Abstract
Tumor budding refers to single or small cluster of tumor cells detached from the main tumor mass. In colon cancer high tumor budding is associated with positive lymph nodes and worse prognosis. Therefore, we investigated the value of tumor budding as a predictive feature of lymph node status in breast cancer (BC). Whole tissue sections from 148 surgical resection specimens (SRS) and 99 matched preoperative core biopsies (CB) with invasive BC of no special type were analyzed on one slide stained with pan-cytokeratin. In SRS, the total number of intratumoral (ITB) and peripheral tumor buds (PTB) in ten high-power fields (HPF) were counted. A bud was defined as a single tumor cell or a cluster of up to five tumor cells. High tumor budding equated to scores averaging >4 tumor buds across 10HPFs. In CB high tumor budding was defined as ≥10 buds/HPF. The results were correlated with pathological parameters. In SRS high PTB stratified BC with lymph node metastases (p ≤ 0.03) and lymphatic invasion (p ≤ 0.015). In CB high tumor budding was significantly (p = 0.0063) associated with venous invasion. Pathologists are able, based on morphology, to categorize BC into a high and low risk groups based in part on lymph node status. This risk assessment can be easily performed during routine diagnostics and it is time and cost effective. These results suggest that high PTB is associated with loco-regional metastasis, highlighting the possibility that this tumor feature may help in therapeutic decision-making.
Keywords: Breast cancer, No special type, Tumor budding, Vessel invasion, Metastasis
Background
Breast cancer (BC) is considerably heterogeneous both molecularly and histologically. It is presented as a collection of distinct disease subtypes that differ in disease progression, treatment response, and disease-free survival [1–5]. A variety of clinical pathological and genetic factors are routinely used to assess prognosis and determine the most appropriate therapeutic regimen for patients with BC [6, 7]. These include patient age, axillary lymph node status, tumor size, histological grade, lymphatic invasion, hormone receptor status, HER2 status, and evaluation of tumor margins. More recently, molecular tests such as Oncotype DX® and MammaPrint® are being utilized to assess likelihood of treatment response and/or recurrence. However, additional prognosticators are needed to enhance personalized treatment and especially to overcome over and under treatment of patients.
Despite the substantial molecular and morphological differences, all invasive BC subtypes likely share a pathway for invasion, evidenced from morphological changes that occur in the transition to all invasive carcinomas. Normal epithelia, including pre-invasive breast cancers (carcinoma in situ), are physically separated from surrounding stroma and vascular structures by a basement membrane and a layer of myoepithelial cells [3]. Disruption of this barrier is a pre-requisite for invasion in all breast cancers. In addition, the invasive front of breast cancer also includes infiltrates of diverse immune cells including macrophages, neutrophils, and mast cells, and displays angiogenesis, and desmoplasia with deposition of extracellular matrix (ECM), particularly collagen I [8].
In colorectal carcinomas (CRC), morphological studies of the invasive front have revealed the emergence of tumor budding, which reflect detachment of tumor cells into single cells or clusters of up to five cells [9]. Tumor budding is diagnosed at high magnification and should not be mistaken with the tumor border configuration, which is more easily discernable at low magnifications. Biologically, the role of tumor budding includes a mechanism for invasive cells to migrate through peri-tumoral connective tissue, evade the host’s defensive mechanisms and invade lymphatic and blood vessels with the consequence of local and distant metastases [9].
Over the last few years, numerous publications have shown the importance of tumor budding as an independent predictor of lymph node positivity, local and distant relapse, lymphatic invasion, and poor prognosis among patients with CRC of all pathological stages [9]. However, the definite implementation of tumor budding into clinical practice is currently limited by the lack of an internationally standardized scoring system. Whether tumor budding can be considered a histomorphologic features in BC remains to be elucidated. The purpose of the current study was to evaluate the potential clinical value of tumor budding in breast cancer of no special type (NST) as a predictor of lymph node metastasis.
Materials and methods
Patient samples
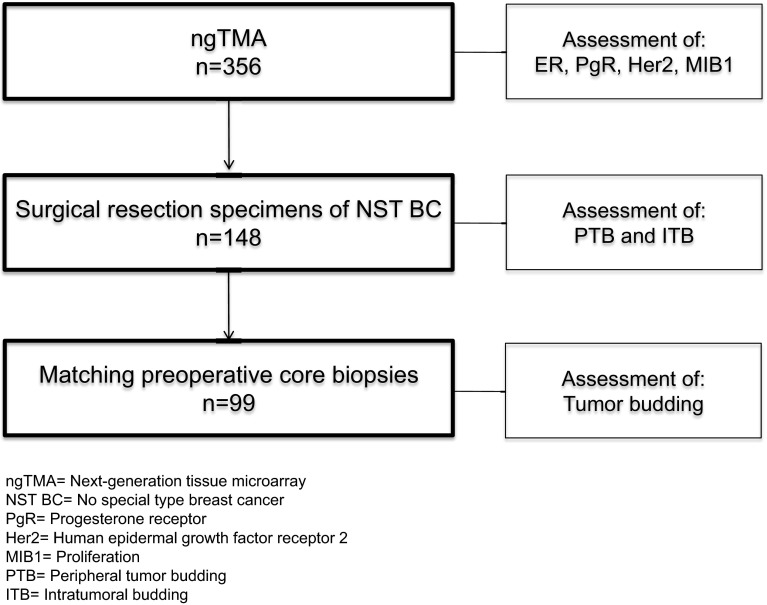
We searched the database at the Institute of Pathology, University of Bern, Switzerland for patients diagnosed with primary BC between January 2005 and December 2011. We identified 356 patients with suitable formalin fixed and paraffin-embedded (FFPE) samples from female patients with therapy naïve, primary, unilateral BC of more than 5 mm in size to construct a next-generation tissue microarray (ngTMA). From these patients, we selected 148 individual surgical resection specimens (SRS) and 99 matching preoperative core biopsies (CB) from patients with invasive BC of no special type (NST) and known lymph node status for tumor budding analysis. The TNM classification was taken from the pathology reports, and the clinical data were extracted from the database of the breast center of the University Hospital Bern, Switzerland. The median age of selected patients at the time of diagnosis was 61 years (range 32–91 years). The clinico-pathological features for the sample cohort are summarized in Table 1, and the study design is presented in Fig. 1.
Table 1.
Patient characteristics (n = 148)
| Feature | Frequency N (%) | |
|---|---|---|
| Age (years) | Median (min, max) | 61 (32, 91) |
| Tumor size (cm) | Median (min, max) | 2.0 (0.6–8.0) |
| ER | Positive | 125 (84.5) |
| Negative | 23 (15.5) | |
| PgR | Positive | 104 (70.3) |
| Negative | 44 (29.7) | |
| Her 2 | Positive | 19 (12.8) |
| Negative | 129 (87.2) | |
| MIB1 | High (>15 %) | 40 (27.0) |
| Low (≤15 %) | 107 (72.3) | |
| No data | 1 (0.7) | |
| Grade | G1 | 18 (12.2) |
| G2 | 69 (46.6) | |
| G3 | 61 (41.2) | |
| pT | T1 | 75 (50.7) |
| T2 | 66 (44.6) | |
| T3 | 3 (2.0) | |
| T4 | 4 (2.7) | |
| pN | pN0 | 68 (45.9) |
| >pN0 | 80 (54.1) | |
| Molecular subtypes | Luminal A | 92 (62.2) |
| Luminal B | 33 (22.3) | |
| Her2 | 3 (2.0) | |
| Triple negative | 20 (13.5) | |
| Lymphatic invasion | No | 89 (60.1) |
| Yes | 58 (39.5) | |
| No data | 1 (0.7) | |
| Venous invasion | No | 135 (91.2) |
| Yes | 12 (8.1) | |
| No data | 1 (0.7) | |
| Perineural invasion | No | 128 (86.5) |
| Yes | 19 (12.8) | |
| No data | 1 (0.7) | |
| Hormone therapy | No | 55 (37.2) |
| Yes | 38 (25.7) | |
| Unknown | 55 (37.1) | |
| Chemotherapy | No | 58 (39.2) |
| Yes | 34 (23.0) | |
| Unknown | 56 (37.8) | |
| Anti-Her2 therapy | No | 81 (54.7) |
| Yes | 5 (3.4) | |
| Unknown | 62 (41.9) | |
| Radiotherapy | No | 32 (21.6) |
| Yes | 106 (71.6) | |
| Unknown | 10 (6.8) |
Fig. 1.
Flow chart study
All BCs were reviewed and the histological subtype and tumor grade were assigned according to the WHO classification 2012. The study was approved by the ethical committee of the University of Bern (Registration 200/2014) and was performed according to the REMARK guidelines [10]. Long-term follow-up data were not available for this cohort.
Next-generation tissue microarray (ngTMA)
For the assessment of the prognostic factors, a ngTMA was constructed including 356 selected BC samples as previously described [11]. In brief, whole tissue sections of the tumors were scanned and uploaded to a digital platform to make annotations for the punching process. The tumor image on the screen allowed an optimal overview of the sample for digital annotations of the regions of intersest, including areas from the tumor center and periphery. The fully automated arraying process synchronized the annotations made on the scanned slide with the image of the donor paraffin block to precisely punch the area of interest. For the majority (97.8 %) of BC samples, six punches of 0.6 mm were integrated into a new recipient block representing the ngTMA.
Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH)
Prior to ngTMA construction, 3 µm whole tissue sections from one selected FFPE tissue block of the 148 NST BCs were cut and incubated with an anti-cytokeratin antibody for better visualization of tumor buds for evaluation. Estrogen (ER) and progesterone (PgR) receptor, Her2 and MIB1 were performed on the ngTMA. The antibodies and the specific conditions used are listed in Table 2.
Table 2.
Specifications of immunohistochemistry and antibodies used
| Antibody | Clone | Cat#/vendor | Dilution | Retrival |
|---|---|---|---|---|
| Estrogen receptor | EP1 | M3643/Dako | 1:50 | Tris pH 8.4 40 min 95 °C |
| Progesterone receptor | 16 SAN27 | NCL-L-PGR-AB/Novocastra | 1:200 | Tris pH 8.4 40 min 95 °C |
| Her2 | Poly | K5207/Dako | Kit | citrat pH6 20 min 100 °C |
| MIB1 | Mib1 | M7240/Dako | 1:200 | Tris pH 8.4 30 min 95 °C |
| Cytokeratin | AE1/AE3 | M351501-2/Dako | 1:300 | Proteinase K 5 min 37 °C |
For ER, PgR, and Her2, the ASCO/CAP guidelines were applied to define positive hormone receptor and Her2 status, respectively [12, 13]. In BC with equivocal immunohistochemistry, fluorescence in situ hybridization (FISH) was performed using a dual-color probe (Abbott Molecular Abbott Park, Illinois, USA).
The molecular subtypes were defined as follows: Luminal A: estrogen (ER) and/or progesterone receptor (PgR) positive, Her2 negative, low proliferative index (≤15 %); Luminal B: hormone receptor positive, Her2 positive/or high proliferative index (>15%); Her2 subtype: Her2 positive and hormone receptor negative, any proliferative index; triple negative: Her2 negative, hormone receptor negative, any proliferative index.
Scoring of tumor buds
Tumor buds were defined as isolated single cancer cells or microscopic cell clusters composed of 1–5 cells. The number of buds was counted on whole tissue sections of surgical resection specimens (SRS; n = 148) and matched preoperative core biopsies (CB; n = 99) stained with a pan-cytokeratin antibody (AE1/AE3) for better visualization. Areas with the highest concentration of buds (“hot-spots”) were selected for scoring. BC samples were selected from the cohort of the ngTMA as described earlier.
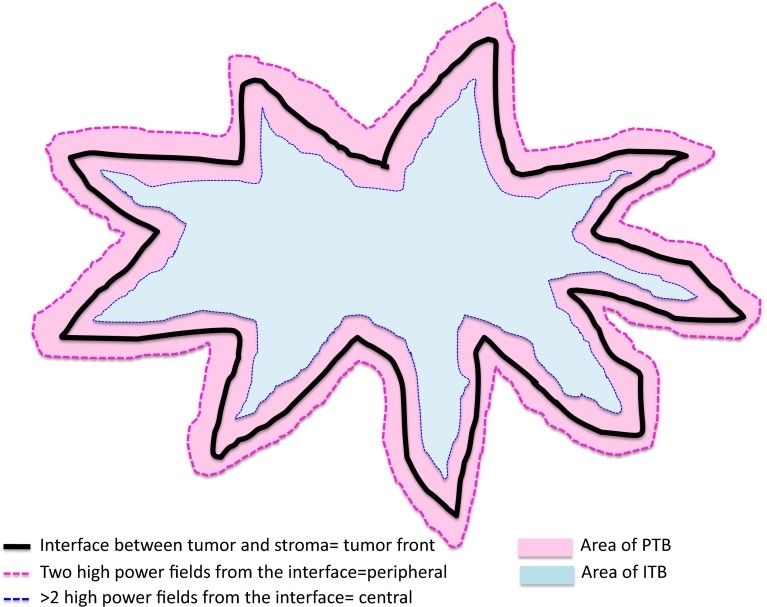
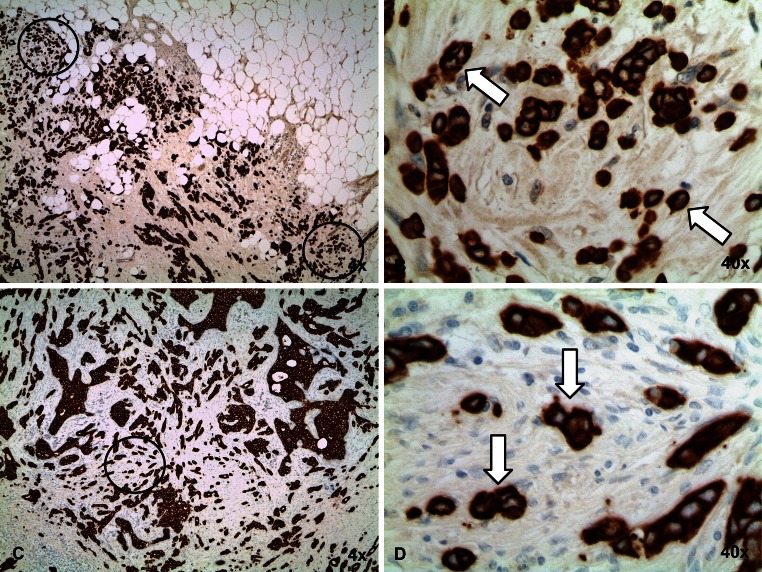
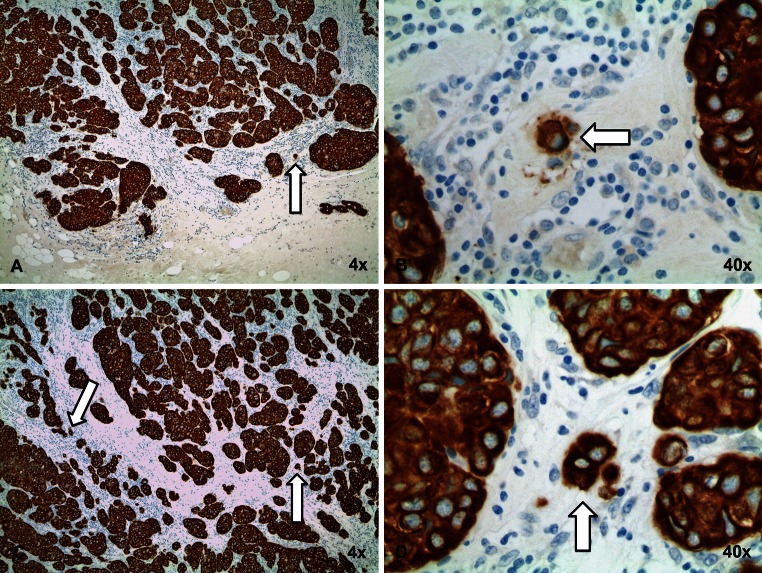
In SRS, tumor buds at the invasive front or within the tumor center were scored separately. Invasive tumor front buds are referred to as peripheral tumor buds (PTB) and were scored within ≈1.1 mm (2 × 1 high-power field (HPF) @0.55 mm2) on either direction of the tumor interface. Tumor buds within the center are referred to as intratumoral buds (ITB) (Fig. 2). We used pre-defined criteria for the assessment of tumor buds in accordance with recent publications describing scoring of tumor buds in colon cancer SRS [14] and cut-off criteria in BC [15]. For SRS, 2 pathologists (CT, MT) scored 10 HPF independently and blindly without knowledge of tumor characteristics (e.g., nodal status) and each other’s results. According to a recent report [15], tumors were considered to have high tumor budding if the average number of tumor buds in 10 HPF >4. In contrast, tumors were considered to have low tumor budding if the average number of buds in 10 HPF was ≤4. Examples of high tumor budding are seen in Fig. 3 and examples of low tumor budding are given in Fig. 4.
Fig. 2.
Peripheral and intratumoral budding
Fig. 3.
NST breast cancer with high tumor budding. Overview (×4) of a high PTB (a) and high ITB (c) breast cancer stained with anti-pan-cytokeratin antibody. b and d A high-power field (×40) with more than four tumor buds
Fig. 4.
NST breast cancer with low tumor budding. Overview (×4) of a low PTB (a) and low ITB (c) breast cancer stained with anti-pan-cytokeratin antibody. b and d A high-power field (×40) with less than four tumor buds
Since CBs are not easily comparable to SRSs, we performed a receiver operating characteristic (ROC) curve analysis to find the appropriate cut-off for high and low tumor budding. In addition, we scored only 1HPF with the highest density of tumor buds as previously reported for preoperative colon biopsies [16]. Furthermore, ITB and PTB cannot be distinguished in CB because they are randomly sampled. One pathologist (CT) scored tumor budding in all CBs. In all SRSs and CBs tumor buds were counted at ×400 magnification (0.55 mm2).
Statistics
The Chi Square test was used to calculate significant differences between categorical variables. Pearson’s correlation coefficient was used to determine the strength of the linear relationship between budding in CB and SRS (ITB and PTB). A p value < 0.05 was considered statistically significant. Interrater reliability was calculated with Cohen’s Kappa test with the following interpretations for the kappa value 0–0.2 = poor; 0.21–0.4 = fair; 0.41–0.6 = moderate; 0.61–0.8 = substantial; >0.8 = excellent [17]. To calculate a cut-off for high and low tumor budding in CB, receiver operating characteristic (ROC) curve analysis was performed. Analyses were carried out using SPSS 21 (IBM, Armonk, US) and SAS (V9.4, The SAS Institute, Cary, NC).
Results
A total of 148 surgical resection specimens (SRS) and 99 matching preoperative core biopsies (CB) with invasive NST BC were evaluated. ER, PgR, Her2, and MIB1 proliferative index were obtained from ngTMA evaluation and are summarized in Table 1. In two patients, Her2 status (1.4 %) and in three patients MIB1 (2.0 %) results were taken from the pathological reports due to insufficient material present for evaluation on the ngTMA.
The concordance of the two pathologist regarding scoring of high and low tumor budding was k = 0.526 for PTB and k = 0.533 for ITB.
Correlation of tumor budding with clinical-pathological characteristics
Significant associations with PTB in surgical resection specimens
The individual scores of pathologists showed a significant correlation of high PTB with positive lymph node status (p = 0.003 and 0.03) and lymphatic invasion (LVI) (p = 0.015 and 0.013), respectively. The only significant result when combining the scoring of both pathologists was the association of high PTB with lymphatic invasion (p = 0.029). In addition, analysis of only ER-positive and low proliferative BCs, respectively, showed the following results: Pathologist 1 showed a significant correlation with lymph node metastasis and high PTB when analyzing only ER-positive BCs (p = 0.007) and in samples with low proliferative BCs (p = 0.006), respectively. Pathologist 2 had no significant results for subgroup analysis and no other significant correlations were seen for high PTB.
Significant associations with ITB in resection specimens
High ITB scoring results of Pathologist 1 showed a significant association with tumor grade (p = 0.036), ER positivity (p = 0.02) and lower proliferation (p = 0.002). The scoring results of Pathologist 2 demonstrated a significant association with negative Her2 status (p = 0.001) and perineural invasion (p = 0.039). The scoring results with significant associations are summarized in Table 3 and the subgroup analysis of PTB are summarized in Table 4.
Table 3.
Summary of significant results and tumor budding
| Features | PTB1 n (%) | p-value | PTB2 n (%) | p-value | |||
|---|---|---|---|---|---|---|---|
| Low 30 (20.3) | High 118 (79.7) | Low 17 (11.5) | High 131 (88.5) | ||||
| pN | Positive | 9 (11.2) | 71 (88.8) | 0.003 | 5 (6.2) | 75 (93.8) | 0.03 |
| Negative | 21 (30.9) | 47 (69.1) | 12 (17.6) | 56 (82.4) | |||
| Lymphatic invasion | Positive | 6 (10.3) | 52 (89.7) | 0.015 | 2 (3.4) | 56 (96.6) | 0.013 |
| Negative | 24 (27) | 65 (73) | 15 (16.9) | 74 (83.1) | |||
| Features | ITB1 n (%) | p-value | ITB2 n (%) | p-value | |||
|---|---|---|---|---|---|---|---|
| Low 30 (20.9) | High 117 (79.1) | Low 24 (16.2) | High 124 (83.8) | ||||
| Grading | G1 | 3 (16.7) | 15 (83.3) | 2 (11.1) | 16 (88.9) | ||
| G2 | 9 (13.0) | 60 (87.0) | 0.036 | 9 (13.0) | 60 (87.0) | 0.364 | |
| G3 | 19 (31.1) | 42 (68.9) | 13 (21.3) | 48 (78.7) | |||
| ER | Positive | 22 (17.6) | 103 (82.4) | 0.02 | 19 (15.2) | 106 (84.8) | 0.434 |
| Negative | 9 (39.1) | 14 (60.9) | 5 (21.7) | 18 (78.3) | |||
| Her2 | Positive | 7 (36.8) | 12 (63.2) | 0.68 | 8 (42.1) | 11 (57.9) | 0.001 |
| Negative | 24 (18.6) | 105 (81.4) | 16 (12.4) | 113 (87.6) | |||
| Molecular subtypes | Luminal A | 11 (12.0) | 81 (88.0) | 11 (12.0) | 81 (88.0) | ||
| Luminal B | 11 (33.3) | 22 (66.7) | 0.007 | 8 (24.2) | 25 (75.8) | 0.038 | |
| Her2 | 1 (33.3) | 2 (66.7) | 2 (66.7) | 1 (33.3) | |||
| Triple negative | 8 (40.0) | 12 (60.0) | 3 (15.0) | 17 (85.0) | |||
| MIB1 | High (>15 %) | 15 (37.5) | 25 (62.5) | 0.002 | 9 (22.5) | 31 (77.5) | 0.216 |
| Low (≤15 %) | 15 (14.0) | 92 (86.0) | 15 (14.0) | 92 (86.0) | |||
| Perineural invasion | Positive | 1 (5.3) | 18 (94.7) | 0.7 | 0 (0.0) | 19 (100) | |
| Negative | 30 (23.4) | 98 (76.6) | 24 (18.8) | 104 (81.2) | 0.039 | ||
PTB1 peripheral tumor budding score from Pathologist 1
PTB2 peripheral tumor budding score from Pathologist 2
ITB1 intratumoral budding score from Pathologist 1
ITB2 intratumoral budding score from Pathologist 2
Bold italic significant results
Table 4.
Subgroup analysis with significant results
| Features | PTB1 n (%) | p-value | PTB2 n (%) | p-value | |||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Only ER positive | pN positive | 7 (10.8) | 58 (89.2) | 5 (7.7) | 60 (92.3) | ||
| pN negative | 18 (30.0) | 42 (70.0) | 0.007 | 10 (16.7) | 50 (83.3) | 0.123 | |
| Only low MIB1 (≤15 %) | pN positive | 4 (7.5) | 49 (92.5) | 0.006 | 3 (5.7) | 50 (94.3) | 0.194 |
| pN negative | 15 (27.8) | 39 (72.2) | – | 7 (13.0) | 47 (87.0) | – | |
Bold italic significant results
Significant associations of tumor budding in preoperative core biopsies
Ninety-nine patients (66.9 %) with CB were identified from the 148 SRSs and evaluated for the presence of tumor budding. A significant positive correlation was found between budding in the CB and ITB (r = 0.28; p = 0.0045) and PTB (r = 0.48; p < 0.0001) of the SRS. In CB, tumor budding was significantly associated with V classification (p = 0.0063). All 35 patients with low-grade tumor budding (<10 buds/HPF) showed no venous invasion while 12/64 (19 %) patients with high-grade budding were diagnosed with venous invasion. No other significant associations were found with other pathological parameters (Table 4
Discussion
Tumor budding is best described as a histologic pattern associated with poor prognosis in early-stage colorectal adenocarcinoma and a predictor of nodal metastasis in T1 colorectal adenocarcinoma [18]. Recently, some of these associations were also found in esophageal carcinoma and pancreatic cancer [19, 20]. Therefore, we investigated the significance of tumor budding and its relationship to known clinico-pathological features of BC such as nodal metastasis, tumor grade, and tumor size. In this retrospective study, we used a semi-quantitative histologic scoring system to categorize 148 surgically resected, primary invasive NST BC’s for the extent of both ITB and PTB. Additionally, 99 matching CBs were analyzed for tumor budding. We selected invasive ductal BC because they are the most prevalent histological subtypes [21] and represent a heterogeneous group with varying tumor features including immunphenotypical and clinical outcomes. Therefore, it would be of great clinical value to find additional prognostic or predictive tumor features for this group.
During the course of our study, a paper was published by Liang et al. which demonstrated that high-grade budding in SRS was significantly associated with the presence of lymphatic invasion (LVI), larger tumor size, and worse clinical outcome [15]. We confirmed a significant association with LVI (p = 0.029) for PTB. Additionally, both pathologists were able to stratify BCs with positive lymph node metastases according to having a high PTB count (p = 0.004 and 0.03). Liang et al. reported that BC patients with high tumor bud scores have worse overall survival. Although long-term follow-up data were not available for the samples in this study, the association of tumor budding with positive lymph nodes suggests that tumor budding is a poor prognostic feature of BC. The limited number of patients and the fact that this is a retrospective study can be regarded as disadvantage. However, the endpint of this study was the lymph node status and not survival.
However, we did not find an association to tumor size or T-category as in the study by Liang et al. Certainly, the differences between the two studies could be related to the manner in which tumor buds were investigated. For our evaluation, we used IHC to better visualize hot-spots of single tumor cells or small tumor clusters and we defined tumor budding to be between 1 and 5 tumor cells and each pathologist scored 10 HPF in SRS. In the Liang study, H&E was predominately used for scoring and tumor buds were defined as 1–4 tumor cells. Pathologists scored 5 “hot-spot” loci. In addition, we investigated ITB, which was not previously described.
Interestingly, in colon cancer, a cut-off of >=10 buds is proposed for high tumor budding [14]. In BC fewer tumor buds are required in SRS to stratify BC with adverse tumor features. The colon mucosa is much more involved in immune defense than breast tissue and for this reason may require a higher budding load to have adverse prognostic effects. Evaluating tumor budding in each tissue and specimen type separately will therefore be necessary to obtain meaningful prognostic data.
Epithelial–mesenchymal transition (EMT), a feature of metastatic cells, has previously been described in the context of tumor budding [22] and could be consistent with the observed association to both LVI and lymph node positivity but requires further analysis. A recent report also demonstrated that estrogen is involved in EMT in breast cancer cell lines with stem cell properties [23] and it was shown that estrogen is involved in disruption of tight-junction and increased cell motility [24]. We suspect that ER-positive tumors with high tumor budding may be undergoing a high degree of EMT and therefore have more metastatic potential. Further, the tumor center, which can have fibrosis, may represent a more hypoxic environment what might contribute to EMT transformation [25]. However, these hypotheses need further investigations.
In our study, we confirmed the presence of tumor budding in BC and its association to known poor prognostic indicators such as lymphatic invasion and positive lymph node status in univariate analysis in SRS. However, information on vascular invasion and lymph node status would be more relevant in the preoperative setting for therapeutic decisions. Therefore, we investigated tumor budding on matching preoperative CBs for the first time in BC. Since CB are randomly sampled and consist of smaller pieces of tissue, tumor buds cannot be classified as PTB or ITB and also require different scoring criteria. In CBs, we identified a significant association of tumor buds with venous invasion, suggesting that CBs with high tumor budding might harbor a tumor cell population with an affinity for vascular invasion. The significance of this finding needs to befurther investigated. The lack of an association with LVI or positive nodal status as is observed in SRSs with high PTB could be explained if CB contain mostly ITB. Another explanation might be that the random capturing of tumor buds amidst a mix of PTB and ITB in CBs dilutes correlations of LVI and positive nodal status.
We demonstrated for the first time in BC that budding can be subdivided into ITB and PTB based on tumor localization, which may have disease implications. More research is needed to better understand the biological and clinical significance of ITB versus PTB in BC. Further, the concordance of scoring between the pathologists is better than the assessment of other tumor markers such as MIB1 [26]. Whether ITB could be used as an additional morphological feature to stratify ER-positive or low proliferative tumors (≤15 %) into a high and low risk category has to be investigated in a larger cohort. In addition, future studies should incorporate MIB1 analysis on whole tissue sections to bypass the reported caveat of obtaining significantly lower proliferation indices when using TMAs [27].
The evaluation of one tissue slide may not be ideal disadvantage, however, most tumor markers are evaluated on a single CB slide randomly selected from the tumor mass and with much less tumor content. Still however, implementation of tumor budding into the diagnostic setting requires further standardization to better define tumor buds and establish scoring parameters and cut-off criteria based on sensitivity and specificity. According to our data, tumor budding is a histological feature in BC with association to a more aggressive tumor phenotype and which may increase potential for metastasis. Our data demonstrate the utility of tumor budding in BC to potentially enhance prognostication in the clinic and warrants further investigation.
Acknowledgments
We thank the tissue bank Bern and the Translational Research Unit (TRU) at the Institute of Pathology, University of Bern, Switzerland for ngTMA construction.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This study was funded by the Claudia von Schilling Foundation, for Breast Cancer Research (Germany).
Contributor Information
Bodour Salhia, Email: bsalhia@tgen.org.
Coya Tapia, Phone: +41(0)31 632 87 63, Email: coya.tapia@hotmail.com.
References
- 1.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K, Hu M. Do myoepithelial cells hold the key for breast tumor progression? J Mammary Gland Biol Neoplasia. 2005;10(3):231–247. doi: 10.1007/s10911-005-9584-6. [DOI] [PubMed] [Google Scholar]
- 4.Sims AH, Howell A, Howell SJ, Clarke RB. Origins of breast cancer subtypes and therapeutic implications. Nat Clin Pract Oncol. 2007;4(9):516–525. doi: 10.1038/ncponc0908. [DOI] [PubMed] [Google Scholar]
- 5.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2001;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 6.Gokmen-Polar Y, Badve S. Breast cancer prognostic markers: where are we now? MLO Med Lab Obs. 2012;44(22):24–25. [PubMed] [Google Scholar]
- 7.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 8.Wessels LF, van Welsem T, Hart AA, van’t Veer LJ, Reinders MJ, Nederlof PM. Molecular classification of breast carcinomas by comparative genomic hybridization: a specific somatic genetic profile for BRCA1 tumors. Cancer Res. 2002;62:7110–7117. [PubMed] [Google Scholar]
- 9.Lugli A, Karamitopoulou E, Zlobec I. Tumour budding: a promising parameter in colorectal cancer. Br J Cancer. 2012;106:1713–1717. doi: 10.1038/bjc.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 11.Zlobec I, Suter G, Perren A, Lugli A. A next-generation tissue microarray (ngTMA) protocol for biomarker studies. J Vis Exp. 2014;23(91):e51893. doi: 10.3791/51893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne KC, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;16:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 14.Karamitopoulou E, Zlobec I, Kölzer V, Kondi-Pafiti A, Patsouris ES, Gennatas K, Lugli A. Proposal for a 10-high-power-fields scoring method for the assessment of tumor budding in colorectal cancer. Mod Pathol. 2013;26:295–301. doi: 10.1038/modpathol.2012.155. [DOI] [PubMed] [Google Scholar]
- 15.Liang F, Cao W, Wang Y, Li L, Zhang G, Wang Z. The prognostic value of tumor budding in invasive breast cancer. Pathol Res Pract. 2013;209(5):269–275. doi: 10.1016/j.prp.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Zlobec I, Hädrich M, Dawson H, Koelzer VH, Borner M, Mallaev M, Schnüriger B, Inderbitzin D, Lugli A. Intratumoral budding (ITB) in preoperative biopsies predicts the presence of lymph node and distant metastases in colon and rectal cancer patients. Br J Cancer. 2014;110(4):1008–1013. doi: 10.1038/bjc.2013.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landis JR, Koch CG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 18.Guzinska-Ustymowicz K. The role of tumour budding at the front of invasion and recurrence of rectal carcinoma. Anticancer Res. 2005;25(2B):1269–1272. [PubMed] [Google Scholar]
- 19.Niwa Y, Yamada S, Koike M, Kanda M, Fujii T, Nakayama G, Sugimoto H, Nomoto S, Fujiwara M, Kodera Y. Epithelial to mesenchymal transition correlates with tumor budding and predicts prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2014;110(6):764–769. doi: 10.1002/jso.23694. [DOI] [PubMed] [Google Scholar]
- 20.Karamitopoulou E, Zlobec I, Born D, Kondi-Pafiti A, Lykoudis P, Mellou A, Genatas K, Gloor B, Lugli A. Tumour budding is a strong and independent prognostic factor in pancreatic cancer. Eur J Cancer. 2013;49(5):1032–1039. doi: 10.1016/j.ejca.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO classification of tumours of the breast. 4e. Lyon: IARC Press; 2012. [Google Scholar]
- 22.Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1(7):651–661. doi: 10.18632/oncotarget.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Wang Y, Fan C, Gao P, Wang X, Wie G, Wie J. Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation. Mol Cancer. 2014;3(13):137. doi: 10.1186/1476-4598-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiménez-Salazar JE, Posadas-Rodriquez P, Lazzarini-Lechuga RC, Luna-Lopez A, Zentella-Dehesa A, Gomez-Quiroz LE, Königsberg M, Dominquez-Gomez G, Damian-Matsumura P. Membrane-initiated estradiol signaling of epithelial-mesenchymal transition-associated mechanisms through regulation of tight junctions in human breast cancer cells. Horm Cancer. 2014;5(3):161–173. doi: 10.1007/s12672-014-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Shen A, Zhang Y, Chen Y, Lin J, Lin W, Sferra T, Peng J. Pien Tze Huang inhibits hypoxia-induced epithelial-mesenchymal transition in human colon carcinoma cells through suppression of the HIF-1 pathway. Exp Ther Med. 2014;7(5):1237–1242. doi: 10.3892/etm.2014.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varga Z, Diebold J, Dommann-Scherrer C, Frick H, Kaup D, Noske A, Obermann E, Oehlschlegel C, Padberg B, Rakozy C, Sancho Oliver S, Schobinger-Clement S, Schreiber-Facklam H, Singer G, Tapia C, Wagner U, Mastropasqua MG, Viale G, Lehr AH. How reliable is Ki-67 immunohistochemistry in grade 2 breast carcinomas? A QA study of the Swiss working group of breast- and gynecopathologists. PLoS One. 2012;7(5):e37379. doi: 10.1371/journal.pone.0037379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutsvik G, Stefansson IM, Aziz S, Arnes J, Eide J, Collett K, Akslen LA. Evluation of Ki67 expression across distinct categories of breast cancer specimens: a population-based study of matched surgical specimens, core needle biopsies and tissue microarrays. PLoS One. 2014;9(11):e112121. doi: 10.1371/journal.pone.0112121. [DOI] [PMC free article] [PubMed] [Google Scholar]