Abstract
Background
Infectious Leptospira colonize the kidneys of reservoir (e.g. rats) and accidental hosts such as humans. The renal response to persistent leptospiral colonization, as measured by urinary protein biosignatures, has not been systematically studied. Urinary exosomes--bioactive membrane-bound nanovesicles--contain cell-state specific cargo that additively reflect formation all along the nephron. We hypothesized that Leptospira-infection will alter the content of urine exosomes, and further, that these Leptospira-induced alterations will hold clues to unravel novel pathways related to bacterial-host interactions.
Methodology/Principal findings
Exosome protein content from 24 hour urine samples of Leptospira-infected rats was compared with that of uninfected rats using SDS-PAGE and liquid chromatography/tandem mass spectrometry (LC-MS/MS). Statistical models were used to identify significantly dysregulated proteins in Leptospira-infected and uninfected rat urine exosomes. In all, 842 proteins were identified by LC-MS/MS proteomics of total rat urine and 204 proteins associated specifically with exosomes. Multivariate analysis showed that 25 proteins significantly discriminated between uninfected control and infected rats. Alanyl (membrane) aminopeptidase, also known as CD13 topped this list with the highest score, a finding we validated by Western immunoblotting. Whole urine analysis showed Tamm-Horsfall protein level reduction in the infected rat urine. Total urine and exosome proteins were significantly different in male vs. female infected rats.
Conclusions
We identified exosome-associated renal tubule-specific responses to Leptospira infection in a rat chronic colonization model. Quantitative differences in infected male and female rat urine exosome proteins vs. uninfected controls suggest that urine exosome analysis identifies important differences in kidney function that may be of clinical and pathological significance.
Author Summary
Leptospirosis is a bacterial disease commonly transmitted from animals to humans. Though this disease affects more than three quarters of a million people every year and takes a disproportionate toll on the poor in in tropical regions, few virulence factors have been identified and very little is known regarding the pathogenesis of leptospirosis. Symptoms vary from fever and fatigue to severe pulmonary hemorrhage and death. Approximately 5–10% of Leptospira infections in humans are chronic (>1 year) and asymptomatic (no overt signs of disease). Nonetheless, very little is known about the clinical significance of these infections. In this report, we show that non-invasive tools namely proteomic analysis of urinary exosomes can be used to identify differences between healthy and Leptospira-infected rat kidney and between Leptospira-infected male and female rat kidney. In future studies, these analyses will be extended to determine clinical significance and extent of renal dysfunction in the asymptomatic human.
Introduction
Leptospirosis is among the world’s most important zoonotic infectious diseases [1,2], characterized by variable manifestations ranging from asymptomatic or self-resolving acute febrile illness to severe disease with a combination of fever, acute kidney injury, jaundice, severe pulmonary hemorrhage syndrome, refractory shock, and aseptic meningitis [3]. Important advances have been made in diverse aspects of leptospirosis including the differential host responses to leptospiral infection [4–9]. Despite these advances, mechanistic details by which end organ damage develops in some individuals but not in others remain to be elucidated. Further, factors that govern host susceptibility to leptospiral infection are not well understood. A recent study by our collaborative group found that approximately 6% of randomly sampled individuals in a highly endemic rural Amazonian village were chronically colonized by Leptospira without any recent clinical evidence of infection [10]. Although renal colonization by leptospires may occur in humans without serological or clinical evidence of infection, the clinical relevance and functional consequences of leptospiral colonization in humans remain to be characterized. New, less invasive, less expensive and more practical tools (compared to kidney biopsy) are needed to study pathologic changes in the kidney in order to understand clinically relevant sequelae of infection. In this report, we use proteomic analysis of urine exosomes in a rat chronic colonization model as a non-invasive window to kidney function in asymptomatic leptospiral infection.
Exosomes are nanovesicles that are released from cells as a mechanism of intercellular communication [11]. Characterization of exosomes from different biological samples has shown the presence of common as well as cell-type specific proteins. The protein content of exosomes has been shown to be modified under pathological or stress conditions [12–14]. Since exosome contents are specifically derived from cellular components, here we tested the hypothesis that urinary exosome protein content from a rat infected with Leptospira would be different from that of the uninfected rat, and that these differences would hold key information about the pathways mediating host responses to Leptospira infection.
After renal colonization, persistent shedding of Leptospira is clearly established in carrier animal hosts, especially rodents. However, they rarely develop symptoms and are not noticeably impaired by infection of their kidneys [1,15,16]. We have recently detected chronic asymptomatic renal colonization by Leptospira in human subjects from a rural Amazonian village [10]. We reasoned that the structural and functional changes in the kidney that arise following asymptomatic Leptospira infection are different from symptomatic disease. These differences between the asymptomatic and symptomatic leptospirotic kidneys can be understood by studying the downstream products of the kidney, such as urine.
Given the nephron cell-state-specific cargo of the urinary exosome, we hypothesized that urine exosome analysis holds key information that is relevant to differences between clinically symptomatic and asymptomatic leptospirosis infection. This report is the first step towards testing this hypothesis. Here we report our preliminary findings from the exosome proteomic analyses of urines from rats infected with Leptospira using uninfected rat urine exosome as controls. We also studied the host-response to Leptospira infection in male and female rats separately.
Materials and Methods
Ethics statement
This work was approved by the Institutional Animal Care and Use Committee of the University of California, San Diego.
Experimental infection of animals and study design
Three-week old Sprague Dawley rats (Charles River Laboratories, USA) (6 male rats and 6 female rats) were housed in cages of 1–2 rats each, with food and water provided ad libidum. Six animals (3 males and 3 females) were inoculated intraperitoneally with 108 mid-log phase L. interrogans serovar Copenhageni strain HAI1026. Uninfected controls were inoculated ip with sterile EMJH. To determine the health status, weight, general body condition, posture, activity, appetite food and water consumption of each animal was monitored daily. Infection was confirmed by serology (microscopic agglutination test) and quantitative polymerase chain reaction (qPCR) of weekly urines, and Warthin-Starry silver staining of kidney sections following necropsy. In order to keep the minimum number of animals for the purpose of statistical calculations, we used this small number of animals. This is also discussed as a limitation of the study under discussion.
Urine sampling and processing
Urine from each animal was collected weekly starting 7 days after post-challenge by placing the animals individually for 20–24 hrs in metabolic cages, and was captured into containers containing Roche Complete Protease Inhibitor, one tablet per 5 mL urine. Urines were separately centrifuged at 3000 x g for 30 min. The supernatant was withdrawn, the pH adjusted to 7, aliquoted and frozen at -70°C until further analysis.
Genomic DNA was extracted from these weekly urines and qPCR was used to assess leptospiruria. In addition, at necropsy, kidneys were harvested, fixed in formalin, paraffin embedded, and infection confirmed by Warthin-Starry silver stain (S1 and S2 Figs.).
Exosome preparation and protein resolution analysis
Exosomes from terminal urine samples from infected and uninfected rats were prepared using an in-house protocol developed based on the solvent exclusion principle using polyethylene glycol (PEG)-induced precipitation. To prevent naturally occurring peptides in the exosome from confounding post in-gel trypsinization peptide information of the full-length proteins, we conducted 1 dimensional SDS-PAGE of the exosome proteins prior to in-gel trypsinization.
Proteomic analysis
Each rat urine sample (only terminal sample) was separately analyzed, without pooling any sample. Each rat urine sample was run in a separate gel lane. Gel slices for each lane were cut to 1 mm x 1 mm cubes and destained 3 times by first washing with 100 μL of 100 mM ammonium bicarbonate for 15 min, followed by addition of the same volume of acetonitrile (ACN) for 15 min [17]. The supernatant was transferred to a clean tube and samples lyophilized and reduced by mixing with 200 μL of 100 mM ammonium bicarbonate-10 mM DTT then incubated at 56°C for 30 min. The liquid was removed and 200 μL of 100 mM ammonium bicarbonate-55mM iodoacetamide was added to gel pieces, which were then incubated at room temperature in the dark for 20 min. After removal of the supernatant and one wash with 100 mM ammonium bicarbonate for 15 min, an equal volume of ACN was added to dehydrate the gel pieces. The solution was then removed and samples lyophilized.
For digestion, ice-cold trypsin (0.01 μg/μL) in 50 mM ammonium bicarbonate solution was added in enough amounts to cover the gel pieces and set on ice for 30 min. After complete rehydration, the excess trypsin solution was removed, replaced with fresh 50 mM ammonium bicarbonate, and left overnight at 37°C. The peptides were extracted twice by the addition of 50 μl of 0.2% formic acid and 5% ACN and vortexed at room temperature for 30 min. The supernatant was removed and saved. A total of 50 μL of 50% ACN-0.2% formic acid was added to the sample, which was vortexed again at room temperature for 30 min. The supernatant was removed and combined with the supernatant from the first extraction. The combined extractions from all the gel slices in a lane pertaining to a single rat urine exosome sample was separately analyzed directly by liquid chromatography (LC) in combination with tandem mass spectroscopy (MS/MS) using electrospray ionization. Thus, multiple slices of gels representing a single rat urine exosome sample was used for mass spectrometry. Two replicates per rat exosome sample were run on the MS.
Trypsin-digested mixtures were analyzed by the Eksigent nanoLC-Ultra 2D System (Eksigent, AB SCIEX Dublin, CA, USA) combined with cHiPLC-nanoflex system (Eksigent) in trap-elute mode. Briefly, samples were first loaded on the cHiPLC trap (200 μm x 500 μm ChromXP C18-CL, 3 μm, 120 Å) and washed in isocratic mode with 0.1% aqueous formic acid for 10 min at a flow rate of 3 μL/min. The automatic switching of cHiPLC ten-port valve then eluted the trapped mixture on a nano cHiPLC column (75 μm x 15 cm ChromXP C18-CL, 3 μm, 120 Å), through a 45 min gradient of 5–50% of eluent B (eluent A, 0.1% formic acid in water; eluent B, 0.1% formic acid in acetonitrile), at a flow rate of 300 nL/min. To preserve system stability, in terms of elution times of components, trap and column were maintained at 35°C.
Mass spectra were acquired using a QExactive mass spectrometer (Thermo Fisher Scientific, San José, CA, USA), equipped with a nanospray ionization source (Thermo Fisher). Nanospray was achieved using a coated fused silica emitter (New Objective, Woburn, MA, USA) (360 μm o.d./50 μm i.d.; 730 μm tip i.d.) held at 1.5 kV. The ion transfer capillary was held at 220°C. Full mass spectra were recorded in positive ion mode over a 400–1600 m/z range and with a resolution setting of 70000 FWHM (@ m/z 200) with 1 microscan per sec. Each full scan was followed by 7 MS/MS events, acquired at a resolution of 17,500 FWHM, sequentially generated in a data dependent manner on the top seven most abundant isotope patterns with charge ≥2, selected with an isolation window of 2 m/z for the survey scan, fragmented by higher energy collisional dissociation (HCD) with normalized collision energies of 30 and dynamically excluded for 30 s. The maximum ion injection times for the survey scan and the MS/MS scans were 50 and 200 ms and the ion target values were set at 106 and 105, respectively.
Data management
All data generated were searched using the Sequest search engine contained in the Thermo Scientific Proteome Discoverer software, version 1.4. The experimental MS/MS spectra were correlated to tryptic peptide sequences by comparison with the theoretical mass spectra obtained by in silico digestion of the Rattus norvegicus protein database downloaded January 2013 from the National Centre for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov). The following criteria were used for the identification of peptide sequences and related proteins: trypsin as enzyme; three missed cleavages per peptide were allowed and mass tolerances of ± 50 ppm for precursor ions and ± 0.8 Da for fragment ions were used. Validation based on separate target and decoy searches and subsequent calculation of classical score-based false discovery rates (FDR) were used for assessing the statistical significance of the identifications. Finally, to assign a final score to proteins, the SEQUEST output data were filtered as follows: 1,5; 2.0; 2.25 and 2.5 were chosen as minimum values of correlation score (Xcorr) for single-; double-; triple- and quadrupole-charged ions, respectively. A high stringency was guaranteed using parameters previously described [18] and the false-positive peptide ratio, calculated through a reverse database, was less than 3%. The output data, protein lists, obtained from the Sequest search were compared using an in-house software, namely, the Multidimensional Algorithm Protein Map (MAProMa) [19].
Normalized spectral abundance factor of the identified urine exosome proteins
The relative abundance of polypeptides was calculated from the normalized spectral abundance factor (NSAF) using the method of Paoletti et al [20] taking into consideration the number of peptides as well as the length of the polypeptide contributing to their respective abundance. To enable comparison of samples from a subject across different time points or across different groups, each animal’s total proteome was normalized to 1. Subsequently the relative contribution of each protein from a given animal from a group was expressed as percentage of the total. Peptide numbers corresponding to a protein were thus more of raw data nature whereas the NSAF number included the peptide number as well as the total length of the protein. The peptide counts data were log-transformed prior to analysis by multivariate partial least squares discriminant analysis (PLSDA), and univariate 1-way ANOVA with unpaired comparisons, Variable Importance in Projection analysis and post hoc correction by Wilcoxon Rank test in MetaboAnalyst [21].
Data processing and statistical analysis
In all analyses, p ≤ 0.05 was considered statistically significant. Analyses including Student’s t-tests, Partial-Least Squares Discriminant Analysis (PLS-DA) and variable importance in projection (VIP) were performed with the MetaboAnalyst 2.0 web portal (www.metaboanalyst.ca) [21]. To reduce systematic variance and to improve the performance for downstream statistical analysis normalization and transformation of raw data were performed before the t-tests, PLS-DA and VIP analysis. Normalization by sum of the spectral count as mentioned previously was used to overcome the variance between the analyzed samples. To make each feature comparable in magnitude to each other, data were transformed by taking the natural log of the concentration values of the analyzed proteins. The data were additionally auto-scaled (mean-centered and divided by the standard deviation of each variable).
Univariate analysis was used to check the differences in the concentrations of the analyzed exosome protein spectral count between the control and infected rat urine samples. The infected rat urines were also split into male rat and female rat categories. Paired Student’s t-test was applied to examine each variable (ratio of individual protein to total concentration in each group considered).
PLS-DA and VIP were used both for the classification and significant feature selection. A VIP plot, which is commonly used in PLS-DA, ranks proteins based on their importance in discrimination between the urinary exosomes from infected and the uninfected rats. The VIP score is a weighted sum of squares of the PLS loadings. The amount of explained Y-variance in each dimension influenced the weights [22]. Protein candidates with a false discovery rate (FDR) of ≤10% were qualified for subsequent validation by Western immunoblotting.
Western immunoblotting and quantification
Antibody against alanyl aminopeptidase was purchased from Proteintech Group, Inc., (Chicago, IL, USA). The THP antibody was from Sigma Chemical Co (St. Louis, MO, USA). HRP-conjugated secondary antibody was from GE Life Sciences (Piscataway, NJ, USA). SDS-PAGE gels (with 10% acrylamide) were used to resolve 100 μg of protein either from exosomes or total urines of male and female rats infected with L. interrogans serovar Copenhageni. After separation the proteins were transferred to nitrocellulose paper, blocked, and incubated with primary antibody overnight before washing with Tris-buffered saline, incubation for 1 h with HRP-secondary antibody conjugate and visualized by developing as described in previous publications from our laboratory [23,24]. The quantification of the Western immunoblot bands was performed using Image J software (NIH) as previously described [24], and plotted using Graphpad Prism software (San Diego, CA, USA).
Results
Urine protein content is different in Lepto-infected and uninfected control rats
Rat urine samples were analyzed for overall protein identification by a combination of SDS-PAGE and mass-spectrometry. We found that the infected rat urine shows an overwhelming increase in both quality and quantity of the protein content, as shown in Panel A of Fig. 1 (156 versus 503 proteins unique to uninfected versus infected urines). A total of 842 proteins were detected, with further classification of subgroups in the infected animals based on gender as shown in Panel B of Fig. 1. In total, 842 proteins were identified from the total urine animals with distribution as shown in Fig. 1 Panel A and B). Importantly, 180 proteins and 272 proteins were unique to the urines of female and male rats infected with L. interrogans serovar Copenhageni respectively, as compared to 156 proteins unique to the uninfected rat urines. These differences in the composition and quantity of proteins from infected and uninfected rats potentially indicates the reactions induced in these animals by leptospiral infection. The identity of each of these proteins is as given in S1 Table.
Fig 1. Panel A: Venn diagram summarizing the total urine proteomics data.
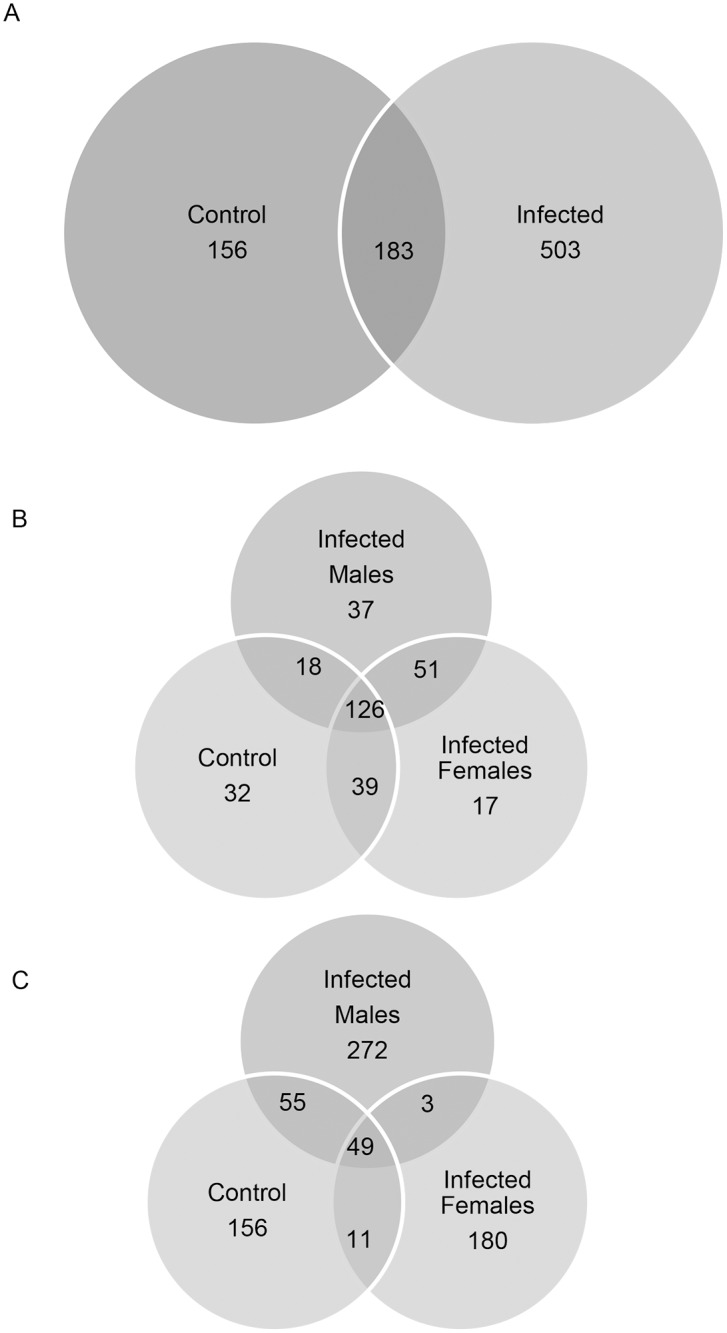
A total of 842 proteins were identified by LC/MS-MS of the rat urines. The Leptospira-infected and uninfected control rat urine samples were used for generating this 2-way split between control rats and infected rats. Panel B: Venn diagram summarizing the total urine proteomics data. The Leptospira-infected male, female and uninfected control rat urine samples were used for generating this 3-way split among control rats, infected male and infected female rats. Panel C: Venn diagram summarizing the urine exosome proteomics data. A total of 204 proteins were identified by LC/MS-MS of the rat urine exosome proteins resolved by 1-d gel electrophoresis and trypsinization of the gel slices.
Urine exosomes from Leptospira-colonized rats show different protein constitution compared to uninfected control rats
Given that the urine exosomes reflect intracellular milieu of all types of various cells lining the nephron in kidney, and emerging evidence from literature that kidney functional alterations are induced due to leptospiral infection, we next focused on exosomes. We found that a total of 204 exosome proteins were identified classifiable into 7 different groups as noted in S2a-S2g Table and summarized in the Venn diagram (Panel C, Fig. 1). The exosome protein constitution also showed increase in the number of proteins expressed in the exosome, viz 32 proteins uniquely present in the uninfected exosome versus 57 unique proteins in the infected rat exosome.
We further conducted the multivariate partial least squares-discriminant analysis (PLS-DA) on these proteins. The analysis depicted in Fig. 2 shows clear separation between uninfected and infected rat urine exosome protein content, suggesting the differences between uninfected and infected rats.
Fig 2. Two-dimensional (2D) partial least squares discriminant analysis separation using protein peptide count concentration-based proteomic measurements in the urine exosome of rats infected with Leptospira vs control rats without Leptospira infection (n = 3).
Clear separation of rat urine exosome proteins for control versus infected is observed, signified by the lack of overlap between the two groups of exosome proteins.
Sex-specific alteration of the rat urine exosome protein content in relation to leptospiral colonization
Urinary exosome proteins in male vs. female infected rat urine were different as determined by PLS-discriminant analysis (Fig. 3). Moreover, the infected male rat urine exosome contents were far more different from those of uninfected rats compared to the infected female rat urine exosome contents. The VIP (Variable Importance in Projection) score of 25 proteins was higher than 1.5 (Fig. 4 and Table 1). Qualitatively, a total of 57 proteins were present among all the infected rats. Of these, only 3 were shared between infected males and females, while 37 were unique to infected males and 17 were unique to infected females. Further, we conducted separate analyses of proteins between proteins of proteins of male infected and female infected rats. Table 2 depicts male infected versus control rat urine exosome analysis. Accordingly, 11 proteins were significantly altered (p < 0.05). Table 3 depicts female infected versus control rat urine exoosme analysis, according to which the number of significantly dysregulated proteins was 9. In the male infected rat urine exosome, the alanyl (membrane) aminopeptidase upregulation not only reached the highest level of significance (p = 0.00019) but also had the lower FDR (3.22%). In the female infected rat however, although this upregulation was significant compared to the uninfected rat, the FDR did not reach the cutoff of <10% (14.37%). Thus both qualitatively and quantitatively, the protein content of exosomes showed gender specificity in infected rats.
Fig 3. Two-dimensional (2D) partial least squares discriminant analysis separation using peptide count concentration-based proteomic measurements in the urine exosome of the rats infected with Leptospira vs control rats without Leptospira infection (n = 3).
Although clear separation between control and infected rat urine exosomes is seen, a fraction of overlap between the infected male and infected female rat urine exosome proteins is observed.
Fig 4. Variable importance in projection (VIP) plot: important features (analyzed serum free amino acids) identified by PLS-DA in a descending order of importance.
The graph represents relative contribution of proteins to the variance between the Leptospira-infected and uninfected control rat urine exosomes. High value of VIP score indicates great contribution of the proteins to the group separation. The green and red boxes on the right indicate whether the protein concentration is increased (green) or decreased (red) in the exosome of the infected rat urine vs. uninfected rat urine samples. For higher n value, a VIP score of 1.5 is considered to enable discrimination between 2 phenotypes. Even with the low n (= 3) per group that is employed in this study, the VIP score of the top 3 proteins is higher than 3, increasing the confidence. Alanyl (membrane) aminopeptidase, also called CD13 is the top protein with a VIP score of 5.72.
Table 1. Top discriminators between control, infected male & infected female rat urine exosome proteins.
| GI # | VIP score | Protein Identification | Serial # |
|---|---|---|---|
| 149057276 | 5.7252 | alanyl (membrane) aminopeptidase | VIP1 |
| 13591914 | 3.7462 | aminopeptidase N precursor | VIP2 |
| 601865 | 3.1009 | aminopeptidase M | VIP3 |
| 392351673 | 2.6881 | PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC287750 | VIP4 |
| 11024674 | 2.1973 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | VIP5 |
| 6981210 | 2.1961 | Neprilysin | VIP6 |
| 25742748 | 2.1827 | glutamate—cysteine ligase catalytic subunit | VIP7 |
| 13592133 | 2.1177 | actin, cytoplasmic 1 | VIP8 |
| 392339909 | 2.0721 | PREDICTED: uncharacterized protein LOC679818 | VIP9 |
| 257467627 | 2.0255 | Fc fragment of IgG binding protein-like precursor | VIP10 |
| 51980580 | 1.9156 | Meprin 1 beta | VIP11 |
| 157817658 | 1.9135 | villin-1 | VIP12 |
| 133777069 | 1.8884 | Serine (or cysteine) peptidase inhibitor, clade A, member 3K | VIP13 |
| 6978769 | 1.8561 | deoxyribonuclease-1 precursor | VIP14 |
| 293348969 | 1.7392 | PREDICTED: keratin, type II cytoskeletal 6A-like | VIP15 |
| 47087085 | 1.6273 | keratin, type I cytoskeletal 17 | VIP16 |
| 13928998 | 1.6155 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | VIP17 |
| 57012352 | 1.6082 | keratin, type II cytoskeletal 75 | VIP18 |
| 38303863 | 1.5999 | Egf protein | VIP19 |
| 530167 | 1.5913 | ventral prostate-specific protein | VIP20 |
| 149022164 | 1.5687 | rCG26871, isoform CRA_c | VIP21 |
| 56013 | 1.5602 | CRP2 | VIP22 |
| 149015740 | 1.5588 | rCG39189, isoform CRA_b | VIP23 |
| 158186649 | 1.5384 | alpha-enolase | VIP24 |
| 119959830 | 1.5203 | beta-actin | VIP25 |
Table 2. Male infected versus Control.
| sl # | gi # | p.value | FDR | % FDR | protein identity |
|---|---|---|---|---|---|
| 1 | 149057276 | 0.00019 | 0.032231 | 3.2231 | alanyl (membrane) aminopeptidase |
| 2 | 13592133 | 0.000636 | 0.053409 | 5.3409 | actin, cytoplasmic 1 |
| 3 | 133777069 | 0.000943 | 0.053409 | 5.3409 | Serine (or cysteine) peptidase inhibitor, clade A, member 3K |
| 4 | 32563565 | 0.024998 | 0.60267 | 60.267 | Serine protease inhibitor |
| 5 | 392339909 | 0.025984 | 0.60267 | 60.267 | PREDICTED: uncharacterized protein LOC679818 |
| 6 | 257467627 | 0.027061 | 0.60267 | 60.267 | Fc fragment of IgG binding protein-like precursor |
| 7 | 149028871 | 0.029327 | 0.60267 | 60.267 | annexin A2, isoform CRA_b |
| 8 | 89573967 | 0.031305 | 0.60267 | 60.267 | isocitrate dehydrogenase 1 |
| 9 | 10198602 | 0.032149 | 0.60267 | 60.267 | collectrin precursor |
| 10 | 392354923 | 0.037988 | 0.60267 | 60.267 | PREDICTED: glyceraldehyde-3-phosphate dehydrogenase-like |
| 11 | 149061924 | 0.041715 | 0.60267 | 60.267 | rCG48611, isoform CRA_c |
Proteins significantly dysregulated between exosomes from urines of control and male infected rats.
Table 3. Female infected versus Control.
| sl # | gi # | p.value | FDR | % FDR | protein identity |
|---|---|---|---|---|---|
| 1 | 58865770 | 0.0001 | 0.00983 | 0.9837 | solute carrier family 7 member 13 |
| 2 | 13592133 | 0.010362 | 0.1437 | 14.37 | actin, cytoplasmic 1 |
| 3 | 149057276 | 0.010425 | 0.1437 | 14.37 | alanyl (membrane) aminopeptidase |
| 4 | 57012358 | 0.010506 | 0.1437 | 14.37 | keratin, type II cytoskeletal 73 |
| 5 | 11024674 | 0.010675 | 0.1437 | 14.37 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 |
| 6 | 57231 | 0.010759 | 0.1437 | 14.37 | unnamed protein product |
| 7 | 119959830 | 0.01163 | 0.1437 | 14.37 | beta-actin |
| 8 | 25742748 | 0.01173 | 0.1437 | 14.37 | glutamate—cysteine ligase catalytic subunit |
| 9 | 347800746 | 0.049478 | 0.34739 | 34.739 | serine protease inhibitor A3K precursor |
Proteins significantly dysregulated between exosomes from urines of control and female infected rats.
Of interest, the top discriminator between control and infected rats, namely alanyl aminopeptidase of the membrane origin,is also known as aminopeptidase neutral (APN) or CD13 [25,26]. Furthermore, CD13 also shows different levels of dysregulation between infected male and infected female rats (Table 1).
The urinary exosome membrane alanyl aminopeptidase or CD13 is a marker of infection
A separate analysis of the proteins dysregulated between control and infected rats showed that 11 proteins were significantly different in urinary exosomes (p <0.05, Table 4). However, only one protein had an FDR of <10%, namely alanyl (membrane) aminopeptidase, also known as CD13. By both multivariate analysis of PLS-DA (Fig. 4) and univariate ANOVA, CD13 is significantly upregulated in the infected rat urine exosome. When the analysis was conducted without gender specificity (Table 4), only CD13 showed an FDR of <10% among these 11 dysregulated proteins. Taking into account gender specificity (Table 2) CD13 was not only significantly upregulated, but also had an FDR of <10%. However, in females, this protein was only significantly upregulated but did not reach the FDR cutoff (Table 3).
Table 4. Proteins significantly dysregulated between exosomes from urines of control rats and infected rats.
| sl # | gi # | p.value | FDR | % FDR | protein identity |
|---|---|---|---|---|---|
| 1 | 149057276 | 3.76E-06 | 0.00076 | 0.0767 | alanyl (membrane) aminopeptidase |
| 2 | 601865 | 0.010627 | 0.55605 | 55.605 | aminopeptidase M |
| 3 | 20806135 | 0.012682 | 0.55605 | 55.605 | galectin-3-binding protein precursor |
| 4 | 133777069 | 0.01489 | 0.55605 | 55.605 | Serine (or cysteine) peptidase inhibitor, clade A, member 3K |
| 5 | 119959830 | 0.023672 | 0.55605 | 55.605 | beta-actin |
| 6 | 57012358 | 0.037372 | 0.55605 | 55.605 | keratin, type II cytoskeletal 73 |
| 7 | 392339909 | 0.038201 | 0.55605 | 55.605 | PREDICTED: uncharacterized protein LOC679818 |
| 8 | 257467627 | 0.039191 | 0.55605 | 55.605 | Fc fragment of IgG binding protein-like precursor |
| 9 | 149028871 | 0.041253 | 0.55605 | 55.605 | annexin A2, isoform CRA_b |
| 10 | 89573967 | 0.043033 | 0.55605 | 55.605 | isocitrate dehydrogenase 1 |
| 11 | 347800746 | 0.048623 | 0.55605 | 55.605 | serine protease inhibitor A3K precursor |
The VIP scores of proteins identified in the rat urine exosomes shows CD13 to be a top discriminant between infected and uninfected rat urine exosome (Table 1). Twenty-five proteins had a VIP score of >1.5, fulfilling criteria to classify a protein as a reliable discriminant. Our analysis shows that this value 5.72 for CD13.
Further, Western immunoblotting of the protein showed that the exosome content of CD13 closely tracked the proteomic data, with a slight increase in the infected female urine exosome, and robust increase in the infected male urine exosome (Fig. 5a). The observed difference was significant (Fig. 5b).
Fig 5. A. Immunoblotting of rat urine exosome for CD13 protein.
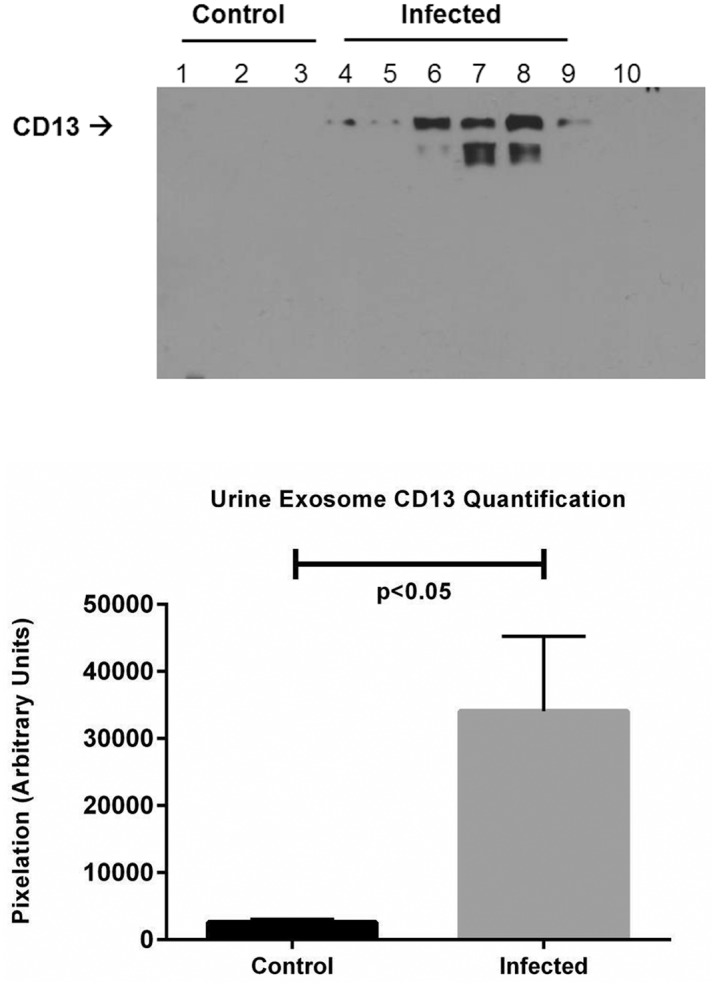
Lanes 1–3: control; lanes 4–6: lepto infected female; lanes 7–9: infected male. B. Quantification of CD13 from immunoblots in A (control, n = 3; infected, n = 6). Data are means ± SEM. p<0.05 rat urine exosome CD13 Leptospira- infected versus uninfected control.
Tamm-Horsfall Protein is significantly decreased in the infected rat urine
Given the tubular location of CD13, we tested the hypothesis that other proteins reflecting the tubular function may be affected in response to leptospiral infection. We chose to study the most abundant protein in the normal urine, namely the Tamm-Horsfall Protein (THP). Western immunoblotting of THP in urine of infected and uninfected rats showed robust THP expression in the uninfected rats and significantly lower THP in infected male and female rats, supporting this hypothesis (Fig. 6).
Fig 6. A. Immunoblotting of rat urines for Tamm-Horsfall Protein (THP).
Left panel Lanes (1–3): Control Uninfected Rat urines; Right panel Lanes (1–6): Leptospira-Infected Rat urines. B. Quantification of THP from immunoblots in A (control, n = 3; infected, n = 6). Data are means ± SEM. p<0.05 rat urine THP, Leptospira-infected versus uninfected control.
Discussion
This proteomic analysis of urinary exosomes in a rat leptospiral colonization model identified important biomarkers of infection, including differentially regulated alanine (membrane) aminopeptidase (CD13) and Tamm Horsfall Protein. Further, there were important sex differences of these biomarkers of leptospiral renal tubular colonization, which is particularly provocative given the male predominance of symptomatic and severe leptospirosis universally found in human clinical studies.
In addition, this study has several other important findings. First, to our knowledge this is the first report on urinary exosome protein analysis in an animal model of leptospiral renal colonization. Second, the overwhelming increase in protein expression quality and quantity indicates a specific consequence of leptospiral infection of the organ of leptospiral transmission and provides an important rationale for pursuing such studies in humans that will allow for further understanding of the clinical and biological consequences of chronic renal colonization. Finally, although renal tubular involvement in leptospiral infection has been well-documented, this study is the first to report increased CD13 and decreased Tamm-Horsfall Protein in infection. Although apparently intuitive and in agreement with the published literature in regard to renal tubular involvement in leptospirosis infection, the biological and clinical significance of these findings remains to be determined further.
Leptospirosis is a globally important tropical infectious disease that takes a disproportionate toll in tropical regions [1]. Important gaps remain in translating the know how to reduce the burden of this infectious disease and its pathogenic mechanisms remain poorly understood [4].
Multiple hypothesis-driven studies as well as unbiased approaches have previously focused on unraveling leptospirosis mechanisms [5,6,27,28] [29–31] have considerably advanced our knowledge of the involvement of renal [32–39], pulmonary hemorrhage [40,41] or cardiovascular system components [42] in leptospirosis. However, the host renal functional changes due to Leptospira infection are yet to be completely characterized. Given the renal failure rates [29,40,43–48] following leptospirosis, exploring these renal consequences is clinically important. In this study, we found that the infected rat urine shows an overwhelming increase in both quality and quantity of the protein content compared to the uninfected rat urine protein both at the total urine as well as the exosome level.
Importantly, urine exosome CD13 upregulation due to leptospiral infection is robust and consistent, in both male and female rats. Previous work has demonstrated expression of CD13/APN on intestinal and kidney epithelial cell brush border [25,49–52]. CD13 is an ectoenzyme with a multitude of functions: (a). enzymatic cleavage of various peptides such as enkephalins, angiotensins or cytokines [53,54] and their biologic process regulation [55]; (b). marker of differentiation for multiple types of immune cells [56,57] and stem cells [58]; (c). various cellular processes such as proliferation [59] and apoptosis [60], motility [61], chemotaxis [62]; (d). critical role in immunogenic, inflammatory and infection pathways such as antigen processing [63,64] and presentation [65], and phagocytosis in defense against pathogens [66], viral receptor on the host cell surface and the subsequent endocytosis of viruses into cell interior [67–71]. Soluble CD13/APN that circulates in the serum is repeatedly reported to be upregulated at inflammation sites [55,72]. Since CD13 represents a cellular potential for activation or inactivation of inflammatory peptides, modulation of its expression by different agents and stimuli may affect inflammatory and immunologic responses [64,73] and antigen processing [63–65].
Recent evidence [56] shows that CD13 is a multifunctional enzyme with at least 3 different types of activities: (a) peptide cleavage, (b) endocytosis, and (c) signaling. In light of our proteomic data which shows robust and reproducible CD13 upregulation in response to leptospirosis infection, it is possible that either one or all three of these CD13 functionalities are employed. Future studies should further delineate how much or if these functions play a role in mediating specific components of host response to leptospirosis infection. This multifunctional property notwithstanding, so far, CD13/APN activity or its upregulation has not been described specifically in the context of Leptospira infection. Nevertheless, relevant to our current findings, a role for the upregulation of APN expression and activity in the membrane is conceivable, and the following features / properties lend this multifunctional enzyme to potentially play a role in Leptospira infection/invasion: the viral receptor function of APN [52,69,74] either requires or results in its internalization [55,71]. Previously, the APN/CD13 molecule has been shown to be the predominant component of the detergent-resistant membrane (DRM) microdomains, also called lipid rafts [71]. CD13/APN is shown to play a critical role in appropriate membrane reorganization and protein distribution after a major reshuffling event [75], and is itself is shown to be localized in lipid rafts of the cell membranes [76,77] and endocytosed through physiological sorting mechanisms [57]. At the time of these reports, the concept of exosomes was not as well-established as it is now. The course taken by regular physiologic endocytosis during internalization of specific viruses [69], the role of CD13/APN as the cellular receptor to facilitate this internalization [70] and its critical dependency on the early edosome-formation [71] and subsequent intracellular sub-plasma membrane molecular events bear close resemblance to the formation of exosomes in the renal tubular epithelial cell and their release into urine, the extracellular milieu modeled by the Knepper group [78]. Our proteomic data shows that APN is the most consistently and significantly upregulated protein molecule on the urine exosome of the infected rat. Renal tubular brush border involvement in leptospirosis infection has been well established in animal models showing leptospiral antigen expression in parallel to kidney changes both by light microscopy and electron microscopy examination [79,80]. Further reinforcement of this view comes from the recent evidence that during phagocytosis, CD13 redistribution to phagosome and further internalization increases the efficiency of the process of phagocytosis.
In view of these previous observations and our proteomic data, mechanistically, it is plausible that the host immune system is activated by leptospiral antigens presented to the host immune system. APN located on the plasma membrane of the renal tubular epithelium brush border would be upregulated due to Leptospira infection; Though its exact mechanism and its details remain to be elucidated, taken together with the literature our data suggest that APN release from brush border via exosomes in infectious condition, may have been part of the host immune response to leptospiral infection, since it is well-established that exosomes are generated from membranes via endocytic mechanisms [81] and further, that under stress, the exosome cargo changes qualitatively [12,13]. However, if leptospiral infection directly causes increased APN packaging into exosome and its subsequent release needs to be addressed in future. It is plausible that due to the stress of leptospiral infection, renal tubular epithelium mediated exosome packaging is qualitatively altered to include APN. We also found that female rats’ response to Leptospira infection is markedly different from that of the male rat. This has important implications and is relevant from the viewpoint of our finding that among asymptomatic human carriers of Leptospira, male and female subjects show different phenotypes [10]. However see below, details on the limitations of using or extrapolating the chronic rodent model to an acute infection or a different species such as humans.
The proteomic data also suggested that the renal functions mapping to tubular epithelium may be affected in the Leptospira infection. Therefore we tested the hypothesis that the expression of the most abundant protein in the normal healthy urine, Tamm-Horsfall Protein (THP) that is exclusively synthesized in renal tubules should be affected in Leptospira infected rats. THP (also called uromodulin) is secreted by the epithelium of thick ascending loop of Henle and the early portion of the distal tubules [82]. One of the phenotypes that a THP knockout mouse model develops is the increased susceptibility to urinary tract infection caused by bacteria [83,84]. THP is a glycosylphosphatidylinositol anchored glycoprotein released from epithelial apical membranes (approximately 120 kDa.) into the tubule fluid by membrane associated phospholipase C [85,86]. Free (urinary) THP has a monomeric weight of approximately 85 kDa with 30% carbohydrate. Our data (Fig. 6) shows that THP expression is reduced in all infected rats. Taken together with the previous work, our data strongly indicate that the tubular epithelial component responsible for THP-synthesis and expression is severely compromised in leptospirosis.
This study design had several limitations. First, the rat model used for our studies is the chronic infection animal model whereas human leptospirosis infection often results in acute infection episodes and often death. The elements of either acute leptospiral infection animal model or human leptospiral infection may yield very different results from those obtained for the model reported here. Even the direction of up/down regulation of significantly different proteins need not necessarily be the same as these chronic animal model findings. However, the methodology that we have employed here namely exosomal analysis, can be eminently applied to study any other model including the acute phases of leptospiral infection in humans or to test the hypothesis that acute infection results in differences in the urine exosome that can be tracked and understood. Secondly, the number of animals analyzed per group is small, reflecting the intensity of this large scale analysis. A higher n value per condition may have increased the robustness of these data further and brought the CD13 differences between uninfected and infected female rats too, to an acceptable FDR of <10%. However this small value of n notwithstanding, these data as such are robust. The difference in protein numbers between infected and uninfected rat urine exosome is remarkably large that it imparted the highest VIP score to CD13. Thirdly, some of the other proteins that feature lower VIP score may have had higher VIP scores by increasing the number of animals. This potentially would have increased our knowledge of what other novel pathways are selected/employed by either leptospires or the host. Finally, we used 1d gel electrophoresis to resolve the exosome proteins prior to LC/MS-MS analysis that resulted in identification of 204 proteins overall. If we had conducted direct exosome protein trypsinization instead of following this method, perhaps the number of identified proteins might have increased. In our experience, exosomes are packaged with non-full length peptides as well, and may have confounded the analysis. By following the gel electrophoresis method, we ensured that we only compare full length protein differences between animals.
In summary, we have characterized host response to leptospiral infection at proteome levels of whole urine and exosomes. Urine is perhaps the most frequently studied biological fluid for exosome proteomics and also one of the most promising biomarker source. Work from the Knepper group shows that many important renal proteins (e.g. aquaporins, polycystins and podocyn) are shed in the urine exosome [87,88]. Our current report adds APN to this group of functionally important renal proteins to be identified in urine exosomes. Our findings suggest that the host response to Leptospira infection is gender-specific, and involves renal tubular elements. This report potentially forms the preliminary level basis for our overarching hypothesis that changes in the kidney function as a result of an external insult can be determined using urine exosome analyses.
Conclusions and future directions
Many conclusions can be drawn from this study:
Although the general quantity of proteins in total urine and exosome component show a similar trend, qualitatively the proteins are different based on identifications, the GI numbers and hence the pathways they belong to.
- Utility of exosome Analysis in characterizing a complex biochemical phenomenon such as the host-response to leptospirosis infection is very high, and has many implications:
- Public health interest: given that nephron-cell specific cargo that urine exosome carries, primarily the pathway employed or dysregulated for infecting the host to bring about the infection phenotype, and secondly, the response that is unique to each host studied potentially opens up the possibilities of disease-specific and severity-specific treatment to leptospirosis.
- Non-invasive: it would be ideal to biopsy every infected individual to understand the various facets of leptospirosis infection in order that our knowledge about Leptospira increases. However, it is neither practical nor ethical, given the invasive nature. Urine exosome analyses is non-invasive, highly specific and provides a window into the organ level structural or functional changes. If implemented quickly enough in a translational research setting, this specificity potentially imparts ability to move the field towards personalized medicine.
- Gender-specific host responses to disease onset: our data shows that this methodology of studying differences between male and female host response to infection can be applied in the setting of human disease. Although we should expect different molecules other than CD13 to by dysregulated, the individual-specific pathways mediating different magnitude responses to the same infection can be accurately measured using exosome markers that are either surrogately or directly linked to a particular host-response event. This assumes importance especially in the setting of sub-clinical leptospirosis infection where in the patient does not display any clinical features/symptoms but continues to play host to the bacterium.
Supporting Information
(DOCX)
(TIF)
(TIF)
All urine samples collected were screened for the presence of pathogenic and intermediate-pathogenic Leptospira using a published qPCR TaqMan assay targeting the leptospiral 16S ribosomal gene (1); this assay has been reported in our previous work. Briefly, this was performed using an Opticon 2 real-time PCR machine (MJ Research, USA). The assay protocol was modified from the published version (2) by using the fluorescent probe at a final concentration of 0.2 mM, primers at a final concentration of 0.5 mM, and a 20 mL reaction volume (3). Standard curves for quantification were made using Leptospira interrogans serovar Copenhageni strain M20. Standards were prepared as follows. Leptospires were counted using a Petroff-Hauser counting chamber (Hauser Scientific, USA) and serially diluted with sterile double-distilled H2O to 108 to 100 leptospires/ ml. Genomic DNA was subsequently prepared using the DNeasy Tissue Kit (Qiagen, USA). Standards were run in triplicate to generate a standard curve with each run. A negative result was assigned where no amplification occurred before 40 cycles. Controls lacking template were extracted and added to qPCR master mix to detect the presence of contaminating DNA. The raw data is shown in S3 Table.
(TIF)
the PCA dendrogram cluster with Ward method was performed as summarized in S4 Fig. All female samples cluster together, and all male samples cluster together indicating the difference between the sexes. While the infected females cluster together at one end of the spectrum followed by control female rat exosomes, one of the control males and one of the infected males cluster differently. This may be due to the outbred nature of the rats wherein the level of infection could be different between animals. This condition is reflected in clinical human infection.
(TIF)
b: 156 Proteins Unique to Control rat urine but absent in infected rat urine. c: 272 Proteins unique to infected male rat urine but absent in control animals and infected female rat samples.
(DOCX)
b: Proteins unique to control rat urine exosomes, but absent in infected rat urine exosomes. C: Proteins unique to Infected male rat urine exosomes, but absent in control animals and infected female rat samples.
(DOCX)
(DOCX)
Acknowledgments
The authors gratefully acknowledge the technical assistance of Sarah Leonhardt for silver staining of infected rat kidney slides.
This paper is dedicated to the memory of Fabrizio Basilico, who prematurely died last year after performing the proteomics experiments, the data from which are included in this manuscript: Fabrizio was a good friend and a great colleague with high scientific integrity and an indomitable spirit.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by NIH grants R01AI108276 and K24AI068903. The NIH-funded UCSD-UAB O’Brien Center for AKI Research (NIH 1P30 DK 079337) supported SPR’s effort. He was also supported by the GSK-SSS funding for biomarker, the UC San Diego CRCHD (NIH/NIMHD-P60 MD000220) and the UC San Diego Academic Senate Health Sciences Research Grant. PM was supported by CNR-Regione Lombardia for project G-care (18094/RCC, ID 17242) and Italian MIUR for Smart Aging project (SCN00_442). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 2. Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N (2008) The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis 12: 351–357. [DOI] [PubMed] [Google Scholar]
- 3. Adler B, de la Pena Moctezuma A (2010) Leptospira and leptospirosis. Vet Microbiol 140: 287–296. 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 4. Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7: 736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Monahan AM, Callanan JJ, Nally JE (2008) Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect Immun 76: 4952–4958. 10.1128/IAI.00511-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nally JE, Monahan AM, Miller IS, Bonilla-Santiago R, Souda P, et al. (2011) Comparative proteomic analysis of differentially expressed proteins in the urine of reservoir hosts of leptospirosis. PLoS One 6: e26046 10.1371/journal.pone.0026046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nature reviews Microbiology 7: 736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chassin C, Picardeau M, Goujon JM, Bourhy P, Quellard N, et al. (2009) TLR4- and TLR2-mediated B cell responses control the clearance of the bacterial pathogen, Leptospira interrogans. Journal of immunology 183: 2669–2677. 10.4049/jimmunol.0900506 [DOI] [PubMed] [Google Scholar]
- 9. Murray GL, Morel V, Cerqueira GM, Croda J, Srikram A, et al. (2009) Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect Immun 77: 810–816. 10.1128/IAI.01293-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, et al. (2010) Asymptomatic renal colonization of humans in the peruvian Amazon by Leptospira . PLoS Negl Trop Dis 4: e612 10.1371/journal.pntd.0000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Looze C, Yui D, Leung L, Ingham M, Kaler M, et al. (2009) Proteomic profiling of human plasma exosomes identifies PPARgamma as an exosome-associated protein. Biochem Biophys Res Commun 378: 433–438. 10.1016/j.bbrc.2008.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, et al. (2008) Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res 7: 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lancaster GI, Febbraio MA (2005) Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem 280: 23349–23355. [DOI] [PubMed] [Google Scholar]
- 14. Lancaster GI, Febbraio MA (2005) Mechanisms of stress-induced cellular HSP72 release: implications for exercise-induced increases in extracellular HSP72. Exerc Immunol Rev 11: 46–52. [PubMed] [Google Scholar]
- 15. Levett PN (2001) Leptospirosis. Clin Microbiol Rev 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turner LH (1967) Leptospirosis. I. Trans R Soc Trop Med Hyg 61: 842–855. [DOI] [PubMed] [Google Scholar]
- 17. Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858. [DOI] [PubMed] [Google Scholar]
- 18. Brambilla F, Lavatelli F, Di Silvestre D, Valentini V, Palladini G, et al. (2013) Shotgun protein profile of human adipose tissue and its changes in relation to systemic amyloidoses. J Proteome Res 12: 5642–5655. 10.1021/pr400583h [DOI] [PubMed] [Google Scholar]
- 19. Mauri P, Deho G (2008) A proteomic approach to the analysis of RNA degradosome composition in Escherichia coli. Methods in enzymology 447: 99–117. 10.1016/S0076-6879(08)02206-4 [DOI] [PubMed] [Google Scholar]
- 20. Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, et al. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A 103: 18928–18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS (2012) MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40: W127–133. 10.1093/nar/gks374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic acids research 37: W652–660. 10.1093/nar/gkp356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramachandra Rao SP, Wassell R, Shaw MA, Sharma K (2007) Profiling of human mesangial cell subproteomes reveals a role for calmodulin in glucose uptake. Am J Physiol Renal Physiol 292: F1182–1189. [DOI] [PubMed] [Google Scholar]
- 24. RamachandraRao SP, Zhu Y, Ravasi T, McGowan TA, Toh I, et al. (2009) Pirfenidone is renoprotective in diabetic kidney disease. J Am Soc Nephrol 20: 1765–1775. 10.1681/ASN.2008090931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riemann D, Kehlen A, Langner J (1995) Stimulation of the expression and the enzyme activity of aminopeptidase N/CD13 and dipeptidylpeptidase IV/CD26 on human renal cell carcinoma cells and renal tubular epithelial cells by T cell-derived cytokines, such as IL-4 and IL-13. Clin Exp Immunol 100: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Look AT, Ashmun RA, Shapiro LH, Peiper SC (1989) Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J Clin Invest 83: 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Natarajaseenivasan K, Shanmughapriya S, Velineni S, Artiushin SC, Timoney JF (2011) Cloning, expression, and homology modeling of GroEL protein from Leptospira interrogans serovar autumnalis strain N2. Genomics Proteomics Bioinformatics 9: 151–157. 10.1016/S1672-0229(11)60018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vedhagiri K, Natarajaseenivasan K, Chellapandi P, Prabhakaran SG, Selvin J, et al. (2009) Evolutionary implication of outer membrane lipoprotein-encoding genes ompL1, UpL32 and lipL41 of pathogenic Leptospira species. Genomics Proteomics Bioinformatics 7: 96–106. 10.1016/S1672-0229(08)60038-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fanton d'Andon M, Quellard N, Fernandez B, Ratet G, Lacroix-Lamande S, et al. (2014) Leptospira Interrogans induces fibrosis in the mouse kidney through Inos-dependent, TLR- and NLR-independent signaling pathways. PLoS Negl Trop Dis 8: e2664 10.1371/journal.pntd.0002664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez SA, Hostutler RA (2014) Distal renal tubular acidosis associated with concurrent leptospirosis in a dog. J Am Anim Hosp Assoc 50: 203–208. 10.5326/JAAHA-MS-5993 [DOI] [PubMed] [Google Scholar]
- 31. Cesar KR, Romero EC, de Braganca AC, Blanco RM, Abreu PA, et al. (2012) Renal involvement in leptospirosis: the effect of glycolipoprotein on renal water absorption. PLoS One 7: e37625 10.1371/journal.pone.0037625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Araujo ER, Seguro AC, Spichler A, Magaldi AJ, Volpini RA, et al. (2010) Acute kidney injury in human leptospirosis: an immunohistochemical study with pathophysiological correlation. Virchows Arch 456: 367–375. 10.1007/s00428-010-0894-8 [DOI] [PubMed] [Google Scholar]
- 33. Barrett-Connor E, Child CM, Carter MJ (1970) Renal failure in leptospirosis. Southern medical journal 63: 580–583. [DOI] [PubMed] [Google Scholar]
- 34. Hostutler RA, DiBartola SP, Eaton KA (2004) Transient proximal renal tubular acidosis and Fanconi syndrome in a dog. J Am Vet Med Assoc 224: 1611–1614, 1605 [DOI] [PubMed] [Google Scholar]
- 35. Khositseth S, Sudjaritjan N, Tananchai P, Ong-ajyuth S, Sitprija V, et al. (2008) Renal magnesium wasting and tubular dysfunction in leptospirosis. Nephrol Dial Transplant 23: 952–958. [DOI] [PubMed] [Google Scholar]
- 36. Kuo HL, Lin CL, Huang CC (2003) Reversible thick ascending limb dysfunction and aseptic meningitis syndrome: early manifestation in two leptospirosis patients. Ren Fail 25: 639–646. [DOI] [PubMed] [Google Scholar]
- 37. Lin CL, Wu MS, Yang CW, Huang CC (1999) Leptospirosis associated with hypokalaemia and thick ascending limb dysfunction. Nephrol Dial Transplant 14: 193–195. [DOI] [PubMed] [Google Scholar]
- 38. Yang CW, Pan MJ, Wu MS, Chen YM, Tsen YT, et al. (1997) Leptospirosis: an ignored cause of acute renal failure in Taiwan. Am J Kidney Dis 30: 840–845. [DOI] [PubMed] [Google Scholar]
- 39. Yang CW, Wu MS, Pan MJ (2001) Leptospirosis renal disease. Nephrol Dial Transplant 16 Suppl 5: 73–77. [DOI] [PubMed] [Google Scholar]
- 40. Monahan AM, Callanan JJ, Nally JE (2009) Review paper: Host-pathogen interactions in the kidney during chronic leptospirosis. Vet Pathol 46: 792–799. 10.1354/vp.08-VP-0265-N-REV [DOI] [PubMed] [Google Scholar]
- 41. Marotto PC, Ko AI, Murta-Nascimento C, Seguro AC, Prado RR, et al. (2010) Early identification of leptospirosis-associated pulmonary hemorrhage syndrome by use of a validated prediction model. J Infect 60: 218–223. 10.1016/j.jinf.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chakurkar G, Vaideeswar P, Pandit SP, Divate SA (2008) Cardiovascular lesions in leptospirosis: an autopsy study. J Infect 56: 197–203. 10.1016/j.jinf.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 43. Daher Ede F, Junior Silva GB, Vieira AP, Souza JB, Falcao Fdos S, et al. (2014) Acute kidney injury in a tropical country: a cohort study of 253 patients in an infectious diseases intensive care unit. Rev Soc Bras Med Trop 47: 86–89. 10.1590/0037-8682-0223-2013 [DOI] [PubMed] [Google Scholar]
- 44. Turhan V, Atasoyu EM, Solmazgul E, Evrenkaya R, Cavuslu S (2005) Anicteric leptospirosis and renal involvement. Ren Fail 27: 491–492. [PubMed] [Google Scholar]
- 45. Turhan V, Polat E, Murat Atasoyu E, Ozmen N, Kucukardali Y, et al. (2006) Leptospirosis in Istanbul, Turkey: a wide spectrum in clinical course and complications. Scand J Infect Dis 38: 845–852. [DOI] [PubMed] [Google Scholar]
- 46. Atasoyu EM, Turhan V, Unver S, Evrenkaya TR, Yildirim S (2005) A case of leptospirosis presenting with end-stage renal failure. Nephrol Dial Transplant 20: 2290–2292. [DOI] [PubMed] [Google Scholar]
- 47. Cetin BD, Harmankaya O, Hasman H, Gunduz A, Oktar M, et al. (2004) Acute renal failure: a common manifestation of leptospirosis. Ren Fail 26: 655–661. [DOI] [PubMed] [Google Scholar]
- 48. Herath NJ, Kularatne SA, Weerakoon KG, Wazil A, Subasinghe N, et al. (2014) Long term outcome of acute kidney injury due to leptospirosis? A longitudinal study in Sri Lanka. BMC Res Notes 7: 398 10.1186/1756-0500-7-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barnes K, Kenny AJ, Turner AJ (1994) Localization of aminopeptidase N and dipeptidyl peptidase IV in pig striatum and in neuronal and glial cell cultures. Eur J Neurosci 6: 531–537. [DOI] [PubMed] [Google Scholar]
- 50. Semenza G (1986) Anchoring and biosynthesis of stalked brush border membrane proteins: glycosidases and peptidases of enterocytes and renal tubuli. Annu Rev Cell Biol 2: 255–313. [DOI] [PubMed] [Google Scholar]
- 51. Sanderink GJ, Artur Y, Siest G (1988) Human aminopeptidases: a review of the literature. J Clin Chem Clin Biochem 26: 795–807. [DOI] [PubMed] [Google Scholar]
- 52. Lendeckel U, Wex T, Reinhold D, Kahne T, Frank K, et al. (1996) Induction of the membrane alanyl aminopeptidase gene and surface expression in human T-cells by mitogenic activation. Biochem J 319 (Pt 3): 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hooper NM (1994) Families of zinc metalloproteases. FEBS Lett 354: 1–6. [DOI] [PubMed] [Google Scholar]
- 54. Hooper NM, Hesp RJ, Tieku S (1994) Metabolism of aspartame by human and pig intestinal microvillar peptidases. Biochem J 298 Pt 3: 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mina-Osorio P (2008) The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med 14: 361–371. 10.1016/j.molmed.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Villasenor-Cardoso MI, Frausto-Del-Rio DA, Ortega E (2013) Aminopeptidase N (CD13) is involved in phagocytic processes in human dendritic cells and macrophages. Biomed Res Int 2013: 562984 10.1155/2013/562984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghosh M, McAuliffe B, Subramani J, Basu S, Shapiro LH (2012) CD13 regulates dendritic cell cross-presentation and T cell responses by inhibiting receptor-mediated antigen uptake. J Immunol 188: 5489–5499. 10.4049/jimmunol.1103490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen L, Gao Z, Zhu J, Rodgers GP (2007) Identification of CD13+CD36+ cells as a common progenitor for erythroid and myeloid lineages in human bone marrow. Exp Hematol 35: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Inoi K, Goto S, Nomura S, Isobe K, Nawa A, et al. (1995) Aminopeptidase inhibitor ubenimex (bestatin) inhibits the growth of human choriocarcinoma in nude mice through its direct cytostatic activity. Anticancer Res 15: 2081–2087. [PubMed] [Google Scholar]
- 60. Wex T, Lendeckel U, Reinhold D, Kahne T, Arndt M, et al. (1997) Antisense-mediated inhibition of aminopeptidase N (CD13) markedly decreases growth rates of hematopoietic tumour cells. Adv Exp Med Biol 421: 67–73. [DOI] [PubMed] [Google Scholar]
- 61. Chang YW, Chen SC, Cheng EC, Ko YP, Lin YC, et al. (2005) CD13 (aminopeptidase N) can associate with tumor-associated antigen L6 and enhance the motility of human lung cancer cells. Int J Cancer 116: 243–252. [DOI] [PubMed] [Google Scholar]
- 62. Tani K, Ogushi F, Huang L, Kawano T, Tada H, et al. (2000) CD13/aminopeptidase N, a novel chemoattractant for T lymphocytes in pulmonary sarcoidosis. Am J Respir Crit Care Med 161: 1636–1642. [DOI] [PubMed] [Google Scholar]
- 63. Larsen SL, Pedersen LO, Buus S, Stryhn A (1996) T cell responses affected by aminopeptidase N (CD13)-mediated trimming of major histocompatibility complex class II-bound peptides. J Exp Med 184: 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gabrilovac J, Breljak D, Cupic B, Ambriovic-Ristov A (2005) Regulation of aminopeptidase N (EC 3.4.11.2; APN; CD13) by interferon-gamma on the HL-60 cell line. Life Sci 76: 2681–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Amoscato AA, Prenovitz DA, Lotze MT (1998) Rapid extracellular degradation of synthetic class I peptides by human dendritic cells. J Immunol 161: 4023–4032. [PubMed] [Google Scholar]
- 66. Tokuda N, Levy RB (1996) 1,25-dihydroxyvitamin D3 stimulates phagocytosis but suppresses HLA-DR and CD13 antigen expression in human mononuclear phagocytes. Proc Soc Exp Biol Med 211: 244–250. [DOI] [PubMed] [Google Scholar]
- 67. Soderberg C, Giugni TD, Zaia JA, Larsson S, Wahlberg JM, et al. (1993) CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J Virol 67: 6576–6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Soderberg C, Larsson S, Bergstedt-Lindqvist S, Moller E (1993) Definition of a subset of human peripheral blood mononuclear cells that are permissive to human cytomegalovirus infection. J Virol 67: 3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, et al. (1992) Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hansen GH, Delmas B, Besnardeau L, Vogel LK, Laude H, et al. (1998) The coronavirus transmissible gastroenteritis virus causes infection after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment. J Virol 72: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nomura R, Kiyota A, Suzaki E, Kataoka K, Ohe Y, et al. (2004) Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J Virol 78: 8701–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Hensbergen Y, Broxterman HJ, Hanemaaijer R, Jorna AS, van Lent NA, et al. (2002) Soluble aminopeptidase N/CD13 in malignant and nonmalignant effusions and intratumoral fluid. Clin Cancer Res 8: 3747–3754. [PubMed] [Google Scholar]
- 73. van Hal PT, Hopstaken-Broos JP, Prins A, Favaloro EJ, Huijbens RJ, et al. (1994) Potential indirect anti-inflammatory effects of IL-4. Stimulation of human monocytes, macrophages, and endothelial cells by IL-4 increases aminopeptidase-N activity (CD13; EC 3.4.11.2). J Immunol 153: 2718–2728. [PubMed] [Google Scholar]
- 74. Delmas B, Gelfi J, L'Haridon R, Vogel LK, Sjostrom H, et al. (1992) Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Petrovic N, Schacke W, Gahagan JR, O'Conor CA, Winnicka B, et al. (2007) CD13/APN regulates endothelial invasion and filopodia formation. Blood 110: 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Navarrete Santos A, Roentsch J, Danielsen EM, Langner J, Riemann D (2000) Aminopeptidase N/CD13 is associated with raft membrane microdomains in monocytes. Biochem Biophys Res Commun 269: 143–148. [DOI] [PubMed] [Google Scholar]
- 77. Riemann D, Hansen GH, Niels-Christiansen L, Thorsen E, Immerdal L, et al. (2001) Caveolae/lipid rafts in fibroblast-like synoviocytes: ectopeptidase-rich membrane microdomains. Biochem J 354: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. van Balkom BW, Pisitkun T, Verhaar MC, Knepper MA (2011) Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int 80: 1138–1145. 10.1038/ki.2011.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cheville NF, Huhn R, Cutlip RC (1980) Ultrastructure of renal lesions in pigs with acute leptospirosis caused by Leptospira pomona. Vet Pathol 17: 338–351. [DOI] [PubMed] [Google Scholar]
- 80. Alves VA, Gayotto LC, Yasuda PH, Wakamatsu A, Kanamura CT, et al. (1991) Leptospiral antigens (L. interrogans serogroup ictero-haemorrhagiae) in the kidney of experimentally infected guinea pigs and their relation to the pathogenesis of the renal injury. Exp Pathol 42: 81–93. [DOI] [PubMed] [Google Scholar]
- 81. Lasser C, Eldh M, Lotvall J (2012) Isolation and characterization of RNA-containing exosomes. J Vis Exp: e3037 10.3791/3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kokot F, Dulawa J (2000) Tamm-Horsfall protein updated. Nephron 85: 97–102. [DOI] [PubMed] [Google Scholar]
- 83. Mo L, Zhu XH, Huang HY, Shapiro E, Hasty DL, et al. (2004) Ablation of the Tamm-Horsfall protein gene increases susceptibility of mice to bladder colonization by type 1-fimbriated Escherichia coli. Am J Physiol Renal Physiol 286: F795–802. [DOI] [PubMed] [Google Scholar]
- 84. Saemann MD, Weichhart T, Horl WH, Zlabinger GJ (2005) Tamm-Horsfall protein: a multilayered defence molecule against urinary tract infection. Eur J Clin Invest 35: 227–235. [DOI] [PubMed] [Google Scholar]
- 85. Cavallone D, Malagolini N, Serafini-Cessi F (2001) Mechanism of release of urinary Tamm-Horsfall glycoprotein from the kidney GPI-anchored counterpart. Biochem Biophys Res Commun 280: 110–114. [DOI] [PubMed] [Google Scholar]
- 86. Fukuoka S, Freedman SD, Yu H, Sukhatme VP, Scheele GA (1992) GP-2/THP gene family encodes self-binding glycosylphosphatidylinositol-anchored proteins in apical secretory compartments of pancreas and kidney. Proc Natl Acad Sci U S A 89: 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pisitkun T, Shen RF, Knepper MA (2004) Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 101: 13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, et al. (2009) Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20: 363–379. 10.1681/ASN.2008040406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
(TIF)
All urine samples collected were screened for the presence of pathogenic and intermediate-pathogenic Leptospira using a published qPCR TaqMan assay targeting the leptospiral 16S ribosomal gene (1); this assay has been reported in our previous work. Briefly, this was performed using an Opticon 2 real-time PCR machine (MJ Research, USA). The assay protocol was modified from the published version (2) by using the fluorescent probe at a final concentration of 0.2 mM, primers at a final concentration of 0.5 mM, and a 20 mL reaction volume (3). Standard curves for quantification were made using Leptospira interrogans serovar Copenhageni strain M20. Standards were prepared as follows. Leptospires were counted using a Petroff-Hauser counting chamber (Hauser Scientific, USA) and serially diluted with sterile double-distilled H2O to 108 to 100 leptospires/ ml. Genomic DNA was subsequently prepared using the DNeasy Tissue Kit (Qiagen, USA). Standards were run in triplicate to generate a standard curve with each run. A negative result was assigned where no amplification occurred before 40 cycles. Controls lacking template were extracted and added to qPCR master mix to detect the presence of contaminating DNA. The raw data is shown in S3 Table.
(TIF)
the PCA dendrogram cluster with Ward method was performed as summarized in S4 Fig. All female samples cluster together, and all male samples cluster together indicating the difference between the sexes. While the infected females cluster together at one end of the spectrum followed by control female rat exosomes, one of the control males and one of the infected males cluster differently. This may be due to the outbred nature of the rats wherein the level of infection could be different between animals. This condition is reflected in clinical human infection.
(TIF)
b: 156 Proteins Unique to Control rat urine but absent in infected rat urine. c: 272 Proteins unique to infected male rat urine but absent in control animals and infected female rat samples.
(DOCX)
b: Proteins unique to control rat urine exosomes, but absent in infected rat urine exosomes. C: Proteins unique to Infected male rat urine exosomes, but absent in control animals and infected female rat samples.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.






