Abstract
Magnaporthe oryzae is a hemibiotrophic fungal pathogen that causes rice blast disease. A compatible interaction requires overcoming plant defense responses to initiate colonization during the early infection process. Nitric oxide (NO) plays important roles in defense responses during host-pathogen interactions. Microbes generally protect themselves against NO-induced damage by using enzymes. Here, we characterized an S-(hydroxymethyl)- glutathione dehydrogenase gene in M. oryzae, MoSFA1, the homologs of which are involved in NO metabolism by specifically catalyzing the reduction of S-nitrosoglutathione (GSNO) in yeasts and plants. As expected from the activities of S-(hydroxymethyl)glutathione dehydrogenase in formaldehyde detoxification and GSNO reduction, MoSFA1 deletion mutants were lethal in formaldehyde containing medium, sensitive to exogenous NO and exhibited a higher level of S-nitrosothiols (SNOs) than that of the wild type. Notably, the mutants showed severe reduction of conidiation and appressoria turgor pressure, as well as significantly attenuated the virulence on rice cultivar CO-39. However, the virulence of MoSFA1 deletion mutants on wounded rice leaf was not affected. An infection assay on barley leaf further revealed that MoSFA1 deletion mutants exhibited a lower infection rate, and growth of infectious hyphae of the mutants was retarded not only in primary infected cells but also in expansion from cell to cell. Furthermore, barley leaf cell infected by MoSFA1 deletion mutants exhibited a stronger accumulation of H2O2 at 24 and 36 hpi. MoSFA1 deletion mutants displayed hypersensitivity to different oxidants, reduced activities of superoxide dismutases and peroxidases, and lower glutathione content in cells, compared with the wild type. These results imply that MoSFA1-mediated NO metabolism is important in redox homeostasis in response to development and host infection of M. oryzae. Taken together, this work identifies that MoSFA1 is required for conidiation and contributes to virulence in the penetration and biotrophic phases in M. oryzae.
Introduction
Magnaporthe oryzae is a hemibiotrophic fungal pathogen that causes rice (Oryza sativa) blast disease. It colonizes plants asymptomatically as a biotroph in susceptible plants before entering its destructive necrotrophic phase [1]. Nitric oxide (NO) produced by the host plays important roles during the plant defense responses [2–4]. Rapid accumulation of NO is induced by recognition of effectors or pathogen-associated molecular patterns (PAMPs), which consequently triggers plant defense response by means of hypersensitive response (HR), phytoalexin biosynthesis and defense gene activation [2, 5–7]. Moreover, NO also has directly antimicrobial activity via cellular damage [8–11]. The excessive NO administration to fungal cells can inhibit the antioxidant enzymes activities, reduce ATP synthesis and increase carbonylation damage [9, 12–13]. The burst of NO in the infected tissues creates a potent antimicrobial environment that conduces to the restriction of the pathogen growth, similar with the burst of reactive oxygen species (ROS). A compatible interaction between plant and pathogen requires overcoming plant defense responses to initiate colonization during the early infection process. However, the means by which pathogens deal with NO is not well understood.
Microbes generally protect themselves against NO-induced damage by using enzymes that convert NO to less toxic molecules. Evidences elaborated that an S-(hydroxymethyl)glutathione dehydrogenase (EC 1.1.1.284), belongs to a class III alcohol dehydrogenase and functions in the glutathione-dependent oxidation of formaldehyde, is involved in NO metabolism though specifically catalyzing the reduction of S-nitrosoglutathione (GSNO) [14], which is the reaction product of glutathione (GSH) and NO, and serves as a naturally occurring mobile reservoir of NO bioactivity [15]. This enzyme is conserved from bacteria to humans, which reduces GSNO to ammonia (NH3) and glutathione disulphide (GSSG) in the presence of NADH and GSH. Deletion of its homologs in several species results in increased hypersensitivity to NO and increased accumulation of cellular S-nitrosothiols (SNOs), subsequently causes nitrosative stress [16–17]. Notable, it is well documented in several human pathogens that this enzyme is required to counteract pathogen-induced NO activity and promote virulence. Expression of adhC gene in bacterial pathogen Streptococcus pneumoniae was strongly induced by GSNO, and adhC deletion mutants exhibited hypersusceptibility to NO stress and decreased the fitness in blood [18]. In Cryptococcus neoformans, although GNO1 deletion mutants proliferated normally in vitro with nitrosative challenge and did not reduce the virulence in vivo, GNO1 can partly promote survival of flavohemoglobin-null mutants in the infected host, which contribute to NO-consuming activity in C. neoformans [19]. Thus, GNO1-mediated GSNO reducing pathway also contribute to virulence of cryptococcal disease. As mentioned above, although NO burst in plants plays crucial roles in defense response, little is known about the roles of S-(hydroxymethyl)glutathione dehydrogenase in plant pathogenic fungi.
In present study, we investigated the function of a S-(hydroxymethyl)- glutathione dehydrogenase gene in M. oryzae, namely MoSFA1, a homolog of Saccharomyces cerevisiae SFA1. Our results showed that MoSFA1 is essential for normal vegetative growth, conidiation and full virulence in M. oryzae. This information provides initial insights into the physiological and biological functions of MoSFA1-associated NO metabolism in the rice blast fungus.
Material and Methods
Fungal strains and growth conditions
The wild type M. oryzae strain Guy11 and its derivative strains were grown on complete medium (CM) plates [20] at 28°C. Conidia were harvested from 9-day-old cultures grown on CM plates. Genomic DNA and total RNA were extracted from 3-day-old cultures grown in liquid CM. Fungal transformants were screened in CM plates supplemented with 200 μg ml−1 of hygromycin B (Roche Applied Science, Mannheim, Germany) or 250 μg ml−1 of glufosinate ammonium (Sigma-Aldrich, St Louis, MO, USA), depending on the selection marker. Evaluations of conidiation and vegetative growth were performed as previously described [21]. And growth rate of M. oryzae was determined by measuring the colony diameter of 6-day-old cultures on CM plates supplemented with chemicals.
Complementation of Saccharomyces cerevisiae Δsfa1 mutant
The total RNA from M. oryzae was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and DNA traces were cleaned up with Turbo DNAfree kit (Ambion, Austin, TX, USA). The CDS of MoSFA1 was produced by RT-PCR using a gene-specific primer set MG125/MG126, which was incorporated in the HindIII and XbaI restriction endonuclease sites, respectively. The PCR product was cloned into the pEASY-T3 vector (Transgen Biotech, Beijing, China) and verified by DNA sequencing. The correct plasmid was digested by HindIII and XbaI, and the fragment was ligated into the pYES2 yeast expression vector (Invitrogen, Carlsbad, CA, USA) to generate pYES2-MoSFA1. After sequencing verification, the pYES2-MoSFA1 plasmid was introduced into the S. cerevisiae Δsfa1 mutant strain YDL168W (BY4741:Mata/his3-1/leu2-0/met15-0/ura3-0/YDL168W::kanMX4) purchased from Thermo Fisher Scientific (Waltham, MA, USA) using the lithium acetate method. For formaldehyde sensitivity assay, yeast cells were incubated in liquid YPD medium (2% glucose, 2% peptone, and 1% yeast extract) with shaking at 28°C for overnight. Cells were collected and washed three times with sterile distilled water. Cell suspensions adjusted to OD600 = 1 were 10-fold serially diluted, and aliquots (5 μl) of 10-fold serial dilution were grown in SC medium (0.67% nitrogen base, 2% raffinose, 2% galactose, 2% agar,) containing 1.1 mM formaldehyde at 28°C for 2 days. Primers used in this study were listed in S1 Table.
GSNO reductase activity assay
GSNO reductase activity was measured spectrophotometrically at 340 nm by the time-dependent oxidation of NADH and reduction of GSNO, as described by Sakamoto et al. [22]. Yeast cells were harvested from the cultures grown in liquid YPD medium to OD600 0.5–1.0 and washed three times with sterile distilled water. The cells were resuspended in 150 μl extract buffer [50 mM Tris-HCl, pH 8.0, and 0.1% (v/v) Tween 20], and mechanically broken with acid-washed glass beads in Fastprep-24 Instrument (MP Biomedicals, Solon, OH, USA). Then samples were centrifuged at 4°C for 10 min at 12,000 x g to remove the beads and insoluble material. The concentration of protein extracts was determined by a Bradford assay, with bovine serum albumin (BSA) as the standard. About 10 μg proteins were incubated in 100 μl assay buffer that contains 20 mM Tris-HCl (pH 8.0), 0.2 mM NADH and 0.5 mM EDTA. The reaction was started through adding GSNO (Cayman Chemical, Ann Arbor, MI, USA) at a final concentration of 400 μM. The absorbance was measured at 340 nm at room temperature using a Thermo Varioskan Flash spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The resultant GSNO reductase activity was expressed as nmol NADH consumed min−1 (mg protein)−1.
Targeted gene replacement of MoSFA1 and complementation
In order to replace MoSFA1 gene, a 1.6-kb upstream flanking sequence fragment and a 1.5-kb downstream flanking sequence were amplified from the genomic DNA of the wild type strain Guy11, with the primer sets MG121/MG122 and MG123/MG124, in which BstXI/EcoRI and XbaI/SalI restriction endonuclease sites were incorporated, respectively. The PCR products were cloned into pEASY-T3 and verified by DNA sequencing. After digesting the correct plasmids with BstXI/EcoRI and XbaI/SalI restriction endonuclease combinations, the upstream and downstream fragments were isolated and orderly inserted in the corresponding sites of p1300-KO [23] to generate the MoSFA1 gene replacement vector pKO-MoSFA1. Agrobacterium tumefaciens strain AGL1 was used to transform M. oryzae as described by Wang et al. [21].
Candidate mutants were screened by PCR with the primer sets and gene deletion mutants were confirmed by Southern blotting. The genomic DNA of candidate mutants and Guy11 were digested by BstXI and blotted on Nylon+ membrane (Roche Applied Science, Mannheim, Germany) and hybridized with a 1.5-kb fragment as the probe, which was amplified from the 3’-end of the MoSFA1 gene with the primer set MG129/MG130 and labeled with the DIG High Prime DNA labeling and detection kit II according to the manufacturer's instructions (Roche Applied Science, Mannheim, Germany).
For complementation of the MoSFA1 deletion mutant, the 3.4-kb fragment containing about 1.8-kb upstream sequence from the start codon ATG, the MoSFA1 gene coding region, and 0.4-kb downstream sequence from the stop codon TAA was amplified with the primers set MG127/MG128, in which EcoRI/XbaI restriction sites were incorporated. This PCR product was cloned into pEASY-T3 and verified by DNA sequencing. The correct plasmid was digested by EcoRI and XbaI, and the fragment was cloned into EcoRI/XbaI-digested p1300BAR, which carried a glufosinate ammonium resistance marker [23]. The resultant plasmid pBAR-MoSFA1R was verified by sequencing and used to transform MoSFA1 deletion mutant by A. tumefaciens-mediated transformation, and fungal transformants were screened on the CM plates supplemented with 250 μg ml-1 glufosinate ammonium.
Pathogenicity assays
For pathogenicity assays, 4-week-old seedlings of rice (Oryzae sativa cv. CO-39) were infected with conidia suspensions prepared in 0.25% gelatin at a concentration of 5×104 conidia ml−1 using an artist's airbrush with high-pressure air. Inoculated plants were placed in a moist chamber at 25°C for 24 hours in the dark. One day after inoculation, rice seedlings were maintained in the moist chamber with a photoperiod of 12 hours under fluorescent lights for additional 5 days to allow the full development of disease symptoms. Disease severity was scaled as described by Fang and Dean [24] to evaluate the virulence of all tested strains. For wounded inoculation, 20-μl aliquots of serial dilutions of conidia suspension were dropped on rice leaf segments wounded with a pin. The experiments were repeated at least three times with triple replications that yielded similar results.
Analysis of infection-related morphogenesis
Conidia harvested from 9-day-old CM plates were suspended in sterile water at 5×104 conidia ml−1. Aliquots (20 μl) of the suspensions were incubated on a plastic coverslip (Thermo Fisher Scientific, Waltham, MA, USA) at 28°C. Conidial germination and appressorium formation were examined at 48 hours post-incubation (hpi). Appressorium turgor pressure was determined using a cytorrhysis assay [25]. Appressoria were allowed to form on a plastic coverslip at 48 hpi, water was then carefully replaced with 20 μl 1–3 M glycerol solution. After 10 min at room temperature, the number of collapsed appressoria was counted under a light microscope. The experiments were replicated three times, and >200 appressoria were observed for each strain. Penetration and infectious hypha growth were assayed in the barley (Hordeum vulgare cv. Golden promise) leaf as described previously [26]. Infected leaves were observed under a light microscope at 36 and 48 hours after inoculation. The experiments were replicated three times, and >50 appressoria were observed for each tested strain.
Detection H2O2 in infected host cells
To observe the accumulation of H2O2 at infected cells, the detached barley leaves were inoculated with conidia suspension at a concentration of 5×104 conidia ml−1 using an artist's airbrush with high-pressure air. The infected barley leaves were stained with 3, 3’-diaminobenzidine (DAB, Sigma-Aldrich, St Louis, MO, USA) as described previously by Daudi et al. [27]. Barley leaf segments were incubated in 1 mg/ml DAB solution in the dark at room temperature for 8 hours and decolorized in boiling ethanol (96%) for 10 min. Samples were observed under a light microscope.
Quantitative RT-PCR
Total RNA (500 ng) was used for reverse transcription and the products were used for the relative gene expression analysis. Quantitative real-time PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Premix Ex Taq kits (Takara, Dalian, China). To compare relative abundance of MoSFA1 transcripts, average threshold cycle (Ct) was normalized to that of β-tubulin gene MGG_00604 as described by Livak & Schmittgen [28]. For the gene MoSFA1, primers GNO1-F3 and GNO1-R3, which amplify a 150-bp fragment from the MoSFA1 coding region, were used. For the gene MGG_00604, the designed primers were Tub-F1 and Tub-R1, which specifically amplified a 156-bp DNA fragment from the β-tubulin coding region. All qPCR experiments were performed with three biological replicates.
Determination of SNOs content
Total SNOs content in M. oryzae was determined using the Saville-Griess assay [29]. All tested strains were cultured in liquid CM for 3 days in the dark. Sodium nitroprusside (SNP), a NO donor, was added to cultures of each strain at a final concentration of 100 μM for an additional day at 2 days after incubation. Cultures were harvested and washed three times with sterile distilled water and ground in liquid nitrogen. The cell extracts were prepared in 50 mM Tris-HCl (pH 8.0) in the dark and incubated for 10 min in solution A (1% sulfanilamide in 0.5 M HCl) or solution B (solution A plus 0.2% HgCl2). The colored azo dye was formed after the incubated solutions reacting with an equal volume of solution C (0.02% (N-(1-naphthyl) ethylenediamine dihydrochloride in 0.5 M HCl) for 10 min. The absorbance was measured at 540 nm in a Thermo Varioskan Flash spectrophotometer. The SNOs content was determined according to the difference in absorbance between the reaction with solution B and that with solution A by comparing with those of a standard curve constructed using GSNO. The concentration of protein extract was determined by a Bradford assay method, with BSA as the standard. The resultant SNOs content was expressed as pmol per mg protein.
Determination of antioxidant enzymes activities and GSH content
To determinate the activities of antioxidant enzymes and GSH content, cultures grown in liquid CM for 3 days were harvested and washed three times with sterile distilled water, then ground in liquid nitrogen. The cell extracts were prepared in 10 mM phosphate buffer (pH 7.4). The activities of superoxide dismutase (SOD, EC 1.15.1.1), catalase (CAT, EC 1.11.1.6), and peroxidase (POD, EC 1.11.1.7) and the content of reduced GSH were detected using commercial assay kits purchased from Nanjing Jiancheng bioengineering institute (Nanjing, China). The concentration of protein extract was determined by a Bradford assay method, with BSA as the standard. The absorbance were read in a Thermo Varioskan Flash spectrophotometer, and enzyme activity and GSH content were calculated according to according to the manufacturer's manual, and expressed as U per mg protein and μmol per mg protein, respectively.
Results
Identification of MoSFA1
The protein sequence of S. cerevisiae SFA1 (P32771) was used to search GFD homologue gene of M. oryzae from Magnaporthe comparative database (http://www.broadinstitute.org/annotation/genome/magnaporthe_comparative/MultiHome.html) with the BlastP program. A predicted gene MGG_06011 exhibited the highest similarity to yeast SFA1. MGG_06011 was annotated as 1333 bp nucleotide length with three exons, and putatively encoded a protein of 381 amino acids. The deduced protein sequence of MGG_06011 showed high similarity to the sequence of S-(hydroxymethyl)glutathione dehydrogenase previously characterized from yeast (67.5% identity with S. cerevisiae SFA1), human (65.5% identity with the Homo sapiens ADH5), and Arabidopsis (61.6% identity with Arabidopsis thaliana ADH2).
According to site-directed mutagenesis studies and crystal structure analysis of H. sapiens ADH5, several highly conserved residues are specifically coordinated to the active site zinc, the binding of substrates and the ligand of binary coenzyme complex, including Cys44, Thr46, Asp55, Glu57, His66, Glu67, Gln111, Arg114, Cys173 and Lys283 [30–34]. Alignment with the sequences from yeast (SFA1, P32771), H. sapiens (ADH5, P11766) and A. thaliana (ADH2, Q96533) revealed that the known active residues were strictly conserved in the protein of MGG_06011, except for Gln111 instead of Gly, but this exception was consistent with yeast and Arabidopsis (S1 Fig.). We therefore, termed MGG_06011 as MoSFA1, a homolog of S. cerevisiae SFA1.
Heterologous expression of MoSFA1
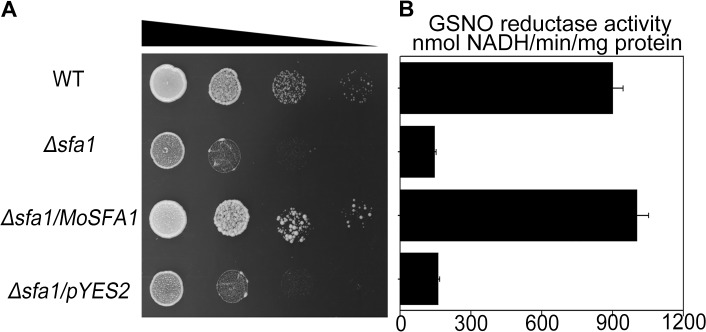
To obtain biochemical evidences of MoSFA1 protein in formaldehyde detoxification and GSNO reductase activity, we have constructed a plasmid pYES2-MoSFA1 containing the CDS of MoSFA1 and consequently transformed the plasmid pYES2-MoSFA1 and empty vector pYES2 into yeast strain Δsfa1, respectively. Cells of yeast strain BY4741 and derivatives strains, Δsfa1, Δsfa1/MoSFA1 with pYES2-MoSFA1, and Δsfa1/pYES2 with pYES2, were serially diluted and plated on SD medium containing 1.1 mM formaldehyde for two days. Consistent with the expected, Δsfa1/pYES2-MoSFA1 regained the resistance to formaldehyde, compared with that of the wild type and of the Δsfa1 strain (Fig. 1A).
Fig 1. Complementation of the yeast Δsfa1 mutant by MoSFA1 and GSNO reductase activity in protein extracts.
(A) Yeast strains, BY4741(WT), Δsfa1, Δsfa1/MoSFA1, and Δsfa1/pYES2 were spotted onto galactose-containing medium with 1.1 mM formaldehyde at 28°C for 2 days. (B) GSNO reductase activity (mean ±SD) was determined in the tested yeast strains. Error bars represent SD.
Simultaneously, protein extracts from yeast were used to determine the GSNO reductase activity of MoSFA1 recombinant protein. Our data showed that GSNO reductase activity of protein extracts from the Δsfa1/MoSFA1 and the wild type were 1002.1 and 899.2 nmol NADH consumed min−1 (mg protein)−1, respectively. However, GSNO reductase activities of protein extracts from both Δsfa1 and Δsfa1/pYES2 were under 160 nmol NADH consumed min−1 (mg protein)−1 (Fig. 1B). Obviously, significant higher GSNO reductase activity of Δsfa1/MoSFA1 strain should be attributed to the MoSFA1 introduction.
Taken together, our data indicated that MoSFA1 gene functionally complemented the defect of Δsfa1 strain, and confirmed that MoSFA1 is the homolog of SFA1 and the encoded protein has formaldehyde detoxification and GSNO reductase activity.
Generation of MoSFA1 deletion mutants
In order to investigate the function of MoSFA1 in M. oryzae, the MoSFA1 gene was disrupted. A MoSFA1 gene replacement vector pKO-MoSFA1 was constructed. Conidia of Guy11 were used to genetic transformation by A. tumefaciens-mediated transformation, and transformants were screened twice in CM plates supplemented with 200 μg ml−1 hygromycin B. Homologous recombination of the MoSFA1 replacement transformants was prescreened by PCR, then Southern blot data showed a single 3.1-kb band for Guy11 and another single 5.8-kb band for candidate transformants (G23 and G24) (S2A-B Fig.). The blotting pattern was consistent with expected gene replacement event at the MoSFA1 gene locus. Thus, the MoSFA1 deletion mutants, G23 and G24, were selected for further functional analysis in this study. We constructed a complementation strain G23R by reintroducing the MoSFA1 genomic DNA sequence, including a 1.8-kb upstream sequence, MoSFA1 gene open reading frame, and 0.4-kb downstream sequence, into the mutant G23. Relative transcript analysis confirmed the gene replacement event in G23 and G24, and that MoSFA1 reintroducing in G23R was functional, with the evidences that expression of MoSFA1 gene in G23R was same as that of Guy11 and MoSFA1 was not transcribed in the gene deletion mutants (S2C Fig.).
MoSFA1 deletion mutants are lethal in formaldehyde-containing medium
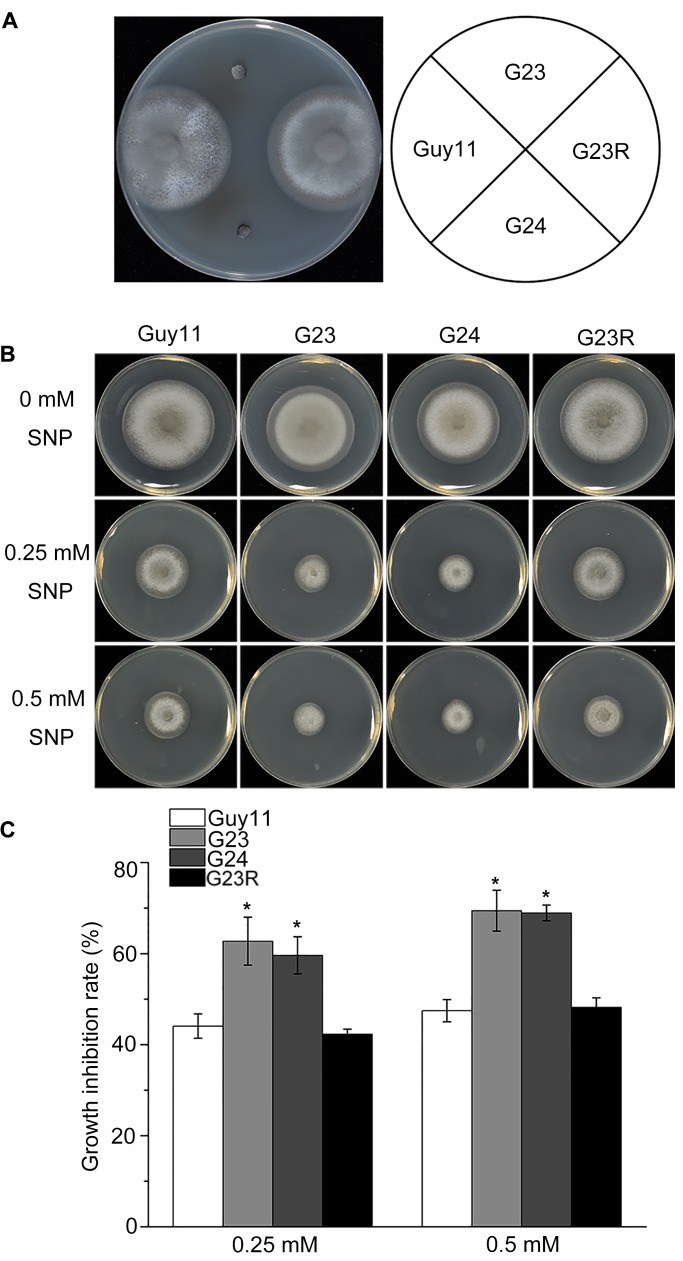
To establish whether MoSFA1 is also involved in formaldehyde metabolism in M. oryzae, formaldehyde sensitivity of MoSFA1 deletion mutants was tested. Growth of MoSFA1 deletion mutants were completely inhibited on CM containing 0.3 mM formaldehyde, compared with that of Guy11 and G23R (Fig. 2A). Loss of MoSFA1 resulted in lethal phenotype in the presence of formaldehyde, indicating that MoSFA1 in M. oryzae is necessary for formaldehyde detoxification.
Fig 2. Effect of formaldehyde and SNP on vegetative growth of the tested M. oryzae strains.
(A) M. oryzae strains were cultured on CM containing 0.3 mM formaldehyde at 28°C for 6 days. (B) Vegetative growth of Guy11, MoSFA1 deletion and reintroduction mutants were grown on CM containing 0.25 or 0.5 mM SNP at 28°C for 6 days. (C) Colony diameters of the tested strains were measured and Growth inhibition rate were evaluated. The experiments were performed in triplicate. ANOVA analysis was performed after growth inhibition rate were arcsine transformed. However, the original percentages were used for presentations. Error bars represent SD of the original percentages. Asterisks in each data column indicate significant differences at p = 0.05.
MoSFA1 deletion mutants shows sensitivity to exogenous NO
The MoSFA1 deletion mutants were biochemically evaluated for the resistance to NO stress in comparison with the wild type. All tested strains were exposed to different concentrations of SNP for 6 days. Growth of all tested strains were reduced in the CM plates with SNP treatment. In particular, MoSFA1 deletion mutants showed more severe growth inhibition (Fig. 2B). The rates of growth inhibition was increased by 17% and 22% in the mutants as compared with Guy11 after exposure to 0.25 and 0.5 mM SNP, respectively (Fig. 2C). Defect of resistance to SNP in MoSFA1 deletion mutants was recovered by reintroducing the gene (Fig. 2B and 2C). These results further confirmed that MoSFA1 is involved in resistance to NO stress.
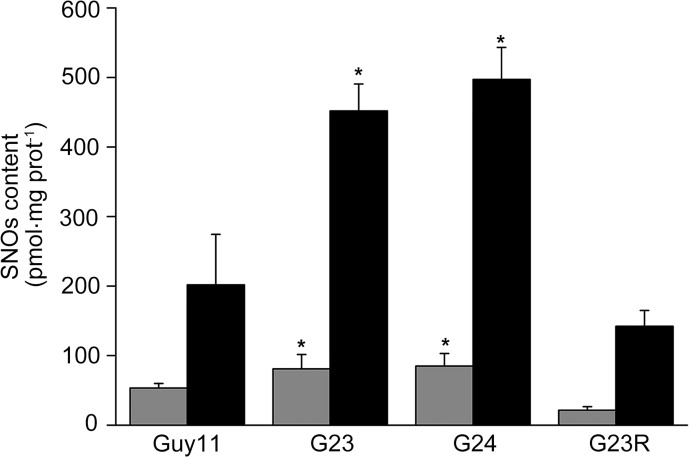
MoSFA1 deletion results in significant increase of SNOs content in cells
In yeast, loss of SFA1 results in increase of cellular SNOs [16, 17]. To compare SNOs content between MoSFA1 deletion mutants and Guy11, protein extracts from the tested strains without or with SNP treatment were used to determine the levels of SNOs. As shown in Fig. 3, the basal levels of SNOs in MoSFA1 deletion mutants reached more than 80 pmol mg−1 protein and were about 1.5 times higher than that of Guy11 [53.4 pmol mg−1 protein] (Fig. 3). With SNP treatment, cellular SNOs contents in all tested strains were significantly increased, against that without SNP treatment. Notably, the SNOs contents in MoSFA1 deletion mutants were at least two fold higher as compared with that of Guy11 [201.8 pmol mg−1 protein]. More interestingly, SNOs content in MoSFA1 reintroduced mutant was reduced regardless of with or without SNP treatment compared with that of gene deletion mutants, and was even lower than that of the wild type. Taken together, loss of MoSFA1 in M. oryzae led to increase of SNOs content, which implied that additional protein S-nitrosylation had occurred to subsequently lead to change in physiological and biological functions in MoSFA1 deletion mutants.
Fig 3. The levels of SNOs in the tested M. oryzae strains.
Total intracellular SNOs content in mycelia cultured in liquid CM in the dark at 28°C for 3 days without (gray bar) or with 100 μM SNP treatment (black bar). The values obtained were compared to a standard curve constructed using GSNO. The results were normalized to the protein content by a Bradford assay. The experiments were performed in triplicate. Error bars represent SD. Asterisks in each data column indicate significant differences at p = 0.05.
MoSFA1 is required for normal vegetative growth and conidiation
The two gene deletion mutants were analyzed in order to detect any possible alteration in normal vegetative growth and conidiation. Our data showed that MoSFA1 deletion mutants grew slowly with whiter colony and looser aerial hyphae, compared with Guy11 (S3A-B Fig.). Moreover, reduction of black pigments in the mutants was more obvious in liquid cultures. Cultures of Guy11 produced more abundant black pigments in the dark after 5 days, compared with the MoSFA1 deletion mutants (S3C Fig.).
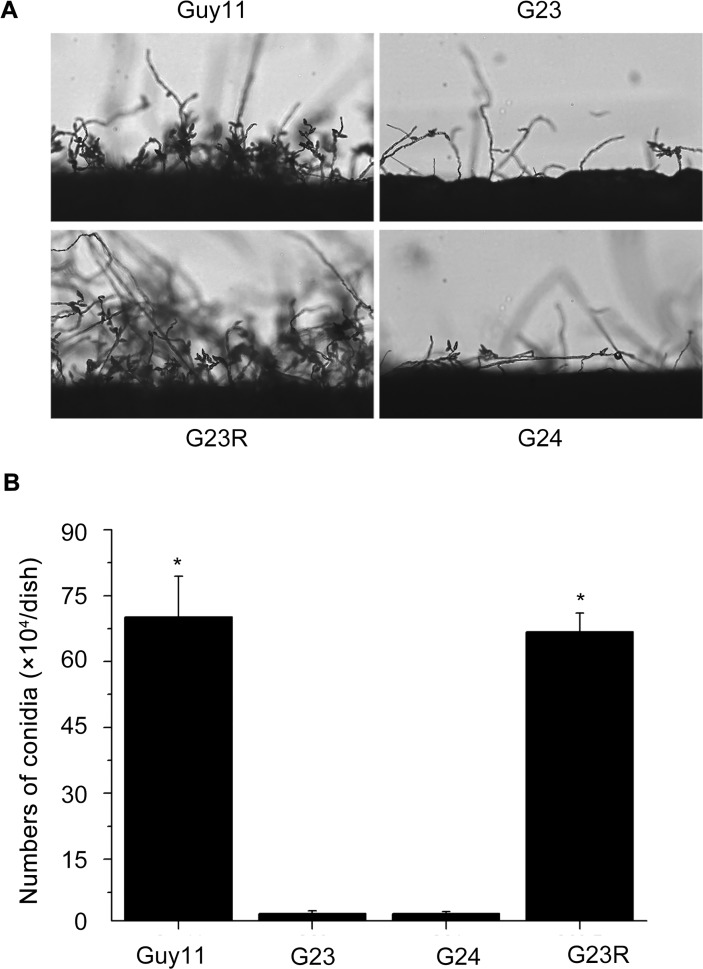
By counting the conidia of the tested strains grown on 9-day-old CM plates with 12 hour light and dark alternation under fluorescent light, conidial production was significantly reduced in the MoSFA1 deletion mutants compared with that of the wild type. As shown in Fig. 4A and 4B, conidia produced in MoSFA1 deletion mutants was only ~2% as compared with the wild type. All defects described above were recovered though reintroducing MoSFA1 gene into the mutant G23. The results suggested that MoSFA1 is involved in normal vegetative growth and conidiation of M. oryzae.
Fig 4. Comparison of the tested M. oryzae strains in conidiation.
(A) Development of conidia on conidiophores on CM. Conidia and conidiophores were observed under light microscopy. (B) Statistical analysis of the number of conidia in each 9-cm-diameter dish. The experiments were performed in triplicate. Error bars represent SD. Asterisks in each data column indicate significant differences at p = 0.05.
MoSFA1 deletion attenuates the virulence of M. oryzae
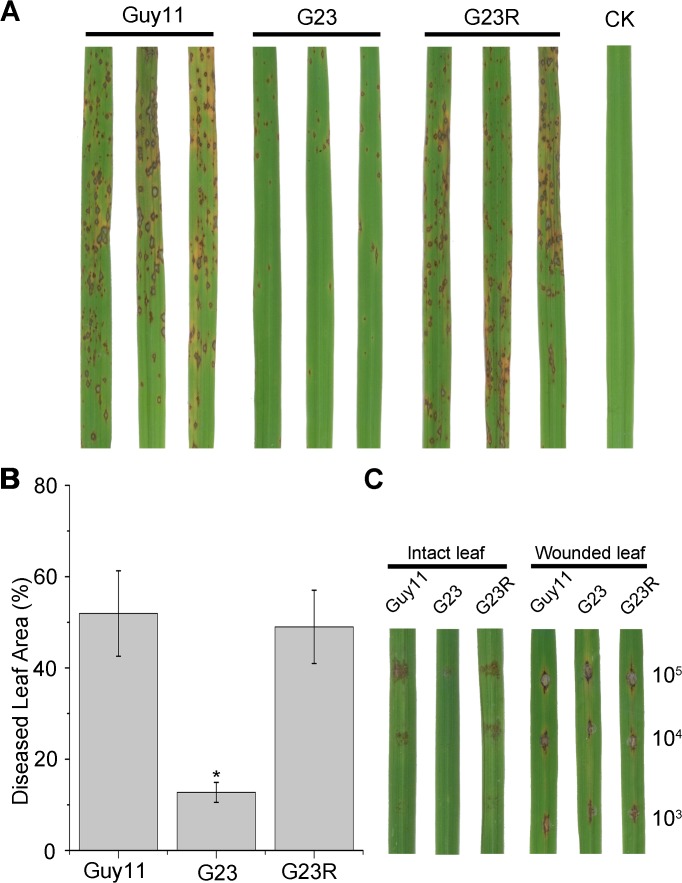
To determine the virulence of the MoSFA1 deletion mutant, 4-week-old susceptible rice seedlings were used for pathogenicity assays. Rice leaves were sprayed with conidial suspensions at the concentration of 5×104 conidia per ml to develop blast lesions. Six days after inoculation, symptoms on rice leaves had fully emerged, and the leaves inoculated with MoSFA1 deletion mutant displayed fewer lesions compared with those inoculated Guy11 (Fig. 5A). The estimated disease lesion area caused by the MoSFA1 mutants occupied about 40% less of the leaf surface than the lesion area caused by the wild-type strain Guy11 (Fig. 5A and 5B). The attenuated virulence of the MoSFA1 mutants could be complemented and fully recovered by reintroducing the MoSFA1 gene. The results suggested that MoSFA1 is required for full virulence of M. oryzae.
Fig 5. Pathogenicity assays on rice leaves.
(A) Spray inoculation with 4-week-old rice seedlings. Rice leaves (O. sativa cv. CO-39) were inoculated with conidia at a concentration of 5×104 conidia ml−1. Representative leaves were photographed 6 days post-inoculation. The experiments were repeated at least three times with triple replications that yielded similar results. (B) Disease severity of each strain was assessed from the percentage of diseased leaf area as described by Fang and Dean (2000). ANOVA analysis was performed after percentages were arcsine transformed. However, the original percentages were used for presentations. Error bars represent SD of the original percentages. Asterisks in each data column indicate significant differences at p = 0.05. (C) Drop inoculation with 20 μl serial dilutions of conidia suspension on intact and wounded rice leaf segments. Representative leaves were photographed 6 days post-inoculation.
In order to determine whether MoSFA1 is involved in the virulence in biotrophic or necrotrophic phase, a wounded inoculation in rice leaf was performed. The results showed no significant difference in virulence among the tested strains (Fig. 5C). Taken together, our finding implies that MoSFA1 significantly contributes to virulence in penetration or biotrophic phases, not in necrotrophic phase.
MoSFA1 deletion results in decreased appressorial turgor pressure and retarded growth of infectious hyphae
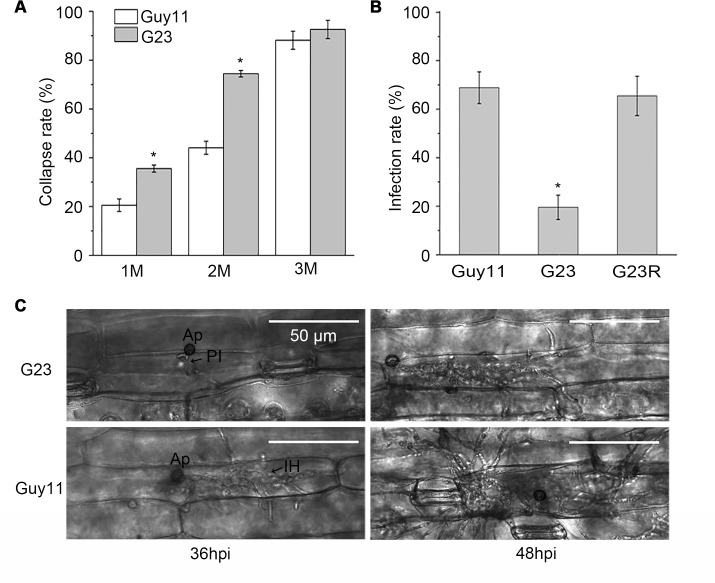
To further explore why MoSFA1 deletion attenuates the virulence of M. oryzae, infection-related morphogenesis of the MoSFA1 deletion mutant was investigated on both artificial hydrophobic interface and barley leaf. Conidia of MoSFA1 deletion mutants normally germinated and differentiated into melanized appressoria, and the rates of conidial germination and appressorial formation were not affected evidently, compared with the wild-type strain (S4 Fig.). However, appressorial collapse rate of the MoSFA1 deletion mutants were significantly higher in 1 M and 2M glycerol solution than of the wild type (Fig. 6A). The fact suggested that loss of MoSFA1 results in reduction of appressorial turgor pressure.
Fig 6. Analysis of infection-related morphogenesis of MoSFA1 deletion mutant.
(A) Cytorrhysis assay using glycerol was performed to compare the appressorial turgor pressure of MoSFA1 mutant and wild-type strain. Different glycerol solutions were given to appressoria at 48 hpi. Appressorial cytorrhysis was counted under an optical microscope. The rate of cytorrhysis was the average of three replications. (B) Infection rate of M. oryzae strains in barley leaf cells at 36 hpi. (C) Growth of infectious hyphae of MoSFA1 deletion mutant was retarded in barley leaf cells. Penetration and infectious hyphae were examined under optical microscopy. Ap, appressorium; PI, primary infectious hyphae; IH, secondary infectious hyphae. Significant difference analysis was performed after percentages were arcsine transformed. The original percentages were used for presentations. Error bars represent SD. Asterisks in each data column indicate significant differences at p = 0.05.
Furthermore, infection rates of MoSFA1 deletion mutants in barley leaf were observed and statistically analyzed. And MoSFA1 deletion mutants significantly caused fewer infection events compared to the wild type. More than 68% of the appressoria of the wild type strain formed primary infectious hyphae at 36 hpi, but the infection rate was only about 20% in MoSFA1 deletion mutant (Fig. 6B).
Meanwhile, we also found that infectious hyphae growth of MoSFA1 deletion mutants were severe delayed in barley cells (Fig. 6C). As shown in Fig. 6C, growth and expansion of infectious hyphae from the point of infection were significantly attenuated at 36 hpi and 48 hpi compared to the wild type strain.
Taken together, our data suggested that MoSFA1 contributes significantly to appressoria maturation and infectious hyphae development in host cells.
Accumulation of H2O2 in cells infected by MoSFA1 deletion mutants
Defense responses of hosts induced by pathogen often associated with the rapid production of ROS, which form a barrier to penetration and inhibit the growth of pathogen directly. Thus, the host ROS burst which responded against the wild type and the mutants were compared. As shown in Fig. 7, the accumulation of H2O2 at infected barley cells by MoSFA1 deletion mutants were stronger when stained with DAB-staining, compared with the infections of Guy11 and G23R (Fig. 7). These data suggest that loss of MoSFA1 gene in M. oryzae resulted in accumulation of H2O2 in infected host cells, which may contribute retarded growth of infectious hyphae in host cells.
Fig 7. Detection of H2O2 accumulation at infected barley cells.
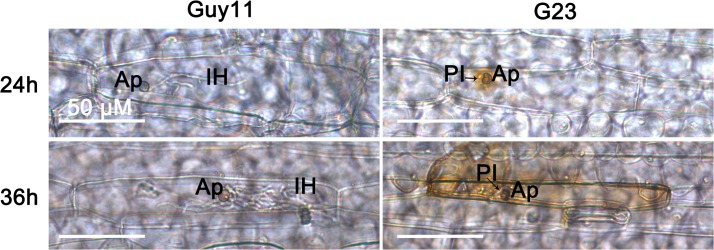
Detached barley leaf were inoculated with the tested strains, stained with DAB at 24 and 36 hpi, and observed under the light microscope. The cells infected by MoSFA1 deletion mutant were strongly stained with DAB, indicating high H2O2 accumulation at the penetration site. Bar is 50 μm. Ap, appressorium; PI, primary infectious hyphae; IH, secondary infectious hyphae.
MoSFA1 deletion mutants shows hypersensitivity to oxidative stress
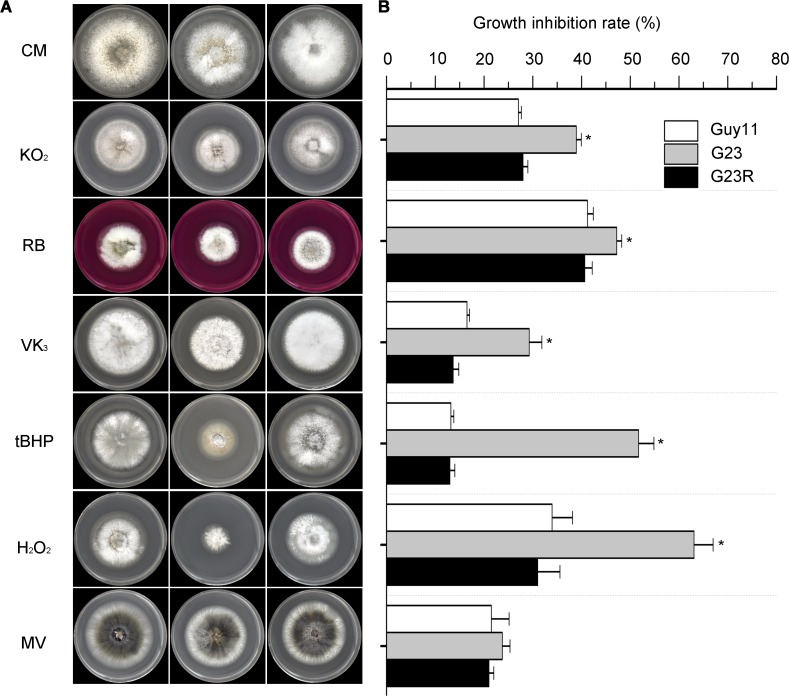
To test a possibility that ROS play a role in the delayed growth of MoSFA1 mutant in host cells, M. oryzae strains were exposed to different oxidants. Growth of MoSFA1 deletion mutants were inhibited on CM containing the tested oxidants, compared with that of Guy11. Growth of MoSFA1 deletion mutant was inhibited by 29.3% to 63.1% in the presence of 100 μM menadione, 50 mM potassium superoxide, 100 μM rose Bengal, 0.001% (v/v) tert-butyl-hydroxyperoxide, and 0.04% (v/v) H2O2, and the growth inhibition rates were significantly more greater than of Guy11 (Fig. 8A and 8B). Methyl viologen (0.25mg/ml) also inhibited growth of the tested strains, but no significant difference in growth inhibition rate was found statistically. The defects to all tested oxidants were recovered by reintroducing MoSFA1 in the mutant G23. Our data suggested that loss of MoSFA1 gene in M. oryzae causes reduced resistance to oxidative stress.
Fig 8. Effect of different oxidants on vegetative growth of the tested M. oryzae strains.
(A) Vegetative growth of Guy11, MoSFA1 deletion and reintroduction mutants were grown on CM containing menadione (VK3, 100 μM), potassium superoxide (KO2, 50 mM), rose bengal (RB, 100 μM), tert-butyl-hydroxyperoxide (tBHP, 0.001%,v/v), and H2O2 (0.04%, v/v) and Methyl viologen (MV, 0.25mg/ml) at 28°C for 6 days. (B) Growth inhibition rate of tested strains exposed to different oxidants. The experiments were performed in triplicate. ANOVA analysis was performed after growth inhibition rate were arcsine transformed. However, the original percentages were used for presentations. Error bars represent SD. Aasterisks in each data column indicate significant differences at p = 0.05.
Loss of MoSFA1 reduces the activities of antioxidant enzymes and GSH content in cells
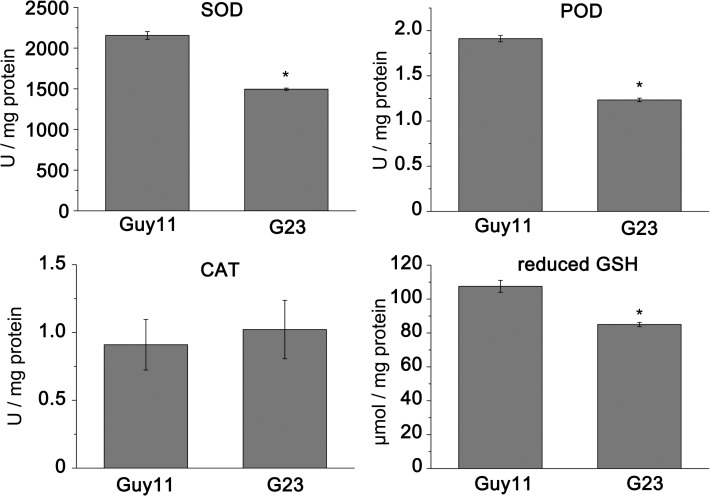
Considering the hypersensitivity of MoSFA1 deletion mutants to oxidants, MoSFA1 were presumed to involve in redox homeostasis in cells. Superoxide dismutases, catalases, and peroxidases provide the cells with highly efficient machinery for detoxifying ROS [35]. GSH acts as a radical scavenger and an electron donor, and the concentration of GSH in cells is important to keep redox homeostasis [36]. Our data showed that the activities of SOD and POD in MoSFA1 deletion mutant had significantly lower levels than those of the wild type, but CAT activity was not different from that of the wild type. In addition, the concentration of reduced GSH in MoSFA1 deletion mutant was significantly diminished in cells (Fig. 9).
Fig 9. The activity of antioxidant enzymes and the content of GSH in tested M. oryzae strains.
Activities of superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) and the content of reduced GSH in the tested strains were determined using assay kits purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The experiments were performed in triplicate. Error bars represent SD. Asterisks in each data column indicate significant differences at p = 0.05.
Discussion
NO produced by the plant plays crucial roles in resistance to pathogen attack, and NO bursts in rice cells has been discovered after treatment with compatible or incompatible blast fungus elicitor, which is required for induction of cell death and defense gene activation [5]. However little is documented about the defense mechanisms that M. oryzae is armed to protect against NO-relative molecules. This work shows that MoSFA1, a homolog of S-(hydroxymethyl)glutathione dehydrogenase gene SFA1 from yeast which specifically catalyze the reduction of GSNO to involve in NO metabolism, is required for conidiation and full virulence.
In present study, MoSFA1 deletion mutants exhibited reduced infection and growth of infectious hyphae (Fig. 6B and 6C). Moreover, the strong accumulation of H2O2 in infected cells were observed by inoculation of MoSFA1 deletion mutant (Fig. 7). However, the induction levels of PR genes in challenged rice tissue by MoSFA1 deletion mutant were not different from those in challenged rice tissue by the wild type (S5 Fig.). It suggests that MoSFA1 in M. oryzae is involved in compromising the host defence-related oxidative burst in compatible interaction. Enzyme activity assay showed that the activities of SOD and POD in MoSFA1 deletion mutant were significantly inhibited, and the concentration of reduced GSH were significantly less than that in wild type (Fig. 9). Thus, the defect in virulence of MoSFA1 deletion mutants is due to the impairment of the ability to maintain redox homeostasis. Firstly, GSH is important for the maintenance of the redox homeostasis [37], and production of GSNO need to consume GSH. Loss of MoSFA1 not only repeals the reduction of GSNO to GSSG but also causes the additional GSH consumption attributing NO administration, which subsequently disturbs with the redox balance of GSH in cells. Recently, GSH-recycling in GSH-dependent antioxidant system of M. oryzae is discovered to be critical in biotrophic colonization of host cells [38]. Loss of GTR1 encoding glutathione reductase significantly retarded growth of infectious hyphae in rice cells and reduced the full virulence. Secondly, GSNO acts as the main NO reservoir in cells and can directly release NO. GSNO accumulation leads to cellular NO stress, and NO can reduce the enzymatic activity to oxidative detoxification [13, 39–42]. Specifically, NO can reversibly inhibit the cytochrome c oxidase enzyme to block respiratory chain and to markedly increase the production rate of ROS [41]. NO administration also reduces the activities of superoxide dismutase and catalase, increases the level of intracellular ROS and secondarily leads to growth retardation in Penicillium expansum [13]. Additionally, NO can rapidly react with oxygen species, such as superoxide [43], to form the peroxynitrite (ONOO-), which is a powerful oxidant and reacts with a wide array of molecules in cells, including DNA, lipids and proteins, leading to cellular damage and cytotoxity [44].
Additionally, GNSO can transfer NO group to other cellular thiols-containing proteins to form S-nitrosothiols, which is one of the most important functional forms of NO-dependent posttranslational modification. Several proteome-wide analyses showed that a large spectrum of proteins are S-nitrosylated, including enzymes, structural proteins, and transcription factors [45–48]. Functional significance of protein S-nitrosylation in plants have been discovered, such as GAPDH (glyceraldehyde-3-phosphate dehydrogenase) [49], NPR1 (nonexpressor of pathogenesis-related gene1), the transcription factor TGA1 (a DNA-binding protein TGA1-like protein) [50, 51], and NADPH oxidase [52], and cytosolic ascorbate peroxidase [41]. In Arabidopsis, S-(hydroxymethyl)glutathione dehydrogenase gene regulates global levels of SNOs, and loss of this gene leads to the increased basal levels of SNOs and causes pleiotropic effects in abiotic or biotic resistances and development, and so on. [53–55]. The facts explain that SNOs are important mediators in the above-mentioned biological process in plants. This explanation also is supported by the findings in S-nitrosylated proteins identified in noe1 rice, which accumulated more SNOs in comparison with wild-type plants and emerges a light-induced leaf cell death. Total 121 nitrosylated proteins were identified from wild-type and noe1 plants, which are involved in different processes, including general metabolism, environmental adaptation, genetic information processing, and redox regulation. Among those, twenty-one and forty-eight proteins were identified only from wild type and noe1 plants, respectively [48]. Similarly, we found that loss of MoSFA1 in M. oryzae leaded to the increased basal level of SNOs, which was ~1.5 times higher than that of the wild type. Moreover, the level of SNOs induced by NO stress was higher in MoSFA1 deletion mutants compared with wild-type strain (Fig. 3). As mentioned above, the difference of S-nitrosylated proteins between MoSFA1 mutants and the wild type strain may result in the changes of gene expression and enzymes activity, which respond to the tested defect of MoSFA1 deletion mutants. It implies that MoSFA1-mediated SNOs homeostasis is important to regulate normal biological function in M. oryzae.
Simultaneously, endogenous NO in fungi is discovered, and found to be involved in conidiation [56, 57], spore germination [58], and formation of infection structure [59]. Recently, Samalova et al. have demonstrated that endogenous NO in M. oryzae is crucial to the development and initiation of infection [60]. According to the findings, endogenous NO was accumulated and consumed during these biological processes. Thus, NO metabolism should be equally important to regulate specific function of NO in fungi.
Collectively, loss of MoSFA1 increases the level of SNOs, which indirectly leads to NO stress. It suggests that MoSFA1-mediated NO metabolism is important in redox homeostasis and protein S-nitrosylation in response to development and host infection of M. oryzae. However, the mechanisms by which proteins is S-nitrosylated and how NO inhibits the antioxidant system in M. oryzae are still needed to understand in details, which may help to reveal the role of NO function in this fungus.
Supporting Information
The amino acid sequence alignment of S-(hydroxymethyl)glutathione dehydrogenase from M. oryzae MoSFA1 (this study), S. cerevisiae (P32771), H. sapiens (P11766), and A. thaliana (Q96533) was performed. Identical amino acids in all sequences are shaded black, conservative replacements are shaded gray. The conserved residues are marked with ▲.
(TIF)
(A) The 2.1-kb fragment including MoSFA1 coding region was replaced with the hph cassette by homologous recombination. A 1.5-kb fragment from the 3’-end of MoSFA1 gene was amplified as the probe for Southern blotting. Scale bar = 1 kb. (B) DNA gel blot analysis of genomic DNA from Guy11 and 2 transformants (G23 and G24) digested with BstXI using the digoxigenin-labeled probes as shown in A. A single 3.1-kb band for Guy11 and another single 5.8-kb band for gene replacement. (C) Transcript levels (mean ±SD) of MoSFA1 in Guy11, MoSFA1 deletion mutants and the reintroduction mutant by quantitative PCR.
(TIF)
(A) Colonies of the strains cultured on CM plates for 9 days at 28°C with 12 h light and dark alternation. (B) Colony diameters of the tested strains were measured and then statistically analyzed. Error bars represent SD. Asterisks in each data column indicate significant differences at p = 0.05. (C) Ten thousand conidia were cultured in liquid CM with shaking at 150 rpm at 28°C for 5 days and then photographed.
(TIF)
The rates of germination and appressoria formation were evaluated at 28°C after 24 hpi, and >200 conidia of each strain were observed for each strain. The experiments were replicated three times. Blank bar for germination rate. Grey bar for appressoria formation rate.
(TIF)
Four-week-old seedlings of rice (O. sativa cv CO-39) were inoculated with suspensions of M. oryzae conidia prepared in 0.25% gelatin at a concentration of 5×104 conidia ml−1 using an artist's airbrush with high-pressure air. Rice leaves were sampled at 24, 48, and 72 hpi. The relative expression levels of PR1a, PR4, PR5, and PR10a in the infected rice was compared using quantitative RT-PCR. Normalization of average threshold cycle (Ct) was performed with that of O. sativa elongation factor 1α gene. Primers were synthesized according to Hao et al. (Plant Physiol Biochem. 2012; 60:150–156).
(TIF)
(PDF)
Acknowledgments
We thank Dr. Bo Dong (Zhejiang Academy of Agricultural Sciences, China), Dr Xiaobin Zhang (Zhejiang Academy of Agricultural Sciences, China) and Dr. Ling Li (Zhejiang Agriculture and Forestry University, China) for their helpfulness.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was supported by the Special Fund for Agro-Scientific Research in the Public Interest (201203014), Zhejiang Academy of Agriculture Science Innovate Found (2013), and State Education Ministry and Key Subject Construction Program of Zhejiang for Modern Agricultural Biotechnology and Crop Disease control (2010DS700124-KF1101; 2010DS700124-KF1203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007; 19: 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mur LA, Carver TL, Prats E. NO way to live; the various roles of nitric oxide in plant-pathogen interactions. J Exp Bot. 2006; 57(3):489–505. [DOI] [PubMed] [Google Scholar]
- 3. Arasimowicz-Jelonek M, Floryszak-Wieczorek J. Nitric oxide: an effective weapon of the plant or the pathogen? Mol Plant Pathol. 2014; 15(4):406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mur LA, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GE, et al. Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants. 2013; 5:pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu X, Neill SJ, Cai W, Tang Z. NO-mediated hypersensitive responses of rice suspension cultures induced by incompatible elicitor. Chinese Sci Bull. 2003; 48(4):358–363. [Google Scholar]
- 6. Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M. Nitric oxide signalling functions in plant-pathogen interactions. Cellul Microbiol. 2004; 6(9):795–803. [DOI] [PubMed] [Google Scholar]
- 7. Hong JK, Yun B, Kang J, Raja MU, Kwon E, Sorhagen K, et al. Nitric oxide function and signalling in plant disease resistance. J Exp Bot. 2008; 59(2):147–154. [DOI] [PubMed] [Google Scholar]
- 8. Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential application of gaseous nitric oxide as a topical antimicrobial agent. Nitric Oxide. 2006; 14:21–29. [DOI] [PubMed] [Google Scholar]
- 9. Lazar EE, Wills RBH, Ho BT, Harris AM, Spohr LJ. Antifungal effect of gaseous nitric oxide on mycelium growth, sporulation and spore germination of the postharvest horticulture pathogens, Aspergillus niger, Monilinia fructicola and Penicillium italicum . Lett Appl Microbiol. 2008; 46: 688–692. 10.1111/j.1472-765X.2008.02373.x [DOI] [PubMed] [Google Scholar]
- 10. Martinez LR, Han G, Chacko M, Mihu MR, Jacobson M, Gialanella P, et al. Antimicrobial and healing efficacy of sustained release nitric oxide nanoparticles against Staphylococcus aureus skin infection. J Invest Dermatol. 2009; 129:2463–2469. 10.1038/jid.2009.95 [DOI] [PubMed] [Google Scholar]
- 11. Macherla C, Sanchez DA, Ahmadi MS, Vellozzi EM, Friedman AJ, Nosanchuk JD, et al. Nitric oxide releasing nanoparticles for treatment of Candida albicans burn infections. Front Microbiol. 2012; 3:193 10.3389/fmicb.2012.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haque MM, Pooja Manzoor N, Khan LA, Basir SF. Effect of sodium nitroprusside on H+-ATPase activity and ATP concentration in Candida albicans . Indian J Exp Biol. 2005; 43:873–879. [PubMed] [Google Scholar]
- 13. Lai T, Li B, Qin G, Tian S. Oxidative damage Involves in the Inhibitory effect of nitric oxide on spore germination of Penicillium expansum . Curr Microbiol. 2011; 62:229–234. 10.1007/s00284-010-9695-1 [DOI] [PubMed] [Google Scholar]
- 14. Thompson CM, Ceder R, Grafström RC. Formaldehyde dehydrogenase: beyond phase I metabolism. Toxicol Lett. 2010; 193:1–3. 10.1016/j.toxlet.2009.11.023 [DOI] [PubMed] [Google Scholar]
- 15. Wang Y, Yun BW, Kwon E, Hong JK, Yoon J, Loake GJ. S-Nitrosylation: an emerging redox-based post-translational modification in plants. J Exp Bot. 2006; 57:1777–1784. [DOI] [PubMed] [Google Scholar]
- 16. Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001; 410:490–494. [DOI] [PubMed] [Google Scholar]
- 17. Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. A genetic analysis of nitrosative stress. Biochemistry. 2009; 48:792–799. 10.1021/bi801813n [DOI] [PubMed] [Google Scholar]
- 18. Stroeher UH, Kidd SP, Stafford SL, Jennings MP, Paton JC, McEwan AG. A pneumococcal MerR-like regulator and s-nitrosoglutathione reductase are required for systemic virulence. J Infect Dis. 2007; 196(12):1820–1826. 10.1086/523107 [DOI] [PubMed] [Google Scholar]
- 19. de Jesús-Berríos M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003; 13:1963–1968. [DOI] [PubMed] [Google Scholar]
- 20. Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell. 1993; 5:1575–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J, Zhang Z, Wang Y, Li L, Chai R, Mao X, et al. PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae . PLoS One. 2013; 8, e55554 10.1371/journal.pone.0055554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakamoto A, Ueda M, Morikawa H. Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 2002; 515:20–24. [DOI] [PubMed] [Google Scholar]
- 23. Li L, Wang J, Zhang Z, Wang Y, Liu M, Jiang H, et al. MoPex19, which Is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus. PLoS ONE. 2014; 9, e85252 10.1371/journal.pone.0085252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fang EG, Dean RA. Site-directed mutagenesis of the magB gene affects growth and development in Magnaporthe grisea . Mol Plant-Microbe Interact. 2000; 13:1214–1227. [DOI] [PubMed] [Google Scholar]
- 25. Dixon KP, Xu JR, Smirnoff N, Talbot NJ. Independent signalling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea . Plant Cell. 1999; 10:2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balhadère PV, Talbot NJ. PDE1 encodes a P-type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea . Plant Cell. 2001; 13(9):1987–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012; 24(1):275–287. 10.1105/tpc.111.093039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2-ΔΔCT method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 29. Schonhoff CM, Benhar M. Analysis of protein S-nitrosylation In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT, editors. Current protocols in protein science, John Wiley and Sons, Inc., New York; 2011. pp. 14.6.1–14.6.21. [DOI] [PubMed] [Google Scholar]
- 30. Engeland K, Hoog JO, Holmquist B, Estonius M, Jornvall H, Vallee BL. Mutation of Arg-115 of human class III alcohol dehydrogenase: a binding site required for formaldehyde dehydrogenase activity and fatty acid activation. Proc Natl Acad Sci. 1993; 90:2491–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang ZN, Bosron WF, Hurley TD. Structure of human χχ alcohol dehydrogenase: a glutathione-dependent formaldehyde dehydrogenase. J Mol Biol. 1997; 265:330–343. [DOI] [PubMed] [Google Scholar]
- 32. Sanghani PC, Bosron WF, Hurley TD. Human glutathione-dependent formaldehyde dehydrogenase structural changes associated with ternary complex formation. Biochemistry. 2002; 41:15189–15194. [DOI] [PubMed] [Google Scholar]
- 33. Sanghani PC, Robinson H, Bennett-Lovsey R, Hurley TD, Bosron WF. Structure-function relationships in human Class III alcohol dehydrogenase (formaldehyde dehydrogenase). Chem Biol Interact. 2003; 143–144:195–200. [DOI] [PubMed] [Google Scholar]
- 34. Sanghani PC, Davis WI, Zhai L, Robinson H. Structure-function relationships in human glutathione-dependent formaldehyde dehydrogenase. role of Glu-67 and Arg-368 in the catalytic mechanism. Biochemistry. 2006; 45:4819–4830. [DOI] [PubMed] [Google Scholar]
- 35. Belozerskaya TA, Gessler NN. Reactive oxygen species and the strategy of antioxidant defense in fungi: a review. Appl Biochem Microbiol. 2007; 43(5):506–515. [PubMed] [Google Scholar]
- 36. Penninckx MJ. An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res. 2002; 2(3):295–305. [DOI] [PubMed] [Google Scholar]
- 37. Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol. 2001; 39:533–541. [DOI] [PubMed] [Google Scholar]
- 38. Fernandez J, Wilson RA. Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS One. 2014; 9(1):e87300 10.1371/journal.pone.0087300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Asahi M, Fujii J, Suzuki K, Seo HG, Kuzuya T, Hori M, et al. Inactivation of glutathione peroxidase by nitric oxide. Implication for cytotoxicity. J Biol Chem. 1995; 270:21035–21039. [DOI] [PubMed] [Google Scholar]
- 40. Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of human manganese superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J Biol Chem. 1998; 273:14085–14089. [DOI] [PubMed] [Google Scholar]
- 41. Carreras MC, Franco MC, Peralta JG, Poderoso JJ. Nitric oxide, complex I, and the modulation of mitochondrial reactive species in biology and disease. Mol Aspects Med. 2004; 25:125–139. [DOI] [PubMed] [Google Scholar]
- 42. de Pinto MC, Locato V, Sgobba A, Romero-Puertas MC, Gadaleta C, Delledonne M, et al. S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol. 2013; 163(4):1766–1775. 10.1104/pp.113.222703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis KL, Martin E, Turko IV, Murad F. Novel effects of nitric oxide. Annu Rev Pharmacol Toxicol. 2001; 41:203–236. [DOI] [PubMed] [Google Scholar]
- 44. Szabo C. Multiple pathways of peroxynitrite cytotoxicity. Toxicol Lett. 2003; 140–141:105–112. [DOI] [PubMed] [Google Scholar]
- 45. Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis . Plant Physiol. 2005; 137(3):921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Missall TA, Pusateri ME, Donlin MJ, Chambers KT, Corbett JA, Lodge JK. Posttranslational, translational, and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: Implications for virulence. Eukaryot Cell. 2006; 5:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maldonado-Alconada AM, Echevarría-Zomeño S, Lindermayr C, Redondo-López I, Durner J, Jorrín-Novo JV. Proteomic analysis of Arabidopsis protein S-nitrosylation in response to inoculation with Pseudomonas syringae . Acta Physiologiae Plantarum. 2011; 33(4):1493–1514. [Google Scholar]
- 48. Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, et al. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012; 158(1):451–464. 10.1104/pp.111.184531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zaffagnini M, Morisse S, Bedhomme M, Marchand CH, Festa M, Rouhier N, et al. Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana . J Biol Chem. 2013; 288(31):22777–22789. 10.1074/jbc.M113.475467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, et al. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008; 321(5891):952–956. 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lindermayr C, Sell S, Müller B, Leister D, Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell. 2010; 22(8):2894–2907. 10.1105/tpc.109.066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011; 478(7368):264–268. 10.1038/nature10427 [DOI] [PubMed] [Google Scholar]
- 53. Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci. 2005; 102:8054–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis . Plant Cell. 2008; 20:786–802. 10.1105/tpc.107.052647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rustérucci C, Espunya MC, Díaz M, Chabannes M, Martínez MC. S-Nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 2007; 143:1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gong X, Fu Y, Jiang D, Li G, Yi X, Peng Y. l-Arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans . Fungal Genet Biol. 2007; 44:1368–1379. [DOI] [PubMed] [Google Scholar]
- 57. Li B, Fu Y, Jiang D, Xie J, Cheng J, Li G, et al. Cyclic GMP as a second messenger in the nitric oxide-mediated conidiation of the mycoparasite Coniothyrium minitans . Appl Environ Microbiol. 2010; 76:2830–2836. 10.1128/AEM.02214-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J, Higgins VJ. Nitric oxide has a regulatory effect in the germination of conidia of Colletotrichum coccodes . Fungal Genet Biol. 2005; 42:284–292. [DOI] [PubMed] [Google Scholar]
- 59. Prats E, Carver TLW, Mur LAJ. Pathogen-derived nitric oxide influences formation of the appressorium infection structure in the phytopathogenic fungus Blumeria graminis . Res Microbiol. 2008; 159:476–480. 10.1016/j.resmic.2008.04.001 [DOI] [PubMed] [Google Scholar]
- 60. Samalova M, Johnson J, Illes M, Kelly S, Fricker M, Gurr S. Nitric oxide generated by the rice blast fungus Magnaporthe oryzae drives plant infection. New Phytol. 2013; 197:207–222. 10.1111/j.1469-8137.2012.04368.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The amino acid sequence alignment of S-(hydroxymethyl)glutathione dehydrogenase from M. oryzae MoSFA1 (this study), S. cerevisiae (P32771), H. sapiens (P11766), and A. thaliana (Q96533) was performed. Identical amino acids in all sequences are shaded black, conservative replacements are shaded gray. The conserved residues are marked with ▲.
(TIF)
(A) The 2.1-kb fragment including MoSFA1 coding region was replaced with the hph cassette by homologous recombination. A 1.5-kb fragment from the 3’-end of MoSFA1 gene was amplified as the probe for Southern blotting. Scale bar = 1 kb. (B) DNA gel blot analysis of genomic DNA from Guy11 and 2 transformants (G23 and G24) digested with BstXI using the digoxigenin-labeled probes as shown in A. A single 3.1-kb band for Guy11 and another single 5.8-kb band for gene replacement. (C) Transcript levels (mean ±SD) of MoSFA1 in Guy11, MoSFA1 deletion mutants and the reintroduction mutant by quantitative PCR.
(TIF)
(A) Colonies of the strains cultured on CM plates for 9 days at 28°C with 12 h light and dark alternation. (B) Colony diameters of the tested strains were measured and then statistically analyzed. Error bars represent SD. Asterisks in each data column indicate significant differences at p = 0.05. (C) Ten thousand conidia were cultured in liquid CM with shaking at 150 rpm at 28°C for 5 days and then photographed.
(TIF)
The rates of germination and appressoria formation were evaluated at 28°C after 24 hpi, and >200 conidia of each strain were observed for each strain. The experiments were replicated three times. Blank bar for germination rate. Grey bar for appressoria formation rate.
(TIF)
Four-week-old seedlings of rice (O. sativa cv CO-39) were inoculated with suspensions of M. oryzae conidia prepared in 0.25% gelatin at a concentration of 5×104 conidia ml−1 using an artist's airbrush with high-pressure air. Rice leaves were sampled at 24, 48, and 72 hpi. The relative expression levels of PR1a, PR4, PR5, and PR10a in the infected rice was compared using quantitative RT-PCR. Normalization of average threshold cycle (Ct) was performed with that of O. sativa elongation factor 1α gene. Primers were synthesized according to Hao et al. (Plant Physiol Biochem. 2012; 60:150–156).
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.