Abstract
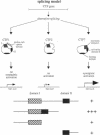
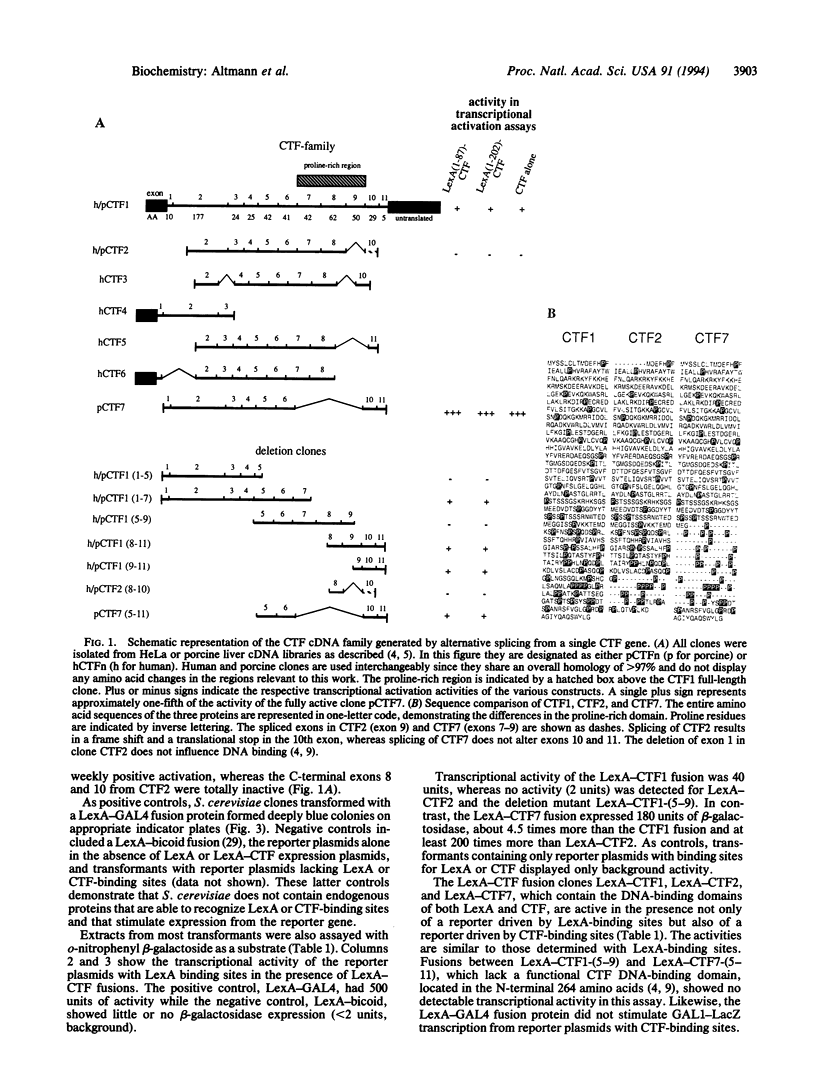
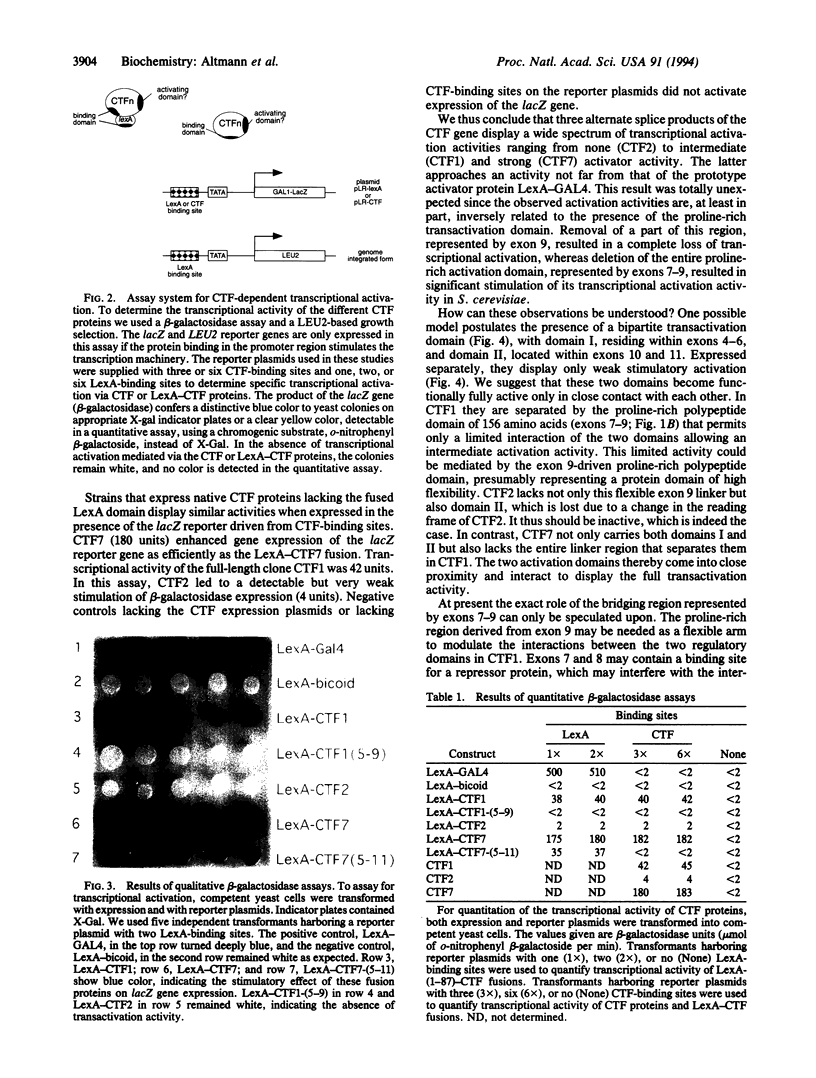
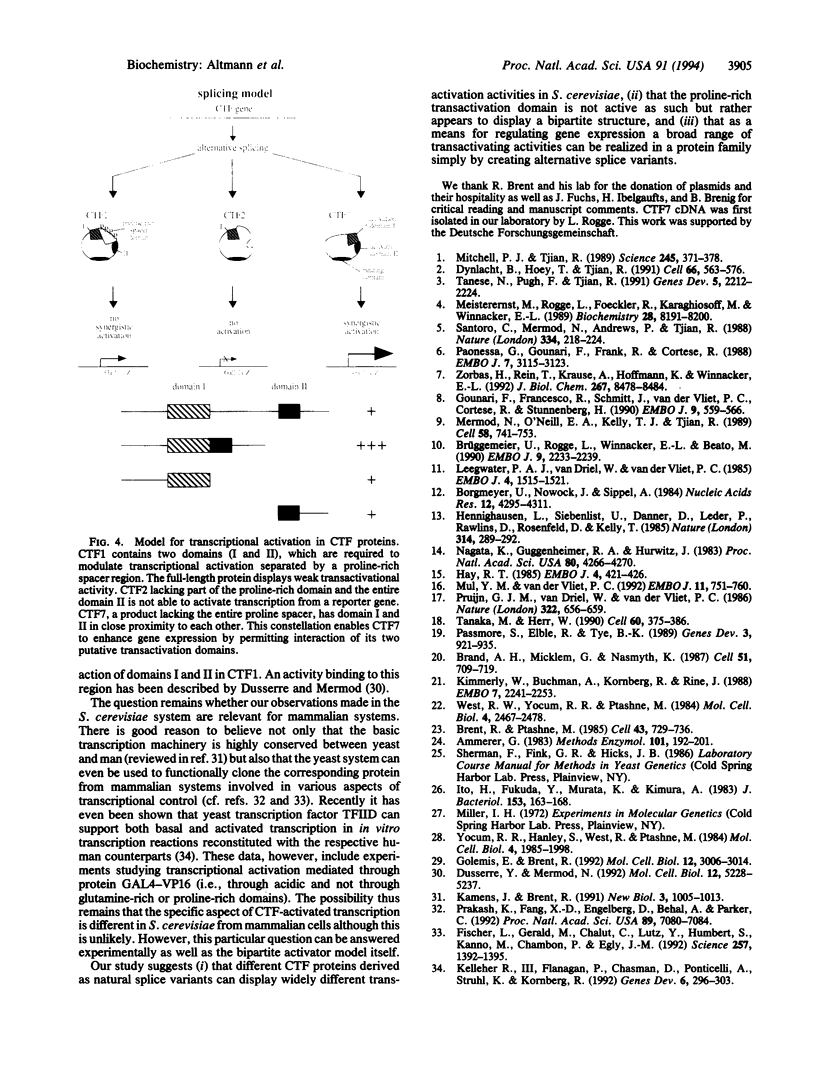
Members of the CCAAT-binding transcription factor (CTF) family of proteins stimulate the initiation of adenovirus DNA replication and act as transcriptional activators. To investigate the mechanisms underlying CTF-mediated transactivation patterns, we expressed several natural CTF variants in Saccharomyces cerevisiae and determined their transactivating activities in enzymatic assays. CTF7, which lacks the entire proline-rich region previously thought to mediate transcriptional activation by CTF proteins, enhances transcription to a greater degree than full-length CTF1, which contains the putative activation domain. CTF2, which contains a partially deleted proline-rich activation region, does not stimulate transcription at all. These findings indicate that the proline-rich region of CTF proteins is not essential for transcriptional activation in yeast. Our studies also suggest a bipartite two-domain structure of CTF-type transcriptional activation domains.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Borgmeyer U., Nowock J., Sippel A. E. The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res. 1984 May 25;12(10):4295–4311. doi: 10.1093/nar/12.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Brüggemeier U., Rogge L., Winnacker E. L., Beato M. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. EMBO J. 1990 Jul;9(7):2233–2239. doi: 10.1002/j.1460-2075.1990.tb07393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusserre Y., Mermod N. Purified cofactors and histone H1 mediate transcriptional regulation by CTF/NF-I. Mol Cell Biol. 1992 Nov;12(11):5228–5237. doi: 10.1128/mcb.12.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Fischer L., Gerard M., Chalut C., Lutz Y., Humbert S., Kanno M., Chambon P., Egly J. M. Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science. 1992 Sep 4;257(5075):1392–1395. doi: 10.1126/science.1529339. [DOI] [PubMed] [Google Scholar]
- Golemis E. A., Brent R. Fused protein domains inhibit DNA binding by LexA. Mol Cell Biol. 1992 Jul;12(7):3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounari F., De Francesco R., Schmitt J., van der Vliet P., Cortese R., Stunnenberg H. Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 1990 Feb;9(2):559–566. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R. T. The origin of adenovirus DNA replication: minimal DNA sequence requirement in vivo. EMBO J. 1985 Feb;4(2):421–426. doi: 10.1002/j.1460-2075.1985.tb03645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Siebenlist U., Danner D., Leder P., Rawlins D., Rosenfeld P., Kelly T., Jr High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985 Mar 21;314(6008):289–292. doi: 10.1038/314289a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens J., Brent R. A yeast transcription assay defines distinct rel and dorsal DNA recognition sequences. New Biol. 1991 Oct;3(10):1005–1013. [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Chasman D. I., Ponticelli A. S., Struhl K., Kornberg R. D. Yeast and human TFIIDs are interchangeable for the response to acidic transcriptional activators in vitro. Genes Dev. 1992 Feb;6(2):296–303. doi: 10.1101/gad.6.2.296. [DOI] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater P. A., van Driel W., van der Vliet P. C. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 1985 Jun;4(6):1515–1521. doi: 10.1002/j.1460-2075.1985.tb03811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M., Rogge L., Foeckler R., Karaghiosoff M., Winnacker E. L. Structural and functional organization of a porcine gene coding for nuclear factor I. Biochemistry. 1989 Oct 3;28(20):8191–8200. doi: 10.1021/bi00446a034. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mul Y. M., Van der Vliet P. C. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 1992 Feb;11(2):751–760. doi: 10.1002/j.1460-2075.1992.tb05108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Hurwitz J. Adenovirus DNA replication in vitro: synthesis of full-length DNA with purified proteins. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4266–4270. doi: 10.1073/pnas.80.14.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paonessa G., Gounari F., Frank R., Cortese R. Purification of a NF1-like DNA-binding protein from rat liver and cloning of the corresponding cDNA. EMBO J. 1988 Oct;7(10):3115–3123. doi: 10.1002/j.1460-2075.1988.tb03178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore S., Elble R., Tye B. K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989 Jul;3(7):921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- Prakash K., Fang X. D., Engelberg D., Behal A., Parker C. S. dOct2, a Drosophila Oct transcription factor that functions in yeast. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7080–7084. doi: 10.1073/pnas.89.15.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn G. J., van Driel W., van der Vliet P. C. Nuclear factor III, a novel sequence-specific DNA-binding protein from HeLa cells stimulating adenovirus DNA replication. Nature. 1986 Aug 14;322(6080):656–659. doi: 10.1038/322656a0. [DOI] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990 Feb 9;60(3):375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- West R. W., Jr, Yocum R. R., Ptashne M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol. 1984 Nov;4(11):2467–2478. doi: 10.1128/mcb.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum R. R., Hanley S., West R., Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas H., Rein T., Krause A., Hoffmann K., Winnacker E. L. Nuclear factor I (NF I) binds to an NF I-type site but not to the CCAAT site in the human alpha-globin gene promoter. J Biol Chem. 1992 Apr 25;267(12):8478–8484. [PubMed] [Google Scholar]