed in vivo evidence for smRNA elongation was provided in the study of the RDR6-dependent silencing of a miR171(21nt)-sensor transgene in Arabidopsis [20]. For RDR2-dependent secondary siRNA synthesis, it has been shown using a transgene reporter system that the silencer locus produced primary siRNAs of 21-, 22- and 24-nt [19]. It remains to be determined whether RDR2 recruitment is dependent on the size of the primary siRNA, and to date there is no in vivo evidence of an RDR2 priming activity. Our finding that RDR2-HA and RDR6-HA can elongate self-primed ssRNA templates prompted us to assess their ability to elongate smRNA hybridized to ssRNA templates and to test if this ability was smRNA-size dependent. RDR2-HA was able to fully elongate the template with no preference for smRNA size (Fig. 4A and 4C). RDR6-HA was also able to perform smRNA-primed elongation although with a lower efficiency (Fig. 4B and 4C). These results show that RDR2 and RDR6 have an intrinsic capacity to elongate smRNA in vitro, and that this activity is not dependent on the length of the smRNA in vitro. Priming activity was not detected in previous in vitro studies with recombinant HA-RDR6 [29,30] however different HA tag locations were used (at the C-terminus in our work and at the N-terminus previously). Furthermore, we show that the NTPs concentration in the reaction buffer has a crucial influence on priming activities, and that priming activity is strongly suppressed at the concentrations used in the previous studies (Fig. 4C). Although detectable, we found that in vitro RDR6-HA priming was weak, and we suggest that this is due to the absence of cellular co-factors that promote priming in vivo. One of these co-factors could be the dsRNA-binding protein SGS3 that physically interacts with RDR6 [40] and stabilizes RDR6 templates in vivo [15]. Furthermore, in vitro SGS3 can bind to partially dsRNA with a 5’ overhang [30] that could mimic the in vivo hybridization of an siRNA or miRNA to a target RNA. Similarly, the SGS3-like protein RDM12, involved in RNA-directed DNA methylation and transcriptional gene silencing can be a co-factor of RDR2 [48]. The experimental set up described in the present work will allow the assessment of the roles of SGS3 and RDM12 in modulating RDR6 and RDR2 polymerase activities and particularly their priming activity. To date there is no evidence for an in vivo priming activity of RDR2. Our results clearly show that RDR2 has the intrinsic ability to initiate primed RNA synthesis thus opening up a new field of investigation into the mechanism of RDR2-dependent RNA silencing.
Supporting Information
Purified RDR2-HA and RDR6-HA were incubated with cold NTPs and 5'[32P]-labeled 37-nt RNA template (RNA37A☢). In these conditions, detection of labeled products higher than the template size (>37nt, higher products) correspond only to elongated template. (*) A single star indicates the position of the RNA37A☢ template.
(TIF)
RNA38 is designed to adopt a stable hairpin-like conformation in the 3' terminus region. Purified RDR2-HA and RDR6-HA were incubated with 5'[32P]-labeled 38-nt ssRNA template (RNA38☢), cold NTPs and trace amounts of [α32-GTP]. (*) A single star indicates the position of the RNA38☢ template.
(TIF)
RNA37A template was hybridized to either 5‘[32P]-labeled RNA of 21- (RNA21☢), 22- (RNA22☢) or 24-nt (RNA24☢) sizes oligonucleotides.
(TIF)
Purified RDR2-HA and RDR6-HA were incubated with partial dsRNAs formed with RNA37A hybridized with 5’[32P]-labeled 22-nt ssRNA (RNA22☢). 5'[32P]-labeled RNA37A (RNA37A☢) and 5'[32P]-labeled RNA22 (RNA22☢) are used as a size markers. (*) Simple star indicate the position of the RNA22☢. (**) Double star indicates the position of the full-length elongated RNA template.
(TIF)
(PDF)
(PDF)
Acknowledgments
We thank NASC for rdr2–2 and rdr6–11 mutants, George P. Lomonossoff for pEAQ plasmid and Keith W. Earley for pEarley plasmids. We thank Muriel Reissolet for technical assistance and plant care. We also thank Bruno Coutard and Barbara Selisko for helpful discussions. We thank Benjamin Field for helpful discussions and for assisting in the writing of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by a grant from Agence Nationale de la Recherche, ANR-06-BLAN-0256 (to N.F., B.C. and P.C). A.D and M.F were supported by a PhD fellowship from the Ecole Doctorale des Sciences de la Vie et de la Santé, Aix-Marseille Université, Ministère de l’enseignement supérieur et de la recherche. Funding for open access charge: Commissariat à l’Energie Atomique, Centre National de la Recherche Scientifique, Aix-Marseille Université. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends plant sci. 2008;13: 317–328. 10.1016/j.tplants.2008.05.004 [DOI] [PubMed] [Google Scholar]
- 2. Vazquez F, Legrand, Windels D. The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 2010; 6: 337–345. 10.1016/j.tplants.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 3. Bologna G, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis . Annu Rev Plant Biol. 2014; 65: 473–503. 10.1146/annurev-arplant-050213-035728 [DOI] [PubMed] [Google Scholar]
- 4. Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006; 11: 142–151. [DOI] [PubMed] [Google Scholar]
- 5. Willmann MR, Endres MW, Cook RT, Gregory BD. The functions of RNA-dependent RNA polymerases in Arabidopsis . Arabidopsis Book. 2011; 9: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simon SA, Meyers BC. Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol.2011; 14: 148–155. 10.1016/j.pbi.2010.11.007 [DOI] [PubMed] [Google Scholar]
- 7. Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lelli AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS biology. 2004; 5: e104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006; 16: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007; 5: e5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One. 2009; 4: e4971 10.1371/journal.pone.0004971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000; 101: 533–542. [DOI] [PubMed] [Google Scholar]
- 12. Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004; 16: 69–79. [DOI] [PubMed] [Google Scholar]
- 13. Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes Dev. 2004; 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005; 121: 207–221. [DOI] [PubMed] [Google Scholar]
- 15. Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes Dev. 2005; 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell. 2005; 123: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis . Nat struct mol biol. 2010; 17: 997–1004. 10.1038/nsmb.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010; 107: 15269–15274. 10.1073/pnas.1001738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJ, et al. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 2008; 28:48–57. 10.1038/emboj.2008.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moissard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007; 13: 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaistij FE, Jones L, Baulcombe D. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant cell. 2002; 14: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwab R, Voinnet O. RNA silencing amplification in plants: size matters. Proc Natl Acad Sci USA. 2010; 107: 14945–14946. 10.1073/pnas.1009416107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA. 2012; 109: 2461–2466. 10.1073/pnas.1200169109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajeswaran R, Pooggin MM. RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 2012; 40: 594–599. 10.1093/nar/gkr760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajeswaran R, Aregger M, Zvereva AS, Borah BK, Gubaeva EG, Pooggin MM. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 2012; 40: 6241–6254. 10.1093/nar/gks242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schiebel W, Haas B, Marinković S, Klanner A, Sänger HL. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J Biol Chem. 1993; 268: 11858–11867. [PubMed] [Google Scholar]
- 27. Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003; 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schiebel W, Pélissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, et al. Isolation of a RNA-directed RNA polymerase specific cDNA clone from tomato leaf-tissue mRNA. Plant Cell. 1998; 10: 2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Curaba J, Chen X. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem. 2008; 283: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fukunaga R, Doudna JA. dsRNA with 5’ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009; 28: 545–555. 10.1038/emboj.2009.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, et al. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell. 2012; 48: 811–818. 10.1016/j.molcel.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998; 82: 259–266. [DOI] [PubMed] [Google Scholar]
- 33. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998; 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 34. Sainsbury F, Thuenemann EC, Lomonossoff GP. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009; 7: 682–693. 10.1111/j.1467-7652.2009.00434.x [DOI] [PubMed] [Google Scholar]
- 35. Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, et al. Gateway-compatible vectors for plant functional genomics and proteomic. Plant J. 2006; 45: 616–629. [DOI] [PubMed] [Google Scholar]
- 36. Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003; 33: 949–956. [DOI] [PubMed] [Google Scholar]
- 37. Makeyev EV, Bamford DH. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell. 2002; 10: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 38. Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005; 102: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laurila MR, Salgado PS, Stuart DI, Grimes JM, Bamford DH. Back-priming mode of phi6 RNA-dependent RNA polymerase. J Gen Virol. 2005; 86: 521–526. [DOI] [PubMed] [Google Scholar]
- 40. Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Letters. 2009; 583: 1261–1266. 10.1016/j.febslet.2009.03.055 [DOI] [PubMed] [Google Scholar]
- 41. Ferrari E, He Z, Palermo RE, Huang HC. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J Biol Chem. 2008; 283: 33893–33901. 10.1074/jbc.M803094200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luo Z, Chen Z. Improperly Terminated, Unpolyadenylated mRNA of Sense Transgenes Is Targeted by RDR6-Mediated RNA Silencing in Arabidopsis . Plant Cell. 2007; 19: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herr AJ, Molnàr A, Jones A, Baulcombe DC. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis . Proc Natl Acad Sci USA. 2006; 103: 14994–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009; 21: 367–376. 10.1016/j.ceb.2009.01.025 [DOI] [PubMed] [Google Scholar]
- 45. Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis . PLoS Genet. 2010; 6: e1001141 10.1371/journal.pgen.1001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007; 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana . Proc Natl Acad Sci USA. 2010; 107: 484–489. 10.1073/pnas.0904086107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng Z, Xing Y, He XJ, Li W, Hu Y, Yadav SK, et al. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis . Plant J. 2010; 62: 92–99. 10.1111/j.1365-313X.2010.04130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purified RDR2-HA and RDR6-HA were incubated with cold NTPs and 5'[32P]-labeled 37-nt RNA template (RNA37A☢). In these conditions, detection of labeled products higher than the template size (>37nt, higher products) correspond only to elongated template. (*) A single star indicates the position of the RNA37A☢ template.
(TIF)
RNA38 is designed to adopt a stable hairpin-like conformation in the 3' terminus region. Purified RDR2-HA and RDR6-HA were incubated with 5'[32P]-labeled 38-nt ssRNA template (RNA38☢), cold NTPs and trace amounts of [α32-GTP]. (*) A single star indicates the position of the RNA38☢ template.
(TIF)
RNA37A template was hybridized to either 5‘[32P]-labeled RNA of 21- (RNA21☢), 22- (RNA22☢) or 24-nt (RNA24☢) sizes oligonucleotides.
(TIF)
Purified RDR2-HA and RDR6-HA were incubated with partial dsRNAs formed with RNA37A hybridized with 5’[32P]-labeled 22-nt ssRNA (RNA22☢). 5'[32P]-labeled RNA37A (RNA37A☢) and 5'[32P]-labeled RNA22 (RNA22☢) are used as a size markers. (*) Simple star indicate the position of the RNA22☢. (**) Double star indicates the position of the full-length elongated RNA template.
(TIF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.