Abstract
Background
Chronic periodontitis (CP) is a common oral disease characterized by inflammation in the supporting tissue of the teeth ‘the periodontium’, periodontal attachment loss, and alveolar bone loss. The disease has a microbial etiology; however, recent findings suggest that the genetic factors, such as vitamin D receptor (VDR) gene polymorphisms, have also been included.
Aim
Investigation of the relationship between VDR gene polymorphisms and CP among Libyans.
Materials and methods
In this study, we examined 196 unrelated Libyans between the ages of 25 and 65 years, including 99 patients and 97 controls. An oral examination based on Ramfjord Index was performed at different dental clinics in Tripoli and information were collected using a self-reported questionnaire. DNA was extracted from buccal swabs; the VDR ApaI, BsmI, and FokI polymorphisms were genotyped using polymerase chain reaction and were sequenced using Sanger Method.
Results
A significant difference in the newly detected ApaI SNP C/T rs#731236 was found (p=0.022), whereas no significant differences were found in ApaI SNP G/T rs#7975232, BsmI SNP A/G rs#1544410, and FokI SNP A/G rs#2228570 between patients and controls (p=0.939, 0.466, 0.239), respectively.
Conclusion
VDR ApaI SNP C/T rs#731236 may be related to the risk of CP in the Libyan population.
Keywords: chronic periodontitis, vitamin D receptor, gene, polymorphisms, variations, SNP
The normal periodontium provides the support necessary to maintain teeth in function. It consists of four distinct components: gingiva, periodontal ligament, cementum, and alveolar bone. Each of these periodontal components is different in location and biochemical composition, but all function together as a single unit (1). Chronic periodontitis (CP) is the most frequent form of periodontal diseases, and it is the principal reason for tooth loss in the elderly (2, 3). It is defined as an oral disease which is characterized by inflammation in the supporting tissue of the teeth ‘the periodontium’, periodontal attachment loss, and alveolar bone loss (4).
Characteristic clinical features in patients with untreated CP may include gingival inflammation, plaque accumulation (often associated with calculus formation), periodontal pocket formation, periodontal attachment loss, alveolar bone loss, occasional suppuration, as well as halitosis and tooth mobility in advanced cases. CP is a slowly progressing disease and usually painless. The primary initiating agent in the etiology of CP is the plaque which accumulates on tooth and gingival surfaces at the dentogingival junction. Attachment and bone loss are associated with an increase in the proportion of gram-negative bacteria in the subgingival plaque. The mechanisms by which this occurs have not been clearly explained, but these bacteria may have a local effect on the cells of the inflammatory response and the tissue of the host, resulting in a local, site-specific disease process. Many factors are involved in the disease which results in disturbance in the balance between microbial plaque and host response (1). Genetic factors may affect the host's response to infection and could lead to noticeable changes in its clinical severity (5).
Vitamin D (1.25-dihydroxyvitamin D3) is a fat-soluble steroid hormone that interacts with its nuclear receptor, vitamin D receptor (VDR), to regulate different biological processes, such as bone metabolism and immune response modulation. In addition, VDR plays a role in the action of vitamin D by regulating its gene expression (6). Several epidemiological studies have shown positive relations between osteoporosis and alveolar bone and tooth loss, which suggests that bad bone quality is a risk factor for CP (7, 8). VDR is a family member of transcriptional regulatory factors and has a sequence similar to the steroid and thyroid hormone receptors (9). VDR is encoded by the VDR gene, which contains nine exons, spans approximately 75 kb, localized to chromosome 12q13.11 and expressed in the intestine, thyroid gland, and kidney (10). The term gene polymorphism is used in genetics to explain the multiple forms of a single gene. At the nucleotide level, the gene encoding a specific protein can have a number of differences in sequence. These polymorphisms do not change the entire product significantly enough to result in a new protein, but may play a role in substrate specificity and particular activity, binding effectiveness, or other functions which might have effects on bone diseases including CP (11, 12). Many polymorphisms have been identified in the VDR gene, and most of them are recognized by biallelic variation in restriction enzyme sites. ApaI (13), BsmI (14), and FokI (15, 16) are examples of single nucleotide polymorphisms (SNPs) in the VDR gene.
Results of previous studies relating to associations of ApaI, BsmI, and FokI VDR gene polymorphisms with CP in various population groups are inconsistent (17–19) . Factors including wide separated areas and genetic effect may be different in variant ethnic groups (20–22) . Therefore, and due to the wide spread of CP, we investigated the relationship between the risk of the disease and ApaI, BsmI, and FokI VDR gene polymorphisms in the Libyan population because this relationship was unknown in Libya.
Materials and methods
Participants
In total, 196 male and female Libyan citizens from many governmental and private dental clinics in Tripoli participated in the study. The participants were examined between January and March 2012, and all of them gave informed consent for participation.
The study comprised patients and control participants between 25 and 65 years of age who were divided into: group one (25–35 years), group two (36–45 year), group three (46–55 years), and group four (56–65 years). All information were obtained using a self-reported questionnaire.
A dental examination was conducted for each participant. The diagnosis of periodontal disease was established according to the clinical parameters of the Periodontal Disease Index (PDI), also known as Ramfjord Index. For the PDI assessment, six teeth were evaluated: the upper left central, first premolar and right first molar; and the lower right central, first premolar, and left first molar were measured. Examination method was applied by using University of Michigan ‘0’ probe with William's markings at 1, 2, 3, 5, 7, 8, 9, and 10 mm. Each sextant was designed as either healthy (score 0); mild to moderate gingivitis, but changes not extending all around the tooth (score 1); mild to moderate gingivitis extending all around the tooth (score 2); severe gingivitis (marked redness, bleeding tendency, and ulceration) (score 3); a probing depth less than 3 mm (score 4); a pocket depth from 3 to 6 mm (score 5); or a pocket depth exceeding 6 mm (score 6), according to the scores recorded at the index teeth. The mean score was recorded as the PDI score of the participant. CP was defined as oral health status with a PDI score of 5 and 6. Oral health status with PDI scores between 0 and 4 was classified as periodontitis free or controls (23). A buccal swab sample for each subject was obtained after the examination.
Genotype determination
Genomic DNA was extracted from buccal swabs using the protocol of Isohelix Buccal DNA Isolation Kit, which contained solution LS (lysis buffer), solution PK (proteinase K) 1 ml, solution CT (capture buffer), and solution TE (Tris-EDTA), (rehydration buffer) (24). Agarose gel electrophoresis was applied to examine the integrity of the genomic DNA (25). The VDR polymorphisms were genotyped using polymerase chain reaction (PCR). Main characteristics of VDR gene SNPs and primers used are listed in Table 1. About 0.75 µM of each forward and reverse primers were added to a reaction mixture containing 1X (1.5 mM MgCl2)2 GoTaq® Reaction Buffer, PCR nucleotide mix 0.2 mM each dNTP, 1.25 µl of GoTaq® DNA Polymerase (5 U/µl), and nuclease-free water to final volume 50 µl. Template DNA 2 µl in a total volume of 50 µl was added. The PCR program was performed for 35 cycles and consisted of an initial denaturation for 2 min at 95°C, 35 sec at 95°C for denaturation, 52°C for 35 sec for annealing, and 72°C for 35 sec for extension, followed by a final extension for 7 min at 72°C (26). Agarose gel electrophoresis was applied to the PCR products (25), then PCR products were purified using the protocol of the QIAquick Purification Kit™ to remove excess salts, PCR primers, and dNTPs (27).
Table 1.
Main characteristics of VDR gene polymorphisms and primers’ sequencesa
| SNP name | dbSNP rs# | SNP type | SNP site | Primers’ sequences |
|---|---|---|---|---|
| ApaI | 7975232 | G/T | Intron 8 | Forward 5′CGGTCAGCAGTCATAGAGG3′ |
| 731236 | C/T | Reverse 5′CAGTGTGTTGGACAGGCG3′ | ||
| BsmI | 1544410 | A/G | Intron 8 | Forward 5′GTGTGCAGGCGATTCGTA3′ |
| Reverse 5′TACCCTGCCCGCAGAAA3′ | ||||
| FokI | 2228570 | A/G | Exon 2 | Forward 5′CGTTCCGGTCAAAGTCTCC3′ |
| Reverse 5′TTGCTGAGCTCCCTGGTG3′ |
From Naito et al. (17).
After purification of the PCR products, DNA cycle sequencing was performed using of BigDye® Terminator v1.1 Cycle Sequencing Protocol; 0.75 µM of forward ApaI and BsmI, and 0.75 µM of reverse FokI primers were prepared. In the reaction mixture, 6.6 µl of nuclease-free water was placed, followed by 4 µl of ready reaction premix, 2 µl of BigDye Terminator v1.1, sequencing buffer (5X) and 6.4 µl of ApaI, BsmI, and FokI. The PCR program was performed for 25 cycles and consisted of an initial denaturation for 1 min at 96°C, 10 sec at 96°C for denaturation, 50°C for 5 sec for annealing, and 60°C for 4 min for extension. Extension products purification using BigDye® XTerminator™ Purification Method was performed. Sanger sequencing method was applied for capillary electrophoresis; the run was performed on Applied Biosystem (3500xL Genetic Analyzer). Data collection software was applied (28).
Data analyses output were imported to Sequencer Analysis Software version 5.1. Chromatograms were viewed. SNPs in comparison with the reference sequence were examined manually and recorded.
Statistical analysis
All statistical analyses were performed using the SPSS for Windows, version 15.0 (SPSS Inc., Chicago, IL). Frequency differences of ApaI, BsmI, and FokI polymorphisms and departure from Hardy–Weinberg equilibrium were tested by Pearson's chi-square test which was also used to quantify the association between CP and these polymorphisms. The level of significance (p value) less than 0.05 was considered statistically significant for all analyses.
Results
The study included 112 females (57.1%) and 84 males (42.9%). The distribution of the participants according to age groups between group one (25–35 years) and group four (56–65 years) was 15.5, 23.7, 30.9, and 29.9%, respectively. According to PDI, 99 participants (50.0%) were diagnosed with CP and 97 participants (49.5%) were diagnosed as healthy controls. The distribution of the PDI scores between 0 and 6 was 1.0, 2.6, 4.6, 11.2, 26.5, 32.7, and 21.4%, respectively. Cross-tabulation statistical test, which is based on Pearson's chi-square test, was used to analyze the frequency distribution, in order to find relationship between CP and both sex and age, as well as to find an association between CP and ApaI, BsmI, and FokI VDR SNPs. Statistically significant association between CP and both sex (p=0.000) and age (p=0.001) was detected (Table 2).
Table 2.
Association between chronic periodontitis and age and sex
| Parameter | Distribution | No. CP patients | No. Controls | p |
|---|---|---|---|---|
| Age | 25–35 | 6 | 24 | 0.001 |
| 36–45 | 21 | 24 | ||
| 46–55 | 33 | 25 | ||
| 56–65 | 39 | 24 | ||
| Sex | F | 43 | 69 | 0.000 |
| M | 56 | 28 |
The level of significance (p<0.05).
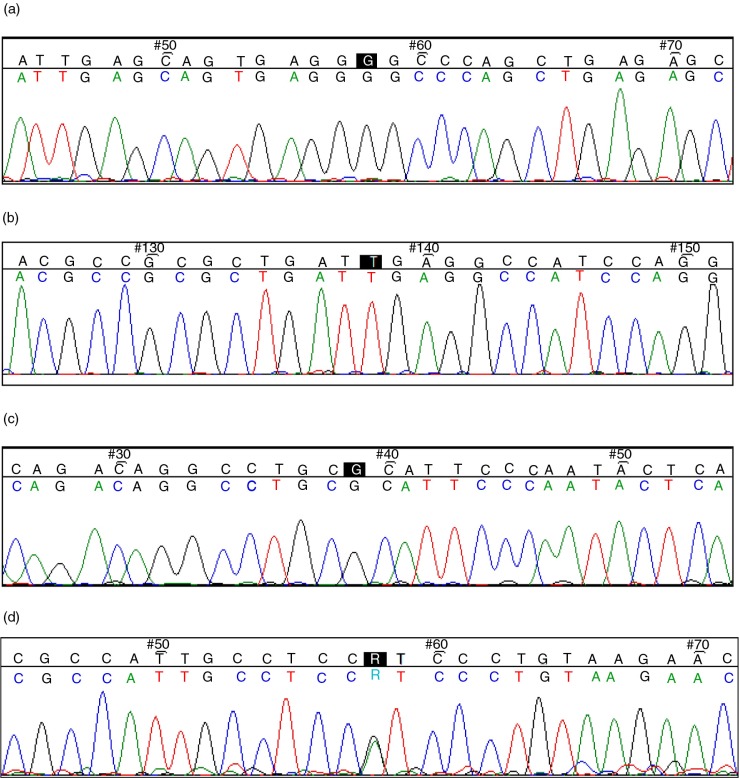
The flanking regions by ApaI, BsmI, and FokI primers were analyzed; the lengths of the PCR products were 156, 106, and 146 bases, respectively. Figure 1a shows a part of the sequence of the region flanked by ApaI primer including G/T SNP rs#7975232. In this study, the variant, C/T rs#731236 SNP was detected in the region flanked by ApaI primers (Fig. 1b). Figure 1c shows part of the sequence of the region flanked by BsmI primers including A/G SNP rs#1544410. Figure 1d shows part of the sequence of the region flanked by FokI primers including A/G SNP rs#2228570.
Fig. 1.
(a) ApaI G/T SNP (highlighted in black, ambiguity code: K). (b) ApaI C/T SNP (highlighted in black, ambiguity code: Y). (c) BsmI A/G SNP (highlighted in black, ambiguity code: R). (d) FokI A/G SNP (highlighted in black, ambiguity code: R).
The analysis of genotypic and allelic frequencies of the analyzed polymorphisms showed that a significant association p =0.022 and p=0.023, respectively, was foundbetween CP and ApaI C/T SNP rs#731236, whereas no statistically significant associations were observed in genotypic frequencies between CP and ApaI G/T rs#7975232 (p=0.939), BsmI A/G rs#1544410 (p=0.466), and FokI A/G rs#2228570 (p=0.239) SNPs. Tables 3–7 show the numbers of the PCR products (varied between 93 and 122 samples) that have been successfully sequenced and read for the analyzed SNPs.
Table 3.
Genotypic frequencies of vitamin D receptor gene ApaI G/T rs#7975232 SNP
| CP patients | Controls | ||||
|---|---|---|---|---|---|
| VDR genotypes | N | % | N | % | p |
| TT | 10 | 16.3 | 7 | 15.5 | |
| GT | 43 | 70.4 | 33 | 73.3 | 0.939 |
| GG | 8 | 13.1 | 5 | 11.1 | |
| Total | 61 | 57.5 | 45 | 42.4 | |
The level of significance (p<0.05).
Table 7.
Genotypic frequencies of vitamin D receptor gene FokI A/G rs#2228570 SNP
| CP patients | Controls | ||||
|---|---|---|---|---|---|
| VDR genotypes | N | % | N | % | p |
| GG | 38 | 57.5 | 21 | 56.7 | |
| GA | 27 | 40.9 | 13 | 35.1 | 0.239 |
| AA | 1 | 1.5 | 3 | 8.1 | |
| Total | 66 | 64.0 | 37 | 35.9 | |
The level of significance (p<0.05).
Table 4.
Genotypic frequencies of vitamin D receptor gene ApaI C/T rs#731236 SNP
| CP patients | Controls | ||||
|---|---|---|---|---|---|
| VDR genotypes | N | % | N | % | p |
| TT | 12 | 22.6 | 15 | 37.5 | |
| CT | 30 | 56.6 | 24 | 60.0 | 0.022 |
| CC | 11 | 20.7 | 1 | 2.5 | |
| Total | 53 | 56.9 | 40 | 43.0 | |
The level of significance (p<0.05).
Table 5.
Allelic frequencies of vitamin D receptor gene ApaI C/T rs#731236 SNP
| CP patients | Controls | ||||
|---|---|---|---|---|---|
| VDR allele | N | % | N | % | p |
| T | 54 | 50.9 | 54 | 67.5 | |
| C | 52 | 49.5 | 26 | 32.5 | 0.023 |
| Total | 106 | 56.9 | 80 | 43.0 | |
The level of significance (p<0.05).
Table 6.
Genotypic frequencies of vitamin D receptor gene BsmI A/G rs#1544410 SNP
| CP patients | Controls | ||||
|---|---|---|---|---|---|
| VDR genotypes | N | % | N | % | p |
| GG | 21 | 28.0 | 18 | 38.2 | |
| GA | 47 | 62.6 | 26 | 55.3 | 0.466 |
| AA | 7 | 9.3 | 3 | 6.3 | |
| Total | 75 | 61.4 | 47 | 38.5 | |
The level of significance (p<0.05).
Discussion
CP is frequently seen among family members and across different generations within a family, suggesting a genetic basis for the susceptibility to periodontal disease (1). In addition, studies of twins suggest a genetic contribution to CP in general, but the effects of bacterial transmission and environmental influences make it difficult to explain a complex interaction (1, 29). For instance, SNPs of interleukin (IL-) 1α, IL-1β, located in different regions of cytokine genes have also been shown to affect the risk of the disease in several populations (30, 31). Because CP is a multifactorial disease (gene–environment interaction), several genetic factors may play a role in the disease development by alteration of the structural component of the periodontal tissues, whereas other genetic factors play a role in regulation of inflammatory response; many gene products interact with each other with environmental factors and the mechanism in which this interaction can increase the risk of CP is incompletely interpreted (32).
A genome wide association study on CP has been done by Divaris et al. (33) and VDR gene has been implicated as a candidate gene. A finding by Naito et al. (17) suggests that the chemical reactions involving bone mineral density-mediated effects are essential for the development of CP, but the effect of the VDR gene polymorphisms in total on CP have not been confirmed (2). Our results showed a significant association between CP and newly detected ApaI C/T SNP #rs731236, which is located in intron 8 of the VDR gene. This finding suggests an essential parameter that generally has not been considered on the prevention of diseases related to bone mineral density such as CP. In the present study no significant association was found between CP and ApaI G/T SNP rs#7975232, BsmI A/G SNP rs#1544410, and FokI A/G SNP rs#2228570. Our findings agreed with studies on Brazilian, Turkish, Japanese, and Chinese populations by de Souza et al. (6), Gunes et al. (11), Naito et al. (17), and Wang et al. (34), respectively, whereas other studies showed significant association between these VDR gene polymorphisms and the periodontal disease (19, 35). To our knowledge, the present study is the first study to examine the association between CP and VDR gene polymorphisms in North Africa.
In this study, we also investigated the association between CP and age. Our results showed a significant association between CP and age, which agrees with a study on by Amarasena et al. (36) and Demetriou et al. (37). It is suggested that as the length of time increases, the chronic plaque accumulation increases, followed by increase in periodontal tissues destruction (1).
This study also showed higher percentage of CP in men than in women. The causes of these sex differences have not been explained clearly, but suggest that men have poorer oral hygiene, less positive thinking toward oral health and dental-visit interaction, which agrees with a study by Nazish et al. On the other hand, women still have varied periodontal problems due to hormonal disturbance in various decades of life. In addition, not enough studies have been done in developing countries which might have different results as compared to developed countries (38). Other studies of hormonal effect on periodontal diseases suggested that estrogen hormone in females can affect tooth retention by preventing the alveolar bone resorption (39–41) . 17β-Estradiol, the primary female sex hormone, may also play a role in promoting periodontal regeneration in an experimental periodontitis model (42).
Therefore, diagnostic periodontal risk assessments like VDR polymorphisms may be useful in the detection of the individuals susceptible for CP. More genetic studies in the future, including analysis of VDR haplotype alleles, are necessary in the prediction of the disease. Clearly, studies with enough funding, larger sample size, and in more geographic regions in the country will be beneficial due to the importance of the VDR gene and its relation to the CP and many other human diseases.
Conclusions
In the present study, we investigated ApaI C/T SNP rs#731236 that confirmed the importance of the VDR gene in the Libyan population. Instead of restriction enzyme technique, which was performed in previous studies, we used the sequencing method which is more advanced, more expensive, and capable of revealing more SNPs in the VDR gene.
Acknowledgement
Thanks to Genetic Engineering Department at Biotechnology Research Center, Tripoli, Libya, for supplying genetic analysis facilities.
Conflict of interest and funding
The authors state that no conflict of interest exists.
References
- 1.Newman M, Takei H, Klokkevold P, Carranza F. 10th ed. Philadelphia: Elsevier; 2007. Carranza's clinical periodontology; p. 68. 494–5, 497–9. [Google Scholar]
- 2.Holla I, Jurajda M, Fassmann A, Dvorakova N, Znojil V, Vacha J. Genetic variations in the matrix metalloproteinase-1 promoter and risk of susceptibility and/or severity of CP in the Czech population. J Clin Periodontol. 2004;31:685–90. doi: 10.1111/j.1600-051X.2004.00547.x. [DOI] [PubMed] [Google Scholar]
- 3.Tachi Y, Shimpuku H, Nosaka Y, Kawamura T, Shinohara M, Ueda M. Vitamin D receptor gene polymorphism is associated with CP. Life Sci. 2003;73:3313–21. doi: 10.1016/j.lfs.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Sexton W, Lin Y, Kryscio R, Dawson D, Ebersole J, Miller C. Salivary biomarkers of periodontal disease in response to treatment. J Clin Peridontal. 2011;38:434–41. doi: 10.1111/j.1600-051X.2011.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalowicz S, Aeppli D, Virag G. Periodontal findings in adult twins. J Peridontal. 1991;62:293–9. doi: 10.1902/jop.1991.62.5.293. [DOI] [PubMed] [Google Scholar]
- 6.de Souza M, Braosi P, Luczyszyn M, Avila R, de Brito B, Jr, Ignacio A, et al. Association between vitamin D receptor gene polymorphisms and susceptibility to chronic kidney disease and periodontitis. Blood Purif. 2007;25:411–19. doi: 10.1159/000109235. [DOI] [PubMed] [Google Scholar]
- 7.Payne B, Reinhardt A, Nummikoski V. Longtudinal alveolar bone loss in postmenopausal osteoporotic/osteopenic women. Osteoporos Int. 1999;10:34–40. doi: 10.1007/s001980050191. [DOI] [PubMed] [Google Scholar]
- 8.Dervis E. Oral implications of osteoporosis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100:349–56. doi: 10.1016/j.tripleo.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto K, Kesterson A, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S, et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol. 1997;11:1165–79. doi: 10.1210/mend.11.8.9951. [DOI] [PubMed] [Google Scholar]
- 11.Gunes S, Sumer P, Keles G, Kara N, Koprulu H, Bagci H, et al. Analysis of vitamin D receptor gene polymorphisms in patients with chronic periodontitis. Indian J Med Res. 2008;172:58–64. [PubMed] [Google Scholar]
- 12.Ayala F, Fitch W. Genetics and the origin of species: an introduction. PNAS. 1997;94:7691–7. doi: 10.1073/pnas.94.15.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faraco H, Morrison A, Baker A. ApaI dimorphism at the human vitamin D receptor gene locus. Nucleic Acids Res. 1989;17:2150. doi: 10.1093/nar/17.5.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison A, Yeoman R, Kelly J. Contribution of trans-acting factor alleles to normal physiological variability: vitamin D receptor gene polymorphism and circulating osteocalcin. Proc Natl Acad Sci. 1992;89:6665–9. doi: 10.1073/pnas.89.15.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross C, Eccleshall R, Malloy J. The presence of a polymorphism at the translation initiation site of the vitamin D receptor gene is associated with low bone mineral density in postmenopausal Mexican-American women. J Bone Miner Res. 1996;11:1850–5. doi: 10.1002/jbmr.5650111204. [DOI] [PubMed] [Google Scholar]
- 16.Saijo T, Ito M, Takeda E. A unique mutation in the vitamin D receptor gene in three Japanese patients with vitamin D-dependent rickets type II: utility of single-strand conformation polymorphism analysis for heterozygous carrier detection. Am J Hum Genet. 1991;49:668–73. [PMC free article] [PubMed] [Google Scholar]
- 17.Naito M, Miyaki K, Naito T, Zhang L, Hoshi K, Hara A, et al. Association between vitamin D receptor gene haplotypes and chronic periodontitis among Japanese men. Int J Med Sci. 2007;4:216–22. doi: 10.7150/ijms.4.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Geng O, Ma B. Association of vitamin receptor gene polymorphisms with the susceptibility to chronic periodontitis of Han nationality. Zhonghua Kou Qiang Yi Xue Za Zhi. 2005;40:50–3. [PubMed] [Google Scholar]
- 19.de Brito B, Jr, Scarel-Caminaga M, Trevilatto C, de Souza P, Barros P. Polymorphisms in the vitamin D receptor gene are associated with periodontal disease. J Periodontol. 2004;75:1090–5. doi: 10.1902/jop.2004.75.8.1090. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Meng H, Cao C, Tachi Y, Shinohara M, Ueda M. Relationship between vitamin D receptor gene polymorphism and periodontitis. J Periodontal Res. 2002;37:263–7. doi: 10.1034/j.1600-0765.2002.01605.x. [DOI] [PubMed] [Google Scholar]
- 21.Armitage G, Wu Y, Wang H, Sorrell J, di Giovine S, Duff W. Low prevalence of a periodontitis-associated interleukin-1 composite genotype in individuals of Chinese heritage. J Periodontol. 2000;71:164–71. doi: 10.1902/jop.2000.71.2.164. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Zhou X, Meng X, Liu G, Xing X, Liu H. Polymorphisms of vitamin D receptor gene and its association with bone mineral density and osteocalcin in Chinese. Chin Med J. 1997;110:366–71. [PubMed] [Google Scholar]
- 23.Ramfjord P. The Periodontal Disease Index (PDI) J Periodontol. 1967;38:602–10. doi: 10.1902/jop.1967.38.6.602. [DOI] [PubMed] [Google Scholar]
- 24.Instructions for Isohelix DNA Isolation kits: DDK-3/DDK-50. Isohelix. 2012. [accessed on 2013 October 9]. Available from: http://www.isohelix.com/PDF/DDK_Instructions.pdf.
- 25.Agarose Gel Electrophoresis. Addgene. 2012. [accessed on 2013 October 12]. Available from: https://www.addgene.org/plasmid_protocols/gel_electrophoresis/
- 26.Promega Corporation; 2012. Promega PCR master mix protocol. [accessed on 2013 October 19]. Available from: http://www.ebiotrade.com/buyf/productsf/promega/9pim300.pdf. [Google Scholar]
- 27.Qiagen Corporation; 2008. QIAquick PCR Purification Kit Protocol. [accessed on 2013 October 25]. Available from: http://2012.igem.org/wiki/images/a/a3/QIAquick_PCR-purification.pdf. [Google Scholar]
- 28.Applied Biosystems Corporation; 2010. BigDye®Terminator v1.1 Cycle Sequencing Kit Protocol. [accessed on 2013 November 2]. Available from: http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_041330.pdf. [Google Scholar]
- 29.Taba M, de Souza S, Mariguela V. Periodontal disease: a genetic perspective. Braz Oral Res. 2012;26:1806–8324. doi: 10.1590/s1806-83242012000700006. [DOI] [PubMed] [Google Scholar]
- 30.Karimbux Y, Saraiya M, Elangovan S, Allareddy V, Kinnunen T, Kornman K, et al. Interleukin-1 gene polymorphisms and chronic periodontitis in adult whites: a systematic review and meta-analysis. J Peridontal. 2012;83:1407–19. doi: 10.1902/jop.2012.110655. [DOI] [PubMed] [Google Scholar]
- 31.Noack B, Görgens H, Lorenz K, Ziegler A, Hoffmann T, Schackert HK. TLR4 and IL-18 gene variants in aggressive periodontitis. J Clin Periodontol. 2008;35:1020–6. doi: 10.1111/j.1600-051X.2008.01334.x. [DOI] [PubMed] [Google Scholar]
- 32.Needleman H, Karimbux N, Van Kayke T, Armitage G, Mariotti A, Delima A. 1st ed. UK: Martin Duntiz; 2001. Peridontal and gingival health and diseases: children, adolescents and young adults; pp. 190–1. [Google Scholar]
- 33.Divaris K, Monda K, North K, Olshan AF, Reynolds LM, Hsueh WC, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet. 2013;22:2312–24. doi: 10.1093/hmg/ddt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Pan P, Teng D, Zhao J, Lin L. The relativity between chronic Periodontitis and the genetic polymorphisms of vitamin D receptor and estrogen receptor. Chin J Stomatol. 2008;43:236–9. [PubMed] [Google Scholar]
- 35.Zhang C, Geng O, Ma B, Huang P, Pang Y, Zhang H. Association of vitamin receptor gene polymorphisms with the susceptibility to chronic periodontitis of Han nationality. Chin J Stomatol. 2005;40:50–3. [PubMed] [Google Scholar]
- 36.Amarasena N, Ikeda N, Win K, Yamaguchi Y, Takehara T, Miyazaki H. Factors associated with severe periodontitis in rural Cambodia. Asia Pac J Public Health. 2004;16:50–3. doi: 10.1177/101053950401600109. [DOI] [PubMed] [Google Scholar]
- 37.Demetriou N, Parashis A, Tsami-Pandi A. Relationship between age and clinical symptoms of periodontal disease. Stomatologia. 1990;47:231–41. [PubMed] [Google Scholar]
- 38.Nazish A, Mishra P, Chandrasekaran S. Gender basis of periodontal diseases. Indian J Basic Appl Med Res. 2012;2:128–35. [Google Scholar]
- 39.Krall A, Wehler C, Garcia I, Harris S, Dawson-Hughes B. Calcium and vitamin D supplements reduce tooth loss in the elderly. Am J Med. 2001;111:452–6. doi: 10.1016/s0002-9343(01)00899-3. [DOI] [PubMed] [Google Scholar]
- 40.Krall A, Garcia I, Dawson-Hughes B. Increase risk of tooth loss in related to bone loss at the whole body, hip, and spine. Calcif Tissue Int. 1996;59:433–7. doi: 10.1007/BF00369206. [DOI] [PubMed] [Google Scholar]
- 41.Norderyd M, Grossi G, Machtei E. Periodontal status of women taking postmenopausal estrogen supplementation. J Periodontol. 1993;64:957–62. doi: 10.1902/jop.1993.64.10.957. [DOI] [PubMed] [Google Scholar]
- 42.Nuñez J, Sanz-Blasco S, Vignoletti F, Muñoz F, Caffesse RG, Sanz M, et al. 17beta-estradiol promotes cementoblast proliferation and cementum formation in experimental periodontitis. J Periodontol. 2010;81:1064–74. doi: 10.1902/jop.2010.090678. [DOI] [PubMed] [Google Scholar]