Abstract
Defining phenotypes according to molecular features would promote the knowledge of functional traits like behaviour in both human and animal research. Beside physiological states or environmental factors, an innate predisposition of individual coping strategies was discussed, including the proactive and reactive pattern. According to backtest reactivity, animals assigned as high-resisting (proactive) and low-resisting (reactive) were immune challenged with tetanus toxoid in a time course experiment. Using the Affymetrix platform and qPCR, individual coping characteristics were reflected as gene expression signatures in porcine peripheral blood mononuclear cells (PBMC) at naïve state (day 0) and in response to the model antigen (day 14, day 28, and day 140). Further, the blood cell count was analysed at all stages. On the transcriptional level, processes acting on cell communication, vasculogenesis, and blood coagulation were highlighted in high-resisting animals at naïve state (day 0), temporarily blurred due to immune challenge (day 14) but subsequently restored and intensified (day 28). Notably, similar amounts of white and red blood cells, platelets and haematocrit between high-resisting and low-resisting samples suggest coping-specific expression patterns rather than alterations in blood cell distribution. Taken together, the gene expression patterns indicate that proactive pigs might favour molecular pathways enabling an effective strategy for defence and recovery. This corroborates the previously suggested belief, that proactive animals are prone to an increased number of injuries as an evolutionary inherited mechanism. In contrast to previous assumptions, coping-specific immunity in pigs lacks inherited shifts between cellular and humoral immune responses.
Introduction
Phenotypic characteristics of functional traits like health, fertility, longevity, and behaviour have remained difficult to define as measurable molecular features despite extensive performance testing and novel insights through quantitative genetics [1,2]. In particular, substantial and reliable data on the impacts of genes on functional traits like behaviour, ‘disease resistance’, and ‘disease tolerance’ are scarce. However, interest in developing molecular signatures (e.g., gene expression profiles) is now growing exponentially, boosted by intentions to establish ethical husbandry and resource conservation in animal-source food production. Defining phenotypes according to molecular features would promote the knowledge of functional traits like behaviour in both human and animal research. Inter-individual variability provides some clues that will aid the identification of molecular signatures. For instance, individuals differ in the ways they cope with environmental challenges [3]. Coping styles have been shaped by evolution to form general adaptive response patterns. Two different coping styles have been distinguished [4]. The proactive, or active, pattern is characterized by a fight-or-flight response, high levels of aggression, and territorial control [5]. The reactive, or passive, pattern involves a conservation-withdrawal response, immobility, and low levels of aggression [6]. Interestingly, these distinct coping styles differ at the physiological and neuroendocrinological levels [7]. Animals with an active coping pattern have high sympathetic activity and moderate hypothalamic-pituitary-adrenal (HPA)-axis reactivity, probably resulting from preparation for action. In contrast, animals with a passive coping pattern have higher parasympathetic reactivity and high HPA axis activity. As the autonomic nervous system (i.e., the sympathetic branch) and the HPA axis communicate between the brain and the immune system, a differential reactivity of these systems may cause inter-individual differences in the immune response [8].
Coping styles have been studied in animals through behavioural tests. In particular, Hessing and collaborators applied the ‘backtest’, in which a pig is placed in a supine position for 60 seconds and monitored for struggling; they hypothesized that this simple method could detect coping styles in domestic pigs [9,10]. Subsequent applications of the backtest demonstrated individual consistency in behavioural responses over time [11,12] and linked neuroendocrine and physiological characteristics [13–15]. Indeed, reactivity to the backtest is believed to reflect basal molecular differences that correspond to immune features comprising both cellular and humoral immunity [15–17]. In particular, aggressive and resistant pigs (i.e., animals showing an active coping pattern) appeared to predominantly rely on cellular immune response, whereas non-aggressive and non-resistant pigs (i.e., animals showing a passive coping pattern) appeared to predominantly rely on humoral immune response [15,16].
However, evidence also suggests that primary behavioural differences and related physiological features between individuals appear particularly due to stressful situations [18]. In this context, the molecular responses producing phenotypes of interest can be investigated through immune challenges, which are, by nature, stressful. For example, vaccination with tetanus toxoid, used as model antigen, causes long-lasting cellular (Th1) and humoral (Th2) responses [19,20] via CD4+ MHC class II-restricted T helper cells [21]. The tetanus toxoid introduces a signal that initiates a coordinated program of gene expression in porcine peripheral blood mononuclear cells (PBMC) [22,23]. In a recent time-course experiment, tetanus toxoid vaccination produced phenotype-specific transcriptional differences in both immune features and metabolic pathways in animals divergent for lean growth performance [24]. These findings demonstrate that the stress of the immune challenge is associated with particular molecular signatures.
Here, we used temperament-dependent behaviour differences, resulting in either proactive (high-resisting) or reactive (low-resisting) coping patterns, to identify phenotypes related to immune response following tetanus toxoid vaccination. In this time-course experiment, molecular effects of an immune stimulation were investigated in vivo, focusing on piglets classified as either high or low-resisting according to latency, total duration, and frequency of struggling bouts in a backtest [12]. A recently-described whole genome microarray platform [25] was used to analyse gene expression in porcine PBMC before and after immune stimulation. We aimed to derive trait-associated molecular signatures applicable for selection according to (i) basal conditions regarding the experimental groups, and (ii) reactivity-dependent immune responses after vaccination. Changes in gene expression were quantified in porcine PBMC at 4 distinct time points.
Materials and Methods
Animals, performance tests, and vaccination
Animals were provided by the Leibniz Institute for Farm Animal Biology (FBN). Animal care, vaccination, and blood collection procedures followed the guidelines of the German Law of Animal Protection. The experimental protocol was approved by the Animal Care Committee of the Leibniz Institute of Farm Animal Biology and the State Mecklenburg-Western Pomerania (Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei; LALLF M-V/TSD/7221.3–2.1–020/09).
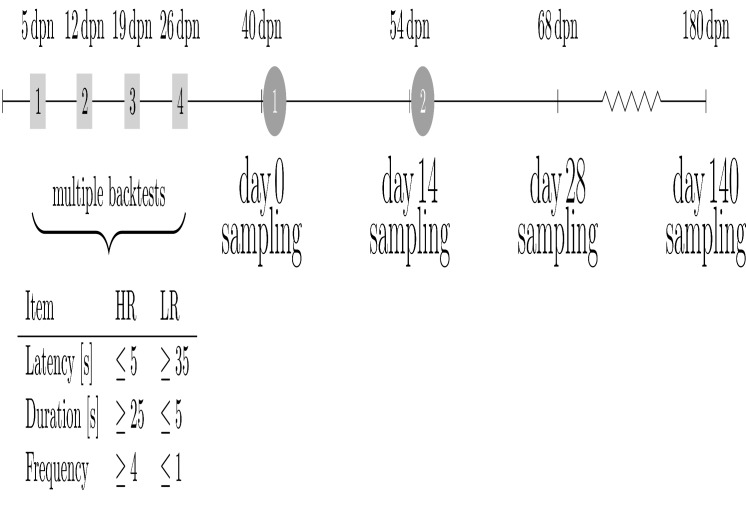
The experimental design is detailed in Fig. 1. German Landrace piglets were part of a larger study and were subjected to a backtest as described [12]. In brief, the backtest was performed on 3,555 piglets (1,759 males and 1,796 females derived from 223 sows and 42 boars) at four time points [5 days post-natum (dpn), 12 dpn, 19 dpn, 26 dpn]. Each backtest lasted 60 seconds. Several criteria were used to classify the investigated piglets as high-resisting (HR), doubtful (D), or low-resisting (LR) animals [13]: (i) latency, or time until first response (HR: ≤ 5s; LR: ≥ 35s); (ii) total duration, or cumulative time interval of responses (HR: ≥ 25s; LR ≤ 5s); and (iii) frequency of struggling bouts (HR: ≥ 4; LR ≤ 1). Regarding to the four measurement periods, each of the individual 12 parameters was considered for classification into HR, D, or LR [12]. In total, 417 piglets were identified as HR animals (~12%) and 784 piglets were identified as LR animals (~22%).
Fig 1. Experimental design.
Pigs were subjected to the backtest at 5 dpn, 12 dpn, 19 dpn, and 26 dpn as indicated by the grey rectangles, resulting in 12 individual parameters considered for classification into HR and LR [12]. The grey circles indicate 1st and 2nd (booster) tetanus toxoid vaccinations shortly after day 0 and day 14 sampling points. Blood (n = 48) was collected at four sampling points: in naïve state (day 0), 14 days after 1st vaccination (day 14), 14 days after 2nd vaccination (day 28), and at slaughter (day 140); dpn = days post-natum.
In general, weaning appears to be a stressful period in life during which an impaired immune responsiveness occurs in pigs [26,27]. Piglets were weaned at 28 dpn. Hence, to eliminate eventual effects, the immune challenge started at approximately 40 dpn when a subset of the classified five-week-old piglets (HR: 145; LR: 107; litters obtained in 2009 and 2010) was subcutaneously vaccinated (day 0) with one dose (1 mL) of tetanus vaccine, comprising tetanus toxoid and aluminium hydroxide as adjuvant (Equilis Tetanus-Vaccine, Intervet, Unterschleißheim, Germany). After two weeks, a booster vaccination was given (day 14). Note, males were castrated at 4 dpn.
Blood samples
Blood samples were collected from jugular veins into EDTA-treated tubes immediately before the first (day 0) and second vaccinations (day 14) as well as two weeks after the booster vaccination (day 28). Blood parameters (leucocytes, lymphocytes, monocytes, neutrophils, eosinophils, basophils, platelets, erythrocytes, and haematocrit) were analysed at day 0, day 14, and day 28 (ABX Pentra 60, HORIBA ABX SAS, Montpellier, France). Additionally, trunk blood was collected at slaughtering (day 140), when pigs were weighed and slaughtered by electronarcosis followed by exsanguination in the experimental slaughterhouse of FBN. Trunk blood was supplemented with EDTA and stored on ice.
RNA isolation, target preparation, and hybridization
For expression analysis a subset of animals was selected aiming for a balanced design in terms of gender, batch and ancestry. Accordingly, twelve castrated males and twelve females per coping group were selected for subsequent analyses, producing 48 individual samples per sampling time point. The PBMCs were isolated from 5 mL blood by centrifugation on a Histopaque density gradient (Sigma-Aldrich, Taufkirchen, Germany), then stored at -80°C. Total RNA was isolated using Qiazol reagent per manufacturer’s directions (Qiagen, Hilden, Germany). Quantification and purification were performed as previously described [24]. All RNA was stored at -80°C until downstream analyses were performed. For the microarray experiments, individual samples (n = 192) were hybridized on genome-wide snowball arrays (Affymetrix, Santa Clara, CA, USA), a platform invented for genome-wide analyses of the pig transcriptome [25]. Processing was performed as previously described [24]. Raw data have been deposited in a MIAME-compliant database, the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) (accession numbers: GSE55418).
Data analyses
In total, 188 of 192 arrays passed the appropriate quality control criteria as proposed by Kauffmann et al. [28]. Data were RMA-normalized (Log2). To improve statistical power [29], inappropriate probe-sets were excluded from further analyses, such as probe-sets with a small logarithmized mean (m<2.5) among all analysed arrays as well as probe-sets with a small standard deviation (SD<0.22) among all arrays included in the analysis. It was assumed that transcriptional differences resulted from genetic factors (paternal effects), time-dependent response to vaccination, and different phenotypes of temperament. Therefore, relative mRNA differences (p≤0.01) including individual and combined effects of coping group, sire (partially confounded to coping group), time, and batch were analysed using a mixed model [Vijkl = μ + coping groupi + timej + batchk + sirel(coping groupi) + (coping group x time)ij + eijkl]. The mixed model was combined with a repeated statement for the time component specified as heterogeneous covariance structure (SAS version 9.3; SAS Institute, Cary, NC, USA). Sampling time points were weighted equally while comparing phenotypes divergent in temperament. Due to multiple testing, p-values were converted to a set of q-values [30]. The level of significance was set at q≤0.25.
Pathway analyses
Annotation data for Affymetrix snowball arrays were obtained from the developers [25]. Gene lists obtained from the PBMC microarray analyses were evaluated with ‘Ingenuity Pathway Analysis’ (IPA, Ingenuity Systems, Redwood City, CA, USA). The significance of association between dataset and pathway/biofunction analyses was calculated (p≤0.05). The top 10 pathways showing the lowest p-values were considered for further analyses. To identify relevant IPA-biofunctions, the z-score was used to discriminate between increased (z > 2; HR > LR) or decreased (z < -2; HR < LR) functional themes.
Quantitative real-time PCR
Selected reference genes were characterized by small standard deviation between coping groups (SD<0.1). Further, their expression values (log2) ranged approximately at the level of the target genes. Total transcript levels of selected target (CD69, GNAZ, ITGA2B) and reference genes (IQGAP1, TSC22D2) were quantified by real-time qPCR (S1 Table). Individual PBMC mRNA samples (n = 44 per sampling day) were analysed in duplicate on a LightCycler 480 system using LightCycler 480 SYBR Green I Master (Roche, Mannheim, Germany). Data were factorial normalized. The statistical analysis included individual and combined effects of coping group, sire (confounded to coping group), time, and batch (SAS version 9.3; SAS Institute, Cary, NC, USA). The level of significance was set at p≤0.05.
Results
The snowball microarray covers 47,845 probe-sets corresponding to 17,964 annotated genes. After filtering as described above, 27,033 probe-sets (~57%) remained for analysis. These probe-sets corresponded to 11,620 annotated genes.
Transcriptional responses in animals divergent in temperament
The comparison of porcine PBMC derived from HR and LR samples revealed a number of probe-sets differing in their mRNA abundance by both coping group and sampling points (S2 Table).
To study expression patterns at naïve state and due to immune challenges in animals divergent in coping style, pathways that showed effects mediated by ‘coping group’ were examined in detail at all sampling points (Table 1, S3 Table). In general, the selected top 10 canonical pathways reflected consistent time-dependent patterns. Analysis of transcriptional differences suggested molecular routes differing between HR and LR samples both at day 0 and day 28 (‘α-Adrenergic Signaling’, ‘G Beta Gamma Signaling’, ‘Thrombin Signaling’, ‘Integrin Signaling’, ‘Protein Kinase A Signaling’, ‘RhoGDI Signaling’), as well as at day 140 (‘P2Y Purigenic Receptor Signaling Pathway’, ‘IL-8 Signaling’, ‘Gap Junction Signaling’, ‘CXCR4 Signaling’). Among those pathways, only ‘CXCR4 Signaling’ was also altered at day 14. A complete list of the altered transcripts associated with the displayed canonical pathways is provided in S3 Table.
Table 1. Top 10 Ingenuity pathways of transcripts with higher and lower expression between HR and LR PBMC samples at four time points (day 0, day 14, day 28, and day 140).
| Canonical pathway | Sampling day | p-value | Number of involved genes |
|---|---|---|---|
| P2Y Purigenic Receptor Signaling Pathway | day 0 | 8.51E-04 | 9 |
| day 14 | 2.44E-01 | 1 | |
| day 28 | 1.51E-05 | 9 | |
| day 140 | 2.24E-02 | 4 | |
| IL-8 Signaling | day 0 | 1.55E-03 | 11 |
| day 14 | 6.76E-02 | 2 | |
| day 28 | 8.32E-05 | 10 | |
| day 140 | 2.40E-02 | 5 | |
| Gap Junction Signaling | day 0 | 4.57E-03 | 9 |
| day 14 | 3.01E-01 | 1 | |
| day 28 | 1.05E-04 | 9 | |
| day 140 | 1.20E-02 | 5 | |
| α-Adrenergic Signaling | day 0 | 4.07E-04 | 8 |
| day 14 | 1.83E-01 | 1 | |
| day 28 | 6.31E-04 | 6 | |
| day 140 | 1.79E-01 | 2 | |
| G Beta Gamma Signaling | day 0 | 7.94E-05 | 9 |
| day 14 | 1.85E-01 | 1 | |
| day 28 | 6.76E-04 | 6 | |
| day 140 | 1.82E-01 | 2 | |
| Thrombin Signaling | day 0 | 5.37E-04 | 12 |
| day 14 | 7.08E-02 | 2 | |
| day 28 | 9.77E-05 | 10 | |
| day 140 | 8.71E-02 | 4 | |
| CXCR4 Signaling | day 0 | 1.15E-03 | 10 |
| day 14 | 4.79E-02 | 2 | |
| day 28 | 4.90E-04 | 8 | |
| day 140 | 4.47E-02 | 4 | |
| Integrin Signaling | day 0 | 1.23E-05 | 15 |
| day 14 | 3.64E-01 | 1 | |
| day 28 | 4.27E-06 | 12 | |
| day 140 | 2.47E-01 | 3 | |
| Protein Kinase A Signaling | day 0 | 1.05E-05 | 22 |
| day 14 | 5.83E-01 | 1 | |
| day 28 | 2.00E-03 | 12 | |
| day 140 | 2.37E-01 | 5 | |
| RhoGDI Signaling | day 0 | 5.01E-05 | 13 |
| day 14 | 5.89E-02 | 2 | |
| day 28 | 4.07E-05 | 10 | |
| day 140 | - | - |
Metadata of involved genes were displayed in S3 Table. HR—High resisting; LR—Low resisting.
To gain insight into functional and structural components, genes with altered mRNA abundances were assigned to IPA-biofunctions. Here, the z-score was used (z > 2 or z < -2) to identify functional themes with respect to stage-specific alterations. The analyses revealed transcriptional differences at day 0 and day 28 only. Biofunctions that appeared to be altered between coping groups at both day 0 and day 28 are listed in Table 2. Moreover, biofunctions revealing a molecular signature specifically for either day 0 or day 28 are shown in Tables 3 and 4, respectively. In general, the addressed biofunctions corresponded to themes like ‘Cell-to-cell Signaling and Interaction’, ‘Cellular Assembly and Organization’, ‘Haematological System Development and Function’, ‘Immune Cell Trafficking’, ‘Inflammatory Response’, ‘Tissue Development’, and ‘Tissue Morphology’. A complete list of the altered transcripts associated with the displayed biofunctions at day 0 and day 28 is provided in S4 Table.
Table 2. Common Ingenuity biofunctions of transcripts with higher and lower expression between HR and LR PBMC samples at day 0 and day 28.
| Biofunction | mRNA abundance | p-value | Number of involved genes |
|---|---|---|---|
| adhesion of endothelial cells | HR > LR; day 0 | 9.37E-03 | 7 |
| HR > LR; day 28 | 2.63E-03 | 6 | |
| aggregation of blood platelets | HR > LR; day 0 | 1.65E-04 | 12 |
| HR > LR; day 28 | 1.09E-07 | 13 | |
| bleeding time | HR < LR; day 0 | 3.58E-05 | 7 |
| HR < LR; day 28 | 6.16E-08 | 8 | |
| quantity of lymphatic system cells | HR < LR; day 0 | 3.88E-03 | 14 |
| HR < LR; day 28 | 1.09E-02 | 9 |
Metadata of involved genes were displayed in S4 Table. HR—High resisting; LR—Low resisting.
Table 3. Specific Ingenuity biofunctions of transcripts with higher and lower expression between HR and LR PRMC samples at day 0.
| Biofunction | mRNA abundance | p-value | Number of involved genes |
|---|---|---|---|
| adhesion of vascular endothelial cells | HR > LR; day 0 | 1.29E-02 | 5 |
| quantity of blood platelets | HR > LR; day 0 | 9.55E-04 | 9 |
| vasculogenesis | HR > LR; day 0 | 9.77E-03 | 27 |
| quantity of megakaryocytes | HR < LR; day 0 | 2.30E-03 | 5 |
| senescence of cells | HR < LR; day 0 | 1.14E-02 | 10 |
Metadata of involved genes are displayed in S4 Table. HR—High resisting; LR—Low resisting.
Table 4. Specific Ingenuity biofunctions of transcripts with higher and lower expression between HR and LR PBMC samples at day 28.
| Biofunction | mRNA abundance | p-value | Number of involved genes |
|---|---|---|---|
| adhesion of blood cells | HR > LR; day 28 | 1.30E-04 | 15 |
| adhesion of granulocytes | HR > LR; day 28 | 5.77E-03 | 5 |
| attachment of cells | HR > LR; day 28 | 3.82E-04 | 7 |
| binding of blood cells | HR > LR; day 28 | 2.77E-03 | 9 |
| binding of blood platelets | HR > LR; day 28 | 1.88E-05 | 6 |
| binding of cells | HR > LR; day 28 | 1.14E-05 | 19 |
| cell movement | HR > LR; day 28 | 8.93E-08 | 55 |
| cell movement of leukocytes | HR > LR; day 28 | 2.31E-03 | 21 |
| cell movement of mononuclear leukocytes | HR > LR; day 28 | 4.01E-03 | 14 |
| cell movement of myeloid cells | HR > LR; day 28 | 5.40E-03 | 15 |
| cell movement of neutrophils | HR > LR; day 28 | 1.18E-02 | 9 |
| cell viability of blood cells | HR > LR; day 28 | 1.09E-02 | 9 |
| cell viability of hematopoietic progenitor cells | HR > LR; day 28 | 2.79E-03 | 5 |
| chemotaxis of cells | HR > LR; day 28 | 2.51E-04 | 17 |
| chemotaxis of leukocytes | HR > LR; day 28 | 2.61E-03 | 12 |
| chemotaxis of myeloid cells | HR > LR; day 28 | 1.16E-02 | 9 |
| engulfment of cells | HR > LR; day 28 | 1.20E-03 | 12 |
| homing of cells | HR > LR; day 28 | 1.85E-04 | 18 |
| homing of leukocytes | HR > LR; day 28 | 1.49E-03 | 13 |
| invasion of cells | HR > LR; day 28 | 3.49E-04 | 20 |
| leukocyte migration | HR > LR; day 28 | 2.22E-04 | 26 |
| MAPKKK cascade | HR > LR; day 28 | 1.75E-03 | 8 |
| microtubule dynamics | HR > LR; day 28 | 4.47E-06 | 27 |
| migration of cells | HR > LR; day 28 | 4.19E-07 | 50 |
| morphology of cells | HR < LR; day 28 | 8.06E-04 | 35 |
| organization of cytoskeleton | HR > LR; day 28 | 4.26E-07 | 34 |
Metadata of involved genes are displayed in S4 Table. HR—High resisting; LR—Low resisting.
Cluster analyses
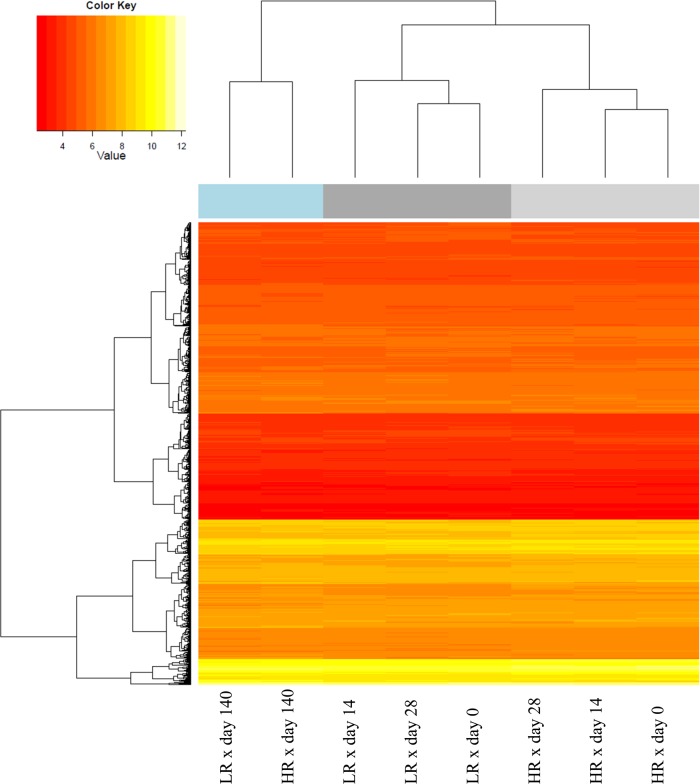
The impact of coping group and time on gene expression was visualized by hierarchical clustering, accounting for significantly altered probe-sets of all investigated subgroups (Fig. 2). Clustering revealed two superior clusters, where coping group appeared to dominate early sampling points (HR x day 0, HR x day 14, HR x day 28, and LR x day 0, LR x day 28, LR x day 14, respectively). The remaining cluster consisted of the subgroups HR x day 140 and LR x day 140, indicating a minor impact of coping group on gene expression at slaughter age. Notably, the final slaughter weight was similar between experimental groups (HR: 110.5 ± 7.6 kg and LR: 112.0 ± 7.6 kg, respectively).
Fig 2. Heatmap displaying probe-sets with significantly altered mRNA abundances.
Effects mediated by coping group appeared to dominate early sampling points (day 0, day 14, day 28). Later, age-specific effects were more pronounced as visualized by young adult subgroups (day 140). Columns = variance component coping group x time; Rows = transcripts showing altered mRNA abundances between HR and LR on at least one time point; HR—High resisting; LR—Low resisting.
Alterations in mRNA abundances of selected transcripts
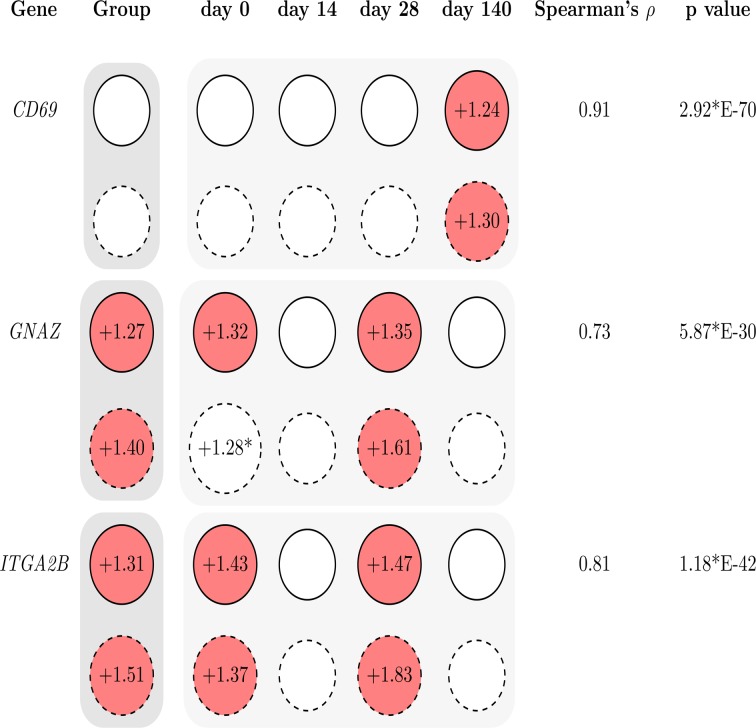
Both microarray and qRT-PCR analyses were correlated to verify differences in mRNA abundance of genes encoding cell surface receptors (CD69, GNAZ, ITGA2B) between coping groups (Fig. 3). The transcripts were analysed at multiple sampling points. The fold-changes revealed a reliable dimension. Between microarray and qPCR data the correlation coefficients were highly significant and ranged between 0.73 and 0.91. Taken together, the qPCR analyses indicate reproducibility of the microarray analysis.
Fig 3. Comparison of microarray and quantitative PCR (qPCR) results for selected transcripts (CD69, GNAZ, ITGA2B) to verify microarray data.
Values were calculated by factorial normalisation on IQGAP1 and TSC22D2 expression values. Fold-changes displayed in red circles indicate significant differences in mRNA abundances between HR and LR at either microarray (solid lined circles) or qPCR data (dashed lined circles). Positive values display increased mRNA abundances in HR (HR > LR). Correlation of normalized expression values was calculated by Spearman (n = 176). * p = 0.06.
Similar cell blood count in HR and LR samples
At day 0, day 14, and day 28, HR and LR samples were unaltered regarding their total leucocyte number, lymphocytes, monocytes, neutrophils, eosinophils, basophils, platelet number, erythrocyte number, and haematocrit (S1 Fig.).
Discussion
Both genetic [11,31,32] and environmental factors [33] appear to contribute to determining coping characteristics in pigs. In particular, extrinsic and intrinsic cues are perceived by the brain, which, in turn, orchestrates appropriate behavioural responses. In this context, due to the linkage between brain and immune system, both efferent and afferent signals enact sensor and effector functions of the immune system [34–36]. Expanding upon and consistent with in vivo situations, the current study investigated transcriptional patterns in peripheral immunocompetent cells obtained from pigs divergent for temperament. The individual coping characteristics were reflected as transcriptional differences at naïve state and in response to the immune challenge. Notably, the unchanged absolute and relative amounts of white blood cells, red blood cells, thrombocytes, and the haematocrit reflect the temperament-specific character of the observed mRNA alterations.
Re-establishing transcriptional differences following the immune challenge in juveniles
Our analysis revealed higher basal gene expression of transcripts associated with cell communication, vasculogenesis, pro-inflammation, and wound healing in HR animals at day 0. Interestingly, the observed differences at naïve stage disappeared at day 14, suggesting that the acute response to the model toxoid is similar in terms of extent and type in both HR and LR animals. Hence, responses due to acute immune challenges do likely dominate subtle coping-specific differences. In contrast, the PBMC signature was characterized by the re-establishment of the naïve patterns at day 28, promoting molecular routes related to vasculogenesis and pro-inflammation. Moreover, the assigned IPA biofunctions highlighted coping specificities for predominantly haematological processes, including cell communication, vasculogenesis, and blood coagulation.
Taken together, our analysis reveals heterogeneous molecular signatures between HR and LR samples, particularly for molecular pathways associated with vasculogenesis and pro-inflammation. Therefore, the temperament-dependent gene expression might reflect differences in the platelet—leukocyte interaction. Indeed, the cross-talk between blood cells has a bidirectional character, highlighting lymphocyte engagement in primary steps of blood coagulation, as reviewed elsewhere [37,38].
Since the divergent expression patterns were more pronounced at day 28, these findings support the growing evidence that challenging situations promote the unravelling of the subtle inherited differences in behaviour-related molecular features [18]. The observed expression patterns suggest that HR animals inherited vigilant basal immune functions that manifest independently of any wounding or injuries. Indeed, animals that are prone to experience stressful events do likely have to deal with an increased number of injuries. Thus, as suggested previously, HR animals might favour an evolutionary inherited mechanism that interrelate a physically active behaviour with the immune system [39]. However, it is of particular scientific interest whether a potential ‘improved’ immune status was developed at the expense of a predisposition for adverse immunological outcomes [40]. Superficially, this would dilate the hypothesis concerning a trade-off between immunity and stress response [41] to a trade-off between immunity and temperament. However, this remains to be scientifically investigated.
Temperament-specific transcriptional differences in young adult animals
At day 140 the analyses revealed stage-specific transcriptional responses, accompanied by a lack of relevant IPA-biofunctions. Hence, both age and coping group influenced the immune response in growing pigs, a finding that corresponds to previous observations made when baseline immune measures were analysed [17,42]. In contrast, no differences in immune properties were found between HR and LR pigs at the age of 12 months [43]. These conflicting observations account, at least in part, for the limited prior success in the discovery of molecular features related to coping groups in young adult animals. Despite reports that coping styles in farm animals such as cattle are stable over years [44], it is conceivable that subtle unnoticed differences in temperament traits become more pronounced by age [45,46]. Further, predicted personality traits such as boldness and docility are considered to be age-related in non-human mammals [47]. Consequently, temperament-specific molecular features such as immune responses might also be modified with age.
Coping style and immunity
HR and LR pigs are reportedly distinguishable by their favoured immune response, preferentially using either cellular or humoral immunity [15,16]. In contrast, HR pigs exhibited an increased lymphocyte proliferation following DNP-KLH-stimulation [17]. However, in our study no distinct shift between Th1 and Th2 immune responses was observed. Further, we found that mRNA abundances of genes associated with α-adrenergic signaling were mostly decreased in HR animals at both day 0 and day 28. Because α-adrenergic signaling is known to mediate parts of the diverse biological effects of the sympathetic branch of the autonomic nervous system, those findings seems contradictory to the established theories. Thus, the polarization into either HR or LR animals featuring (i) an emphasis on cellular immune response, moderate HPA reactivity, and sympathetic activation in HR samples versus (ii) an emphasis on humoral immune response, HPA activity, and parasymthatetic reactivity in LR samples may be rather incomplete. Taken together, the general model relating coping style, autonomous nervous system, and immune properties may lack additional genetic and environmental interactions.
Individual variation in modern breeding systems
Currently, farm animal breeding aims to reduce individual variation by strong selection criteria corresponding to various traits. As a side effect, these intentions may have narrowed the spectrum of principles underlying genotypic variation and phenotypic plasticity [7]. Indeed, in rats the reduction in individual variation due to inbreeding contributes to an altered frequency distribution of behavioural traits, creating a paucity of individuals exhibiting extreme responses [7]. Accordingly, due to accelerated animal breeding and the need to adapt to modern housing systems [48,49] (e.g., decreased space, high temperature, high levels of noxious gases, high stocking density, missing wallow, increased bacterial load) it is very likely that similar results will be found in domesticated pig breeds. However, our data indicate that in terms of coping behaviour a great variation exists among pigs.
Conclusion
In this study, the coping groups high-resisting (HR) and low-resisting (LR) were successfully discriminated according to their molecular features related to cell communication, vasculogenesis, and blood coagulation, although the tetanus toxoid vaccination blurred transcriptional differences between coping groups temporarily (day 14). However, the distinct expression signatures observed at naïve state were even intensified at day 28, as reflected by transcripts associated with platelet—leukocyte interaction. Thus, at juvenile stages it appears that HR pigs might favour molecular pathways enabling an effective strategy for defence and recovery. In contrast to previous assumptions, coping-specific immunity in pigs lacks inherited shifts between Th1 and Th2 immune responses. Notably, the similar amounts of white and red blood cells, platelets and haematocrit between HR and LR samples suggest coping-specific expression patterns rather than alterations in blood cell distribution. Thus, blood could be a suitable tissue to obtain molecular markers for distinct coping styles that might be used to drive genetic selection decisions, to optimize animal management, and for in-depth, molecular phenotyping of animals for further research of the relationships of behaviour, immune traits and production traits.
Supporting Information
(TIF)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
The authors thank Angela Garve, Hannelore Tychsen, Annette Jugert, and Joana Bittner for their excellent technical help.
Data Availability
All relevant data are within the paper and its supporting information files or are deposited in a MIAME-compliant database, the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) (accession numbers: GSE55418).
Funding Statement
This work was partly funded by the Federal Ministry of Education and Research (BMBF; www.bmbf.de) as part of the PHENOMICS project (support code: 0315536F; www.phaenomics.auf.uni-rostock.de). The Leibniz Institute for Farm Animal Biology (FBN) provided own matched funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi B, et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine dgat1 gene with major effect on milk yield and composition. Genome Res. 2002;12(2): 222–31. [DOI] [PubMed] [Google Scholar]
- 2. Winter A, Kraemer W, Werner F, Kollers S, Kata S, Durstewitz G, et al. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-coa:diacylglycerol acyltransferase (dgat1) with variation at a quantitative trait locus for milk fat content. Proc Natl Acad Sci U S A. 2002;99(14): 9300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koolhaas J, Korte S, De Boer S, Van der Vegt B, van Reenen C, Hopster H, et al. Coping styles in animals: current status in behavior and stress-physiology. Neurosci Biobehav Rev. 1999;23(7): 925–35. [DOI] [PubMed] [Google Scholar]
- 4. Henry J, Stephens P. Stress, Health, and the Social Environment A Sociobiology Approach to Medicine. Springer; 1977. [Google Scholar]
- 5.Cannon W. Bodily Changes in Pain, Hunger, Fear and Rage: An Account of Recent Researches into the Function of Emotional Excitement. Appleton New York; 1915.
- 6. Engel G, Schmale A. Conservation-withdrawal: a primary regulatory process for organismic homeostasis. Ciba Found Symp. 1972;8: 57–75. [DOI] [PubMed] [Google Scholar]
- 7. Koolhaas J, de Boer S, Coppens C, Buwalda B. Neuroendocrinology of coping styles: towards understanding the biology of individual variation. Front Neuroendocrinol. 2010;31(3): 307–21. 10.1016/j.yfrne.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 8. Koolhaas J. Coping style and immunity in animals: making sense of individual variation. Brain Behav Immun. 2008;22(5): 662–7. 10.1016/j.bbi.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 9. Hessing M, Hagelso A, van Beek J, Wiepkema P, Schouten W, Krukow R. Individual behavioural characteristics in pigs. Appl Anim Behav Sci. 1993;37: 285–295. [Google Scholar]
- 10. Hessing M, Hagels A, Schouten W, Wiepkema P, van Beek J. Individual behavioral and physiological strategies in pigs. Physiol Behav. 1994;55(1): 39–46. [DOI] [PubMed] [Google Scholar]
- 11. Velie B, Maltecca C, Cassady J. Genetic relationships among pig behavior, growth, backfat, and loin muscle area. J Anim Sci. 2009;87(9): 2767–7. 10.2527/jas.2008-1328 [DOI] [PubMed] [Google Scholar]
- 12. Zebunke M, Nuernberg G, Repsilber D, Puppe B. The backtest in pigs—individual variation in behaviour or behavioural strategy? KTBL-Schrift. 2013;503: 188–197. [Google Scholar]
- 13. Ruis M, te Brake J, van de Burgwal J, de Jong I, Blokhuis H, Koolhaas J. Personalities in female domesticated pigs: behavioural and physiological indications. Appl Anim Behav Sci. 2000;66: 31–47. [Google Scholar]
- 14. Geverink N, Schouten W, Gort G, Wiegant V. Individual differences in aggression and physiology in peri-pubertal breeding gilts. Appl Anim Behav Sci. 2002;77(1): 43–52. [DOI] [PubMed] [Google Scholar]
- 15. Schrama J, Schouten J, Swinkels J, Gentry J, de Vries Reilingh G, Parmentier H. Effect of hemoglobin status on humoral immune response of weanling pigs differing in coping styles. J Anim Sci. 1997;75(10): 2588–96. [DOI] [PubMed] [Google Scholar]
- 16. Hessing M, Coenen G, Vaiman M, Renard C. Individual differences in cell-mediated and humoral immunity in pigs. Vet Immunol Immunopathol. 1995;45(1–2): 97–113. [DOI] [PubMed] [Google Scholar]
- 17. Bolhuis J, Parmentier H, Schouten W, Schrama J, Wiegant V. Effects of housing and individual coping characteristics on immune responses of pigs. Physiol Behav. 2003;79(2): 289–96. [DOI] [PubMed] [Google Scholar]
- 18. Geverink N, Schouten W, Gort G, Wiegant V. Individual differences in behaviour, physiology and pathology in breeding gilts housed in groups or stalls. Appl Anim Behav Sci. 2003;81(1): 29–41. [Google Scholar]
- 19. ElGhazali G, Paulie S, Andersson G, Hansson Y, Holmquist G, Sun J, et al. Number of interleukin-4- and interferon-gamma-secreting human t cells reactive with tetanus toxoid and the mycobacterial antigen ppd or phytohemagglutinin: distinct response profiles depending on the type of antigen used for activation. Eur J Immunol. 1993;23(11): 2740–5. [DOI] [PubMed] [Google Scholar]
- 20. Tierney T, Simpson-Morgan M. The immune response of foetal calves. Vet Immunol Immunopathol. 1997;57(3–4): 229–38. [DOI] [PubMed] [Google Scholar]
- 21. Mayer S, Laumer M, Mackensen A, Andreesen R, Krause S. Analysis of the immune response against tetanus toxoid: enumeration of specific t helper cells by the elispot assay. Immunobiology. 2002;205(3): 282–289. [DOI] [PubMed] [Google Scholar]
- 22. Ponsuksili S, Muráni E, Wimmers K. Porcine genome-wide gene expression in response to tetanus toxoid vaccine. Dev Biol (Basel). 2002;132: 185–95. [DOI] [PubMed] [Google Scholar]
- 23. Adler M, Muráni E, Brunner R, Ponsuksili S, Wimmers K. Transcriptomic response of porcine PBMCs to vaccination with tetanus toxoid as a model antigen. PLoS One. 2013;8(3): e58306 10.1371/journal.pone.0058306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adler M, Muráni E, Ponsuksili S, Wimmers K. PBMC transcription profiles of pigs with divergent humoral immune responses and lean growth performance. Int J Biol Sci. 2013;9(9): 907–916. 10.7150/ijbs.6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freeman T, Ivens A, Baillie J, Beraldi D, Barnett M, Dorward D, et al. A gene expression atlas of the domestic pig. BMC Biol. 2012;10: 90 10.1186/1741-7007-10-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blecha F, Pollmann D, Nichols D. Weaning pigs at an early age decreases cellular immunity. J Anim Sci. 1983;56(2): 396–400. [DOI] [PubMed] [Google Scholar]
- 27. Le Dividich J, Herpin P. Effects of climatic conditions on the performance, metabolism and health status of weaned piglets: a review. Livest Prod Sci. 1994;38(2): 79–90. [Google Scholar]
- 28. Kauffmann A, Gentleman R, Huber W. Arrayqualitymetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics, 2009;25(3): 415–6. 10.1093/bioinformatics/btn647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107(21): 9546–51. 10.1073/pnas.0914005107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16): 9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Erp-van der Kooij E, Kuijpers A, Schrama J, Ekkel E, Tielen M. Individual behavioural characteristics in pigs and their impact on production. Appl Anim Behav Sci. 2000;66(3): 171–185. [DOI] [PubMed] [Google Scholar]
- 32. Turner S, Roehe R, Mekkawy W, Farnworth M, Knap P, Lawrence A. Bayesian analysis of genetic associations of skin lesions and behavioural traits to identify genetic components of individual aggressiveness in pigs. Behav Genet. 2008;38(1): 67–75. [DOI] [PubMed] [Google Scholar]
- 33. Olsson I, de Jonge F, Schuurman T, Helmond F. Poor rearing conditions and social stress in pigs: repeated social challenge and the effect on behavioural and physiological responses to stressors. Behav Proc. 1999;46: 201–15. [DOI] [PubMed] [Google Scholar]
- 34. Zen M, Canova M, Campana C, Bettio S, Nalotto L, Rampudda M, et al. The kaleidoscope of glucorticoid effects on immune system. Autoimmun Rev. 2011;10(6): 305–10. 10.1016/j.autrev.2010.11.009 [DOI] [PubMed] [Google Scholar]
- 35. Webster Marketon J, Glaser R. Stress hormones and immune function. Cell Immunol. 2008;252(1–2): 16–26. 10.1016/j.cellimm.2008.05.006 [DOI] [PubMed] [Google Scholar]
- 36. Maier S. Bi-directional immune-brain communication: Implications for understanding stress, pain, and cognition. Brain Behav Immun. 2003;17(2): 69–85. [DOI] [PubMed] [Google Scholar]
- 37. Cerletti C, de Gaetano G, Lorenzet R. Platelet—leukocyte interactions: multiple links between inflammation, blood coagulation and vascular risk. Mediterr J Hematol Infect Dis. 2010;2(3): e2010023 10.4084/MJHID.2010.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li N. Platelet-lymphocyte cross-talk. J Leukoc Biol. 2008;83(5): 1069–78. 10.1189/jlb.0907615 [DOI] [PubMed] [Google Scholar]
- 39. Dhabhar F. Stress, leukocyte trafficking, and the augmentation of skin immune function. Ann N Y Acad Sci, 2003;992: 205–17. [DOI] [PubMed] [Google Scholar]
- 40. Wilkie B, Mallard B. Selection for high immune response: an alternative approach to animal health maintenance? Vet Immunol Immunopathol. 1999;72(1–2): 231–5. [DOI] [PubMed] [Google Scholar]
- 41. Rauw W. Immune response from a resource allocation perspective. Front Genet. 2012;3: 267 10.3389/fgene.2012.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sutherland M, Rodriguez-Zas S, Ellis M, Salak-Johnson J. Breed and age affect baseline immune traits, cortisol, and performance in growing pigs. J Anim Sci. 2005;83(9): 2087–95. [DOI] [PubMed] [Google Scholar]
- 43. Geverink N, Parmentier H, de Vries Reilingh G, Schouten W, Gort G, Wiegant V. Effect of response to backtest and housing condition on cell-mediated and humoral immunity in adult pigs. Physiol Behav. 2004;80(4): 541–6. [DOI] [PubMed] [Google Scholar]
- 44. Van Reenen C, O'Connell N, Van der Werf J, Korte S, Hopster H, Jones R, et al. Responses of calves to acute stress: individual consistency and relations between behavioral and physiological measures. Physiol Behav. 2005;85(5): 557–70. [DOI] [PubMed] [Google Scholar]
- 45. Trillmich F, Hudson R. The emergence of personality in animals: the need for a developmental approach. Dev Psychobiol. 2011;53(6): 505–9. 10.1002/dev.20573 [DOI] [PubMed] [Google Scholar]
- 46. Graunke K, Nuernberg G, Repsilber D, Puppe B, Langbein J. Describing temperament in an ungulate: a multidimensional approach. PLoS One. 2013;8(9): e74579 10.1371/journal.pone.0074579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petelle M, McCoy D, Alejandro V, Martin J, Blumstein D. Development of boldness and docility in yellow-bellied marmots. Anim Behav. 2013;86: 1147–54. [Google Scholar]
- 48. Chmielowiec-Korzeniowska A, Tymczyna L, Babicz M. Assessment of selected parameters of biochemistry, hematology, immunology and production of pigs fattened in different seasons. Arch Anim Breed. 2012;55: 469–79. [Google Scholar]
- 49. Geverink N, Heetkamp M, Schouten W, Wiegant V, Schrama J. Backtest type and housing condition of pigs influence energy metabolism. J Anim Sci. 2004;82(4): 1227–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its supporting information files or are deposited in a MIAME-compliant database, the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) (accession numbers: GSE55418).