Abstract
Plasmonic nanomaterials as drug delivery or bio-imaging agents are typically introduced to biological systems through intravenous administration. However, the potential for agglomeration of nanoparticles in biological systems could dramatically affect their pharmacokinetic profile and toxic potential. Development of rapid screening methods to evaluate agglomeration is urgently needed to monitor the physical nature of nanoparticles as they are introduced into blood. Here, we establish novel methods using darkfield microscopy with hyperspectral detection (hsDFM), single particle inductively-coupled plasma mass spectrometry (spICP-MS), and confocal Raman microscopy (cRM) to discriminate gold nanoparticles (AuNPs) and their agglomerates in blood. Rich information about nanoparticle agglomeration in situ is provided by hsDFM monitoring of the plasmon resonance of primary nanoparticles and their agglomerates in whole blood; cRM is an effective complement to hsDFM to detect AuNP agglomerates in minimally manipulated samples. The AuNPs and the particle agglomerates were further distinguished in blood for the first time by quantification of particle mass using spICP-MS with excellent sensitivity and specificity. Furthermore, the agglomeration status of synthesized and commercial NPs incubated in blood was successfully assessed using the developed methods. Together, these complementary methods enable rapid determination of the agglomeration status of plasmonic nanomaterials in biological systems, specifically blood.
1. Introduction
Nanomaterial-containing commercial products are under intense development by the pharmaceutical industry for imaging, diagnosis, prevention, and treatment of disease [1]. The safety of these new, nanomaterial-containing products remains a concern to scientists and the public [2]. The toxicity of nanomaterials depends strongly on their physiochemical properties (e.g. size, size distribution, shape, surface charge, crystal structure, hydrophobicity, surface reactivity, solubility, aggregation and purity) and material composition itself [3–5]. The United States Food and Drug Administration (FDA) has developed a nanotechnology regulatory science program to enhance research in nanomaterial characterization, in vitro and in vivo modeling, and product-focused disposition and toxicity [6]. One priority is the development of the analytical tools to detect and characterize nanomaterials in commercial products, food matrices, and biological systems. The challenge facing biomedical research is the poor understanding of the agglomeration status and biological fate of nanomaterials once they are introduced into the blood stream.
Gold nanomaterials (GNMs) are particularly appealing candidates as new diagnostic and therapeutic agents because of their relative bioinertness, tightly controllable morphology, facile surface functionalization, and unique optical properties [7–10]. Some GNM-based medicines are in clinical trials. As an example, Aurimune® is a nanomedicine with tumor necrosis factor (rhTNFα) covalently conjugated to PEGylated gold nanoparticles that has been approved for phase II clinical trials for cancer therapy [11]. AuroLase®, which is also in clinical trials, utilizes gold nanoshells and laser technology as a new photothermal treatment modality for refractory head and neck cancer [12]. In practice, GNM-based medicine is generally administered intravenously, i.e. directly into the bloodstream. Blood is a complex fluid with significant concentrations of proteins, salts, and blood cells. Nanoparticles interact with blood proteins; adsorption onto the materials’ surface results in the formation of a protein corona [13, 14]. The change in surface properties introduced by blood ionic strength can induce nanoparticle agglomeration [15], and thus significantly alter the cellular interactions, biodistribution [16], and toxicity profile of the particles [17]. Additionally, nanomaterials retard cell motility [18], and nanomaterial induced endothelial leakage (NanoEL), wherein nanomaterials bypass the cell membrane by disrupting through adherens junction, has also shown a strong correlation to the hydrodynamic size of the particle [19]. Due to their potential of agglomeration, in situ monitoring of nanoparticles in blood is crucial to fully understand in vivo effects of GNM-materials targeted for human theranostics.
There are many well-developed methods to characterize GNMs; however, characterizing GNMs in blood is complicated by the complex environment, e.g. plasma proteins and various blood cells. Electron microscopy (EM) is considered the most accurate method to measure the size of nanoparticles and the most widely used method of assessing nanoparticle morphology. However, EM requires considerable sample manipulation, which introduces artifacts; it provides only a static image of the GNMs, typically in thin, dry sections of tissues or matrices (e.g. 50–100 nm), and cannot readily distinguish agglomerates from primary particles located in close proximity [20–23]. Recently, cellular uptake of 30 nm, spherical AuNPs has been visualized using scanning transmission electron microscopy (STEM) imaging of liquids, yet at this point the technique suffers from low resolution [24]. Chromatographic techniques, such as size exclusion chromatography, ultracentrifugation, and electrophoresis, have been shown to separate primary and agglomerated nanoparticles based on shape, size, or charge [25]. One considerable disadvantage is that these techniques often require extensive sample preparation, which may affect particle agglomeration status. Dynamic light scattering (DLS) has been used to estimate particle concentration in blood [26]; however, DLS is limited due to (1) its requirement that the blood cells to be lysed prior to measurement, which could affect the agglomeration status of nanomaterials; (2) the presence of proteins and cells in blood interferes with nanoparticle signal; and (3) larger particles which are overrepresented in polydisperse samples [27]. UV-Vis and Raman spectroscopy can readily distinguish primary and agglomerated plasmonic nanoparticles [28, 29], but the signal is compromised by the extremely high optical density and opacity of blood. Accordingly, we are unable to find a simple and robust methodology for in situ characterization of nanoparticle agglomeration in blood and other complex biological environments.
In this work, complementary and rapid in situ methods have been developed to monitor the agglomeration status of plasmonic nanoparticles in ex vivo blood. These methods include darkfield microscopy with hyperspectral imaging (hsDFM), confocal Raman microscopy (cRM), and single-particle inductively-coupled plasma mass spectrometry (spICP-MS). The first two methods are based on the optical properties of GNMs, that is, the localized surface plasmon resonance (LSPR) [30]. The LSPR is extremely sensitive to particle morphology with agglomeration of GNMs resulting in a shift of the LSPR to lower energy [31]. The hsDFM can monitor this shift of scattered photons from GNMs, thereby distinguishing light scattered by the cellular environment from that scattered by nanoparticles [32–36], and this instrument has been used to distinguish macrophage maturity based on silver nanoparticle (AgNPs) uptake [37].
Scattering also gives rise to enhancement of the fluorescence and Raman scattering properties of molecules close to the GNMs [38]. The areas of high curvature or between adjacent AuNPs at agglomeration sites create localized “hot spots” that enhance Raman signals, similar to a roughened metal surface [38], with enhancement factors that can be 108 or greater [39]. Surface enhanced Raman spectroscopy (SERS) has been used to identify circulating tumor cells in blood [40] and detect tumor cells in vivo [41], as well as a wide-range of other chemical detection and sensing applications [42, 43]. We have applied SERS via cRM to directly monitor nanoparticle agglomeration as a label-free detection method in this study.
In addition to the optical techniques, spICP-MS provides a high-resolution technique to detect individual particle events for characterization of a variety of engineered nanomaterials [44]. Unlike traditional ICP-MS, which provides bulk elemental analysis of homogenized metallic species, spICP-MS provides a measure of individual particles in suspension. At sufficiently low particle concentrations, particles are introduced individually for atomization and ionization in the plasma, creating a packet of ions that are detected as a discrete signal. This quantifiable spike in intensity due to single particle events enables individual nanoparticles to be distinguished from the background ionic current [45]. The signal intensity is proportional to the number of atoms in an individual event, so larger particles produce higher signal intensity. Similarly, particle agglomeration can be detected by monitoring the increase in signal intensity [46]. By collecting a large number of data points, the particle size and distribution can be determined [44, 46–48].
In this report, we have validated and applied the spICP-MS technique to analyze the agglomeration status of nanoparticles in blood as a complement to the optical methods. AuNPs and agglomerates were synthesized and characterized using well-developed methods like UV-Vis, Raman spectroscopy, TEM, DLS, nanoparticle tracking analysis (NTA), and discrete dipole approximation (DDA) calculations. Detection of particle agglomeration was validated in a variety of simple biological media. The promising methods were then used to detect agglomerates in blood, with hsDFM and spICP-MS as the primary methodologies. Finally, citrate-capped NPs were incubated in blood, and their agglomeration was monitored using the developed methods. Together, hsDFM, cRM, and spICP-MS provide a rapid and robust means to analyze nanoparticle agglomeration in biological systems with minimal sample preparation. These diverse, label-free methods can distinguish primary particles from agglomerates in blood. Identification of the interactions between nanoparticles and components in biological systems is, in turn, critically important for rational design and implementation of nanomedicine.
2. Materials and methods
2.1. Chemicals
Tetrachloroauric acid trihydrate (HAuCl4·3H2O) and trisodium citrate heptahydrate (Na3-Cit; Na3C6H5O7·7H2O) were acquired from Alfa Aesar (Ward Hill, MA). Phosphate buffered saline (PBS), fetal bovine serum (FBS), and Eagle medium were acquired from Corning Cellgro (Manassas, VA). The reference Au NPs with nominal diameters of 10 nm (RM8010), 30 nm (RM8012), and 60 nm (RM8013), were purchased from the National Institute of Standards and Technology (NIST). Citrate-capped 75-nm AgNPs were obtained from NanoComposix (San Diego, CA). Sodium chloride (NaCl) and bovine serum albumin (BSA) were obtained from Sigma Aldrich (St. Louis, MO). All chemicals were used as received.
2.2. Synthesis of AuNPs and agglomerates
Citrate-capped AuNPs (AuNP-cit) were synthesized via the Turkevich method as has been previously described [49]. Briefly, 10.6 mg HAuCl4 was dissolved in 99 mL 18 MΩ H2O and heated to boiling. To this solution, 0.9 mL of 10 mg/mL Na3Cit was quickly injected. After 20 min, a deep wine-red color was observed, indicating the formation of AuNPs. The suspension was removed from heat and allowed to cool to room temperature. Prior to use, the excess precursors were removed by centrifuging at 20,800 g for 20 min at 20 °C, and the AuNP-cit was redispersed in 18 MΩ H2O.
Agglomerates were generated by adding 0, 2.5, 5, 7.5, 10, 20, or 50 µL of saturated NaCl solution to 1 mL of the AuNP-cit suspension, immediately followed by thoroughly mixing for 5 sec. After 10 min, 50 µL BSA (40 mg/mL, 18 MΩ H2O) was added to arrest agglomeration [50]. The solution was immediately mixed and allowed to incubate without further mixing for 30 min at room temperature. Agglomerates were isolated by centrifugation at 20,800 g for 20 min at 20 °C. The pellet was redispersed in 18 MΩ H2O, centrifuged a second time (20,800 g, 20 min, 20 °C), redispersed in 18 MΩ H2O, and stored at room temperature. Samples were redispersed by brief (<2 sec) bath sonication (Branson 5510, Branson Ultrasonics, Danbury, CT) prior to use. No appreciable changes in the UV-Vis spectra or hydrodynamic diameter of agglomerates used were observed after 1 month of storage at room temperature in the dark.
2.3. Introduction of nanoparticles into blood
Adult Fisher 344 rats were obtained from the National Center for Toxicological Research (NCTR) Breeding Colony (Jefferson, AR). Whole blood samples from the rats were collected via cardiac puncture into plastic whole blood collection tube (Becton, Dickinson, Franklin Lakes, NJ) for the study. The animal procedures followed the guidelines of the “Guide for the Care and Use of Laboratory Animals” and were approved by the NCTR Institutional Animal Care and Use Committee. AuNP-cit (100 µL; 80 ppm) was incubated directly with 1.5 mL fresh whole blood in a blood collection tube. Alternatively, primary AuNPs and agglomerates (80 ppm) were redispersed by bath sonication (~2 sec) prior to use. A 20 µL sample was added to 200 µL of fresh, nanoparticle-free blood, followed by mixing for 2 sec (final concentration ~8 ppm). To prepare samples for imaging, 3 µL of blood was applied to a clean-room-grade glass microscope slide (Schott, Elmsford, NY), smeared, and a coverslip applied immediately prior to acquisition of hsDFM images. An identical process was followed to introduce AgNPs to blood.
2.4. Transmission electron microscopy (TEM)
TEM images were recorded using a JEOL JEM-1400 (JEOL, Tokyo, Japan) with accelerating voltage of 80 keV. Samples (3 µL) were applied onto a 200 mesh carbon-coated copper grid, followed by addition of 3 µL isopropyl alcohol and air dried. Images were acquired using XR41 4mp TEM CCD system (AMT, Woburn, MA) and the particle size was then measured using ImageJ software (National Institute of Health, Bethesda, MD).
2.5. Dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA)
Hydrodynamic diameter (dH) and zeta potential were determined using a ZetaPALS instrument (Brookhaven, Holtzville, NY). The sample was prepared by diluting 20 µL of primary AuNPs or agglomerates to 2 mL with 1 mM of KCl after brief sonication (<5 sec) of the sample (~800 ppb). AuNP-cit was diluted 100-fold in 18 MΩ H2O. The dH was measured by performing three consecutive 60 sec runs while the zeta potential was measured at 10 runs of 5 cycles each. Average hydrodynamic size was determined using nanoparticle tracking analysis (NTA), and was performed using NanoSight LM-10 (Malvern Instruments Inc, MA, USA). The sample was prepared by diluting 10 µL of sample to 10 mL with 18 MΩ H2O (80 ppb) after brief sonication (<5 sec) of the sample. The diameter corresponding to the mode of the sample was used to represent the most prominent nanoparticle size in solution. Triplicate measurements were acquired for each sample.
2.6. UV-Vis spectroscopy
Extinction spectra were acquired on an Agilent HP8453 UV-Vis spectrometer (Agilent Technologies, Santa Clara, CA) using a quartz cuvette with 1-cm path length and 18 MΩ H2O as a blank. Samples were sonicated for <5 sec and diluted 10-fold to 8 ppm with 18 MΩ H2O prior to measurement.
2.7. Confocal Raman microscopy (cRM)
Raman spectra were acquired using a Horiba Jobin Yvon high-resolution LabRam Raman microscope system (Horiba, Edison, NJ) equipped with a charge-coupled detector and a spectrometer with a grating of 600 lines/mm. The systems were set up with a 150-µm entrance slit and a 400-µm pinhole. The 633-nm laser excitation was provided by a HeNe laser operating at 5 mW. Raman shift calibration was performed using the 521 cm−1 line of a silicon wafer. Raman samples was prepared by diluting 100 µL of AuNPs, or agglomerates, to 1 mL with 18 MΩ H2O (8 ppm) after brief sonication (<5 sec) of the sample. The diluted sample was then transferred to a 50 µL, 10 mm path length quartz cuvette. Signal was acquired for 10 sec and three acquisitions were averaged per spectrum to improve signal-to-noise ratio.
2.8. Darkfield microscopy with hyperspectral detection (hsDFM)
Darkfield microscopy images and hyperspectral plots were acquired using enhanced darkfield transmission optical microscope (Olympus BX41) equipped with hyperspectral imaging spectrophotometer (Headwall, CytoViva Inc, Auburn, AL). This system is capable of recording high quality spectra (high signal-to-noise ratio) in the visible and near infrared wavelength range (400–1000 nm). Hyperspectral images were acquired using a 0.25 sec collection time and a white light source. Images were acquired using a 100× oil immersion lens.
2.9. Single particle inductively-coupled plasmon mass spectrometry (spICP-MS)
The nanoparticles or agglomerates were stored at 4 °C in the dark prior to analysis. Samples were warmed to room temperature, then mixed and sonicated briefly prior to dilution to ensure full dispersion without disrupting agglomeration. Nanoparticles or agglomerates (0.1 mg/mL) were briefly incubated with fresh blood, and then diluted into 10 mL 18 MΩ water. Single particle analysis was performed using an Agilent 7700X ICP-MS (Agilent, Santa Clara, CA) which was tuned daily to optimal sensitivity and resolution. A MicroMist glass concentric nebulizer was used with nickel composite sample and skimmer cones. Argon was used as a carrier gas at a flow rate of 1.05 mL/min. Peristaltic pump speed was set at 0.1 rpm. Samples were analyzed in selected time resolved analysis (TRA)/full quantification mode with a sampling period of 0.01 sec and an acquisition time of 180 sec. Between each acquisition, the instrument was first flushed with a solution containing 10% HNO3 and 10% HCl in water at a pump speed of 0.5 rpm for 3 min to eliminate the AuNPs that were trapped inside the sampling tube from prior measurement. Then the system was rinsed with water for another 3 min to purge the acid. Every sample was measured in triplicate. Agilent’s MassHunter Workstation software was used for data analysis.
2.10. Calculation of optical properties
The optical properties were calculated according to the discrete dipole approximation (DDA) using the DDSCAT 7.3 program [51]. In this formalism, the particles are represented by an array of dipole moments residing within the volume of the nanoparticles. The nanoparticles were represented as a dielectric continuum with the complex dielectric response function of bulk Au [52]. The optical efficiency, Q, is reported as the ratio of the respective optical cross section to , where the effective radius, aeff,, is defined as the radius of a sphere whose volume is equal to that of the nanoparticle. The optical cross sections were averaged over two orthogonal polarization directions of the incident light.
3. Results and discussion
3.1. Synthesis and characterization of nanoparticles and agglomerates
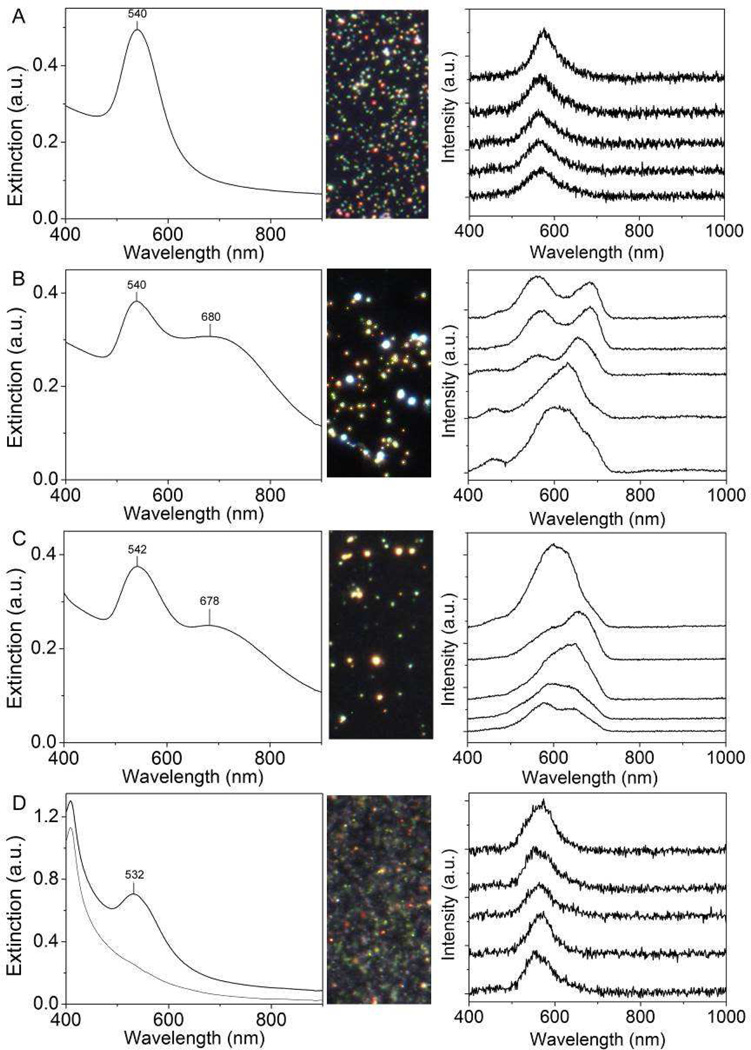
The AuNPs synthesized via the citrate reduction method (AuNP-cit) [49] had a diameter of 44.5 ± 9.2 nm from TEM measurements (Fig. 1A inset, Fig S1). Based on DLS, the dH was 50.1 ± 0.7 nm and the zeta potential was −35.2 ± 0.9 mV. NTA indicated a mode diameter of 51.3 ± 2.3 nm. The size difference between TEM and DLS/NTA measurements is expected because TEM is used to determine the size of the metal core, while DLS and NTA measure dH, which reflects the size of the metal particle and the citrate stabilizer in solution. AuNP-cit exhibited an LSPR maximum at 537 nm (Fig. 1A) and appeared wine-red in color. The simulated spectrum of a 45 nm diameter Au nanosphere showed an extinction peak at 534 nm, similar to that of the measured spectrum (Fig. 1E).
Figure 1.
Characterization of AuNPs and agglomerates synthesized as described in the methods: (left) UV-Vis spectra of samples with inset TEM images (200 nm scale bar) and (right) corresponding spectra simulated by discrete dipole approximation with inset geometric illustrations: (A, E) citrate-capped AuNPs; (B, F) BSA-capped primary AuNPs, (C, G) small agglomerates; and (D, H) large agglomerates. The spectra in (G) are averages from six possible geometries as shown in Figure S3.
Citrate-capped AuNPs have long been known to agglomerate following an increase in the ionic strength in the suspensions [50]. Agglomerates of increasing size were generated by addition of increasing volumes of saturated NaCl to AuNP-cit suspensions. After 10 min reaction, BSA solution (40 mg/mL) was added to the suspension to arrest agglomeration as described previously [17], and the samples in various agglomeration states were collected via centrifugation. The transmitted color of the suspension visibly changed from its initial wine-red to purple and then purple-black. The UV-Vis spectra of primary, BSA-coated AuNPs and agglomerates showed that as the NaCl concentration increased the LSPR red-shifted from 539 to 545 nm and a shoulder, red-shifted from the LSPR, emerged and resolved to a peak in the NIR (Fig. S2). The morphology of the agglomerates was assessed with TEM (Fig. S3).
From these BSA-coated samples, “primary AuNPs,” “small agglomerates,” and “large agglomerates” were defined as the samples agglomerated with 0, 2.5, and 7.5 µL of saturated NaCl, respectively. These samples were used as standards for further experiments. Primary AuNPs (Fig. 1B) showed no agglomeration and had an LSPR maximum at 539 nm. Small agglomerates (Fig. 1C) consisted of trimers to pentamers with an LSPR peak at 541 nm and a visible shoulder at ~600 nm. Large agglomerates (Fig. 1D) were typically composed of between 10 and 25 AuNPs per agglomerate and showed LSPR peaks at 545 and 780 nm. The emergence and separation of these two peaks is often observed during controlled agglomeration of nanoparticles and results from the plasmon resonance of the particle ensembles, while the peak attributed to individual particles remains [53]. The particle size, orientation, and interparticle distance can all affect the position of the LSPR that results from the ensemble of particles [29, 31, 54].
The discrete dipole approximation (DDA) method was used to simulate the optical spectra of primary AuNPs and their agglomerates. The simulated spectra of a BSA-coated primary AuNP shows an extinction peak at 543 nm (Fig. 1F), slightly red-shifted from that of a citrate-coated particle (534 nm) due to the increase of refractive index from 1.33 for citrate to 1.46 for BSA. For small agglomerates, spectra of several configurations were simulated according to visualization in TEM, as shown in Figure S4. These spectra were averaged and plotted in Figure 1G, indicating an extinction peak at ~540 nm and a shoulder at ~670 nm, which is comparable to the measured spectrum. In the case of large agglomerates, a representative configuration was chosen for simulation (Fig. 1H) because it was not practical to integrate all the configurations that 10–25 particles could stochastically adopt in the sample. The simulated spectrum shows two peaks at ~550 nm and ~750 nm with a board shoulder in between, which indicates a similar trend as the measured spectrum despite the discrepancy due to different configurations in the actual sample. It is important to note that the scattering efficiency increases in the order of primary AuNPs, small agglomerates, and large agglomerates.
The trend of increasing agglomeration was further confirmed by DLS, NTA, and zeta potential measurements (Table 1). A rapid increase of hydrodynamic diameter (dH) was observed by DLS during the progression from primary AuNPs (53.7 ± 0.4 nm) to small agglomerates (142.3 ± 0.4 nm), and large agglomerates (176.5 ± 3.1 nm). This result was further confirmed by NTA with respective mode diameters of 63.0 ± 1.0, 86.0 ± 13.0, and 177 ± 27.4 nm. Additionally, the zeta potential changed from -−19.5 ± 1.5 mV for primary AuNPs to −20.8 ± 1.1 and −28.3 ± 1.2 mV, suggesting that the primary AuNPs and agglomerates were coated with BSA, in agreement with other studies under similar conditions [55, 56]. The initial discrepancy between DLS and NTA can be ascribed to the different measurements of the instrument. DLS is weighted by signal intensity, so a greater contribution to the dH is made by larger components than smaller components, which can artificially inflate dH values; NTA measures many particles individually but produces a diameter based on population intensity, rather than signal intensity, which may artificially deflate dH values. Further, agglomerates of 3–5 particles can be expected to show high variability, as their overall geometry can be expected to change the most from the stochastic arrangement (see Fig S4 for some examples). Both of these techniques rely on the velocity of particles in solution, and indicate that as the agglomerates become larger, they travel more slowly through the solution.
Table 1.
Summary of characterization of AuNPs and their agglomerates.
| AuNPs per Agglomerate |
LSPR (nm) |
dH (DLS) (nm) |
dH (NTA) (nm) |
Zeta Potential (mV) |
Designation |
|---|---|---|---|---|---|
| Citrate-capped | 537 | 50.1 ± 0.7 | 51.3 ± 2.3 | −35.2 ± 0.9 | AuNP-Cit |
| 1 | 539 | 53.7 ± 0.4 | 63.0 ± 1.0 | −19.5 ± 1.5 | Primary AuNPs |
| 3–5 | 542 | 142.3 ± 0.4 | 86.0 ± 13.0 | −20.8 ± 1.1 | Small agglomerates |
| 10–25 | 545,780 | 176.5 ± 3.1 | 177.0 ± 27.4 | −28.3 ± 1.2 | Large agglomerates |
3.2 Establishment of the hsDFM method for identification of agglomeration
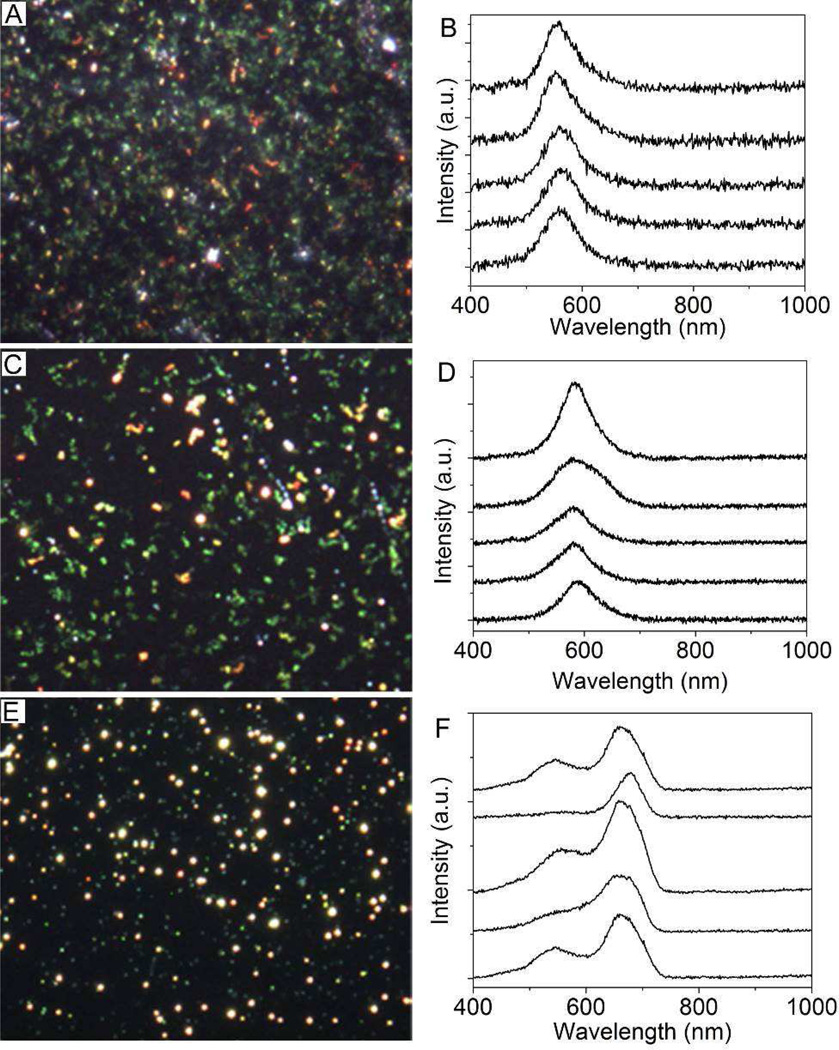
The hsDFM has been used as analytical tool to detect certain nanomaterials in cells [57, 58]. For instance, a recent study demonstrated the combination of the plasmonic Au dimer probes and hyperspectral imaging is capable of quantitatively imaging single mRNA splices in live cells [59]. In this study, primary AuNPs and their agglomerates can be easily seen using hsDFM because of their large scattering cross-sections. Primary AuNPs appeared green on the microscope (Fig. 2A) and moved around extremely quickly due to Brownian motion (i.e. diffusion). The agglomerates appeared yellow or and the particle motion decreased substantially (Fig. 2, C and E). Hyperspectral maps were acquired for the various samples. From these spectra, the most intense spectra were accumulated to represent typical spectral output. The scattering peak of primary AuNPs appeared at ~550–560 nm (Fig. 2B). For small agglomerates, the peak shifted to ~580 nm (Fig. 2D); for large agglomerates peaks were observed at ~550 nm and ~660–700 nm (Fig. 2F). Importantly, the signal intensity increased from primary AuNPs to small and large agglomerates, as would be expected for a scattering-based process. The signal from agglomerates is significantly brighter than that of primary AuNPs, therefore the presence of agglomerates can be readily detected by the relative intensity difference of their scattering spectra. Agglomeration can be qualitatively monitored based on the color of the scattered light as seen in the microscope and by the particle diffusion velocity, which can then be further confirmed using the hyperspectral camera to identify the change in wavelength and intensity.
Figure 2.
Darkfield images and typical hyperspectral scattering spectra of (A, B) primary AuNPs; (C, D) small agglomerates; and (E, F) large agglomerates, which correspond to the samples in Figure 2.
Having demonstrated the capacity to distinguish agglomerates, hsDFM was applied to identify the status of AuNP-cit in different media (i.e., H2O, PBS, cell-culture medium (CCM), and serum) and compared with UV-Vis spectra as shown in Figure 3. In Figure 3 A and C, the hsDFM showed scattering maxima at ~550–560 nm for H2O, and the true color images show mainly green, fast-moving particles, suggesting that the AuNP-cit did not agglomerate in these media. In agreement with these results, the UV-Vis spectra showed no significant changes in either medium. In contrast, hyperspectral plots of AuNP-cit incubated in PBS and CCM showed two typical peaks, one located in the 550–580 nm region and the other in the 630–700 nm region. True color, darkfield imaging displayed primarily bright yellow, slow-moving spots, suggesting that AuNP-cit agglomerates detectably in PBS and CCM. This result was confirmed by UV-Vis spectroscopy, which showed the emergence of a second peak ~680 nm, suggesting the formation of medium to large agglomerates (Figure 3 B and D). These results clearly demonstrate that the hsDFM method can detect and monitor agglomeration of AuNPs in simple media.
Figure 3.
The agglomeration behavior of citrate-capped AuNPs in four aqueous media: (A) H2O; (B) phosphate-buffered saline; (C) cell-culture medium; and (D) fetal bovine serum (black) with and (grey) without AuNPs. (Left) UV-Vis spectra; (Center) darkfield image; and (Right) typical hyperspectral scattering spectra.
3.3. Raman studies of agglomeration
The cRM has been applied as a fast and label-free analytical approach to detect carbon-based nanomaterials in biological system [60]. GNMs’ use as an agent for SERS has attracted much attention in recent years [61]. In this study, cRM was used to characterize agglomeration as a compliment to hsDFM. The surface coatings, citrate and BSA, can serve as indicators of the agglomeration of AuNPs because the gaps between agglomerated AuNPs can enhance Raman signal by up to 108–fold relative to primary particles [62]. Figure 4A shows this same trend with large agglomerates having more intense spectral features than small agglomerates. BSA-coated, primary AuNPs produced the same spectrum as pure H2O with Raman shift at ~1640 cm−1 [63]. As the size of the agglomerates increased, the Raman shift of H2O was replaced by several informative peaks. The peak at ~225 cm−1 may be the result of Au-N bond stretching [64] from the protein adsorbed to the particle surface. Additionally, a number of peaks emerge in the range of 1000–1600 cm−1 that are likely associated with stretching of the citrate carbonyls or aromatic vibrations and carbonyl stretching within the protein structure [65]. The strongest peak to emerge, however, is ~2900 cm−1 and can be attributed to C-H stretching [65]. Monitoring for the emergence of these peaks, particularly the C-H stretch, can be used to track the degree of agglomeration. Interestingly, the intensity of the peaks begins to decrease at higher agglomeration levels (Fig. S5), which could be the result of the agglomerate not being completely irradiated, thereby producing less signal [66], LSPR shift reducing the resonant absorption needed for enhancement [67], or the structure of the agglomerate resulting in less enhancement [68].
Figure 4.
Raman spectra taken from (A) (i) H2O and albumin-capped (ii) primary AuNPs, (iii) small agglomerates, and (iv) large agglomerates and (B) (i) H2O and citrate-capped AuNPs after 10 min incubation in (ii) H2O, (iii) phosphate-buffered saline, (iv) cell-culture medium, and (vi) fetal bovine serum with (v) normal fetal bovine serum as a reference (v).
Raman spectroscopy was used to complement the study of the various media, and the response of AuNP-cit to incubation in H2O, PBS, CCM, and serum was measured (Fig. 4B). Similar to the hsDFM and UV-Vis results, no significant change was seen between solvent spectra and Au containing spectra for water or serum, suggesting that the AuNP-cit remains nonagglomerated. On the other hand, incubation of AuNP-cit in CCM or PBS leads to the emergence of several Raman peaks attributed to citrate, particularly the C=O and C-H stretches. The emergence of these peaks further confirms the agglomeration of AuNP-cit in both of these media, and demonstrates the complementary nature of Raman spectroscopy for detection of agglomeration.
3.4. Detection and evaluation of Au particle agglomeration in blood by hsDFM
The primary AuNPs and their agglomerates were added to whole rat blood to model the behavior of the particles following intravenous injection. The optical density of blood limited the use of Raman and UV-Vis spectroscopy to monitor agglomeration, and the medium’s complexity additionally ruled out the use of many traditional characterization methods such as electron microscopy and light scattering [69]. Figure 5 shows darkfield images and typical hyperspectral plots (hsDFM) of primary AuNPs and large agglomerates following 5 min incubation in blood prior to slide preparation. The primary AuNPs and agglomerates can be visually distinguished using the hsDFM as they appear green and yellow, respectively, as shown by the respective spectra, and with different levels of brightness. The peak does not identically match the peaks observed in simpler media, which may be attributed to the optical interference by the blood. The peak location ~600 nm for primary particles shifts to ~800 nm for agglomerates. These data clearly demonstrate primary AuNPs can be distinguished from agglomerates amongst the blood cells by using the hsDFM technique.
Figure 5.
Characterization of prepared AuNPs in the blood: (A) darkfield image and (B) typical hyperspectral scattering spectra of primary AuNPs; (C) darkfield image and (D) typical hyperspectral scattering spectra of large agglomerates.
The hsDFM method was further used to monitor the agglomeration status of AuNP-cit in blood, as shown in Figure 6. AuNP-cit were incubated in blood for 5 min and 18 h. After 5 min incubation, the hyperspectral plots showed maxima primarily ~600 nm, indicating the presence of primary AuNPs. A slight red-shift to ~620 nm was observed after 18 h incubation, suggesting there may be some slight modification of the particles. This agglomeration could be the result of restructuring of the protein corona during incubation [70] or an artifact of the incubation itself. The typical blood half-life for AuNPs is on the order of hours, though with proper surface coating it can be extended to days [71]. As such, it is likely that the sanguine concentration of AuNPs in vivo would be low after 18 h, which would reduce the likelihood of agglomeration. Nonetheless, the power of hyperspectral imaging of individual particles using darkfield microscopy is demonstrated and can be further explored to identify changes in the surface of the nanoparticles (i.e. agglomeration).
Figure 6.
The agglomeration behavior of the citrate-capped AuNPs in the blood after incubation for different time periods: (A) darkfield image and (B) typical hyperspectral scattering spectra at 5 min; (C) darkfield image and (D) typical hyperspectral scattering spectra at 18 h.
For the most part, AuNP-cit did not agglomerate during incubation in blood, and cell morphology appeared unperturbed, though several anomalies should be remarked upon. First, once the blood sample is prepared, the sample must be imaged quickly to observe an accurate representation of the blood cell morphology. As the specimen remains on the microscope stage, the blood cells begin to deform over the course of 30 min. Presumably this cell disfiguration is the result of evaporation, leading to an increase in salinity and deformation of the blood cells. Blood cells presented typical morphology immediately following sample preparation even after the blood had been stored in a refrigerator for several days. Additionally, slides that had been prepared 30 min prior to imaging began to show evidence of the formation of opalescent particles visible by hsDFM that were likely the initial stages of thrombus formation.
3.5. Detection of Au nanoparticle agglomeration in blood by single particle ICP-MS
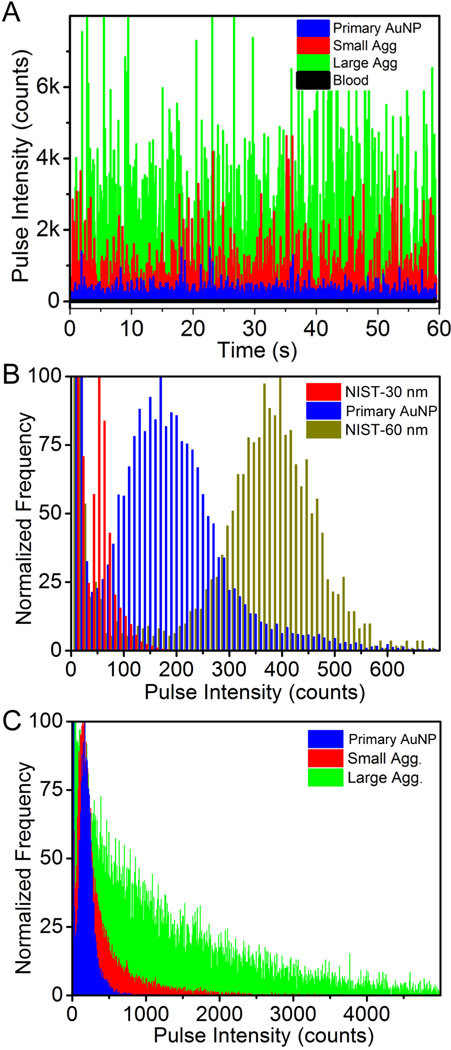
The spICP-MS was applied to discriminate the size of various primary AuNPs in whole rat blood. Whole rat blood was used as a matrix blank to monitor the background counts, and the results showed that majority of the events exhibited counts smaller than 20. Typical pulse intensity versus time output can clearly distinguish primary AuNPs, small agglomerates and large agglomerates in blood (Fig. 7A). Citrate-capped NIST AuNP reference materials with 30 nm and 60 nm diameter were coated with BSA (NIST-30 and NIST-60, respectively) and used as standards against which to compare to the primary AuNPs. The measurements for each sample were performed three times and the results were reported as a summation. Figure 7B clearly shows that a large population of events from our primary AuNPs fell between NIST-30 and NIST-60, consistent with the size of the primary AuNPs (45 nm diameter) obtained from TEM studies. Previous studies have suggested that in spICP-MS, signal intensity is proportional to particle size for diameters < 80 nm [44]. Since the intensity of each event is proportional to the number of detected ions, the intensity ratio of different particle sizes should be the same as their volume ratio. To simplify the process, samples were normalized to the bin with maximum number of events. As a result, the intensity ratio between our primary AuNPs and the NIST standards were 3.40 for NIST-30 and 0.42 for NIST-60, which are very close to their volume ratios using 45 nm particles (V45/V30 = 3.38; V45/V60 = 0.42).
Figure 7.
Single particle ICP-MS analysis of various AuNPs in whole rat blood: (A) Typical pulse intensity vs. time output from ICP-MS in single particle mode for blood without AuNPs (black), or incubated with primary AuNPs (blue), small agglomerates (red), or large agglomerates (green), (B) primary AuNPs in whole rat blood using 30 nm (red) and 60 nm (gold) diameter NIST standard citrate-capped AuNPs stabilized with albumin and in-house synthesized primary AuNPs stabilized with bovine serum albumin (blue); (C) in-house synthesized, albumin stabilized primary AuNPs (blue), small agglomerates (red), and large agglomerates (green) in whole rat blood.
The capability of spICP-MS to distinguish primary AuNPs, small agglomerates and large agglomerates in blood was further examined. Typical measurements of primary particles, small agglomerates, and large agglomerates after incubation in blood are shown in Figure 7C. Relative to primary AuNPs, small agglomerates presented a broader intensity range; a large portion of events had counts higher than 400/event. There were also a considerable number of events with even higher pulse intensities, suggesting a range of agglomerate masses. When large agglomerates were analyzed, the intensity distribution broadened further. As the intensity increased, the frequency of events decreased relative to small agglomerates, further indicating a much greater degree of agglomeration. These results demonstrate that spICP-MS can be readily utilized to monitor agglomeration of AuNPs in blood.
3.6. Evaluation of Ag nanoparticle agglomeration status in blood by hsDFM
Similar to AuNPs, AgNPs are also in development as potential therapeutics with strong plasmonic properties [72]. To test the robustness of the hsDFM method, AgNPs were subjected to similar analysis as AuNPs. Citrate-capped AgNPs were agglomerated by NaCl resulting in a change in LSPR. BSA-capped primary and agglomerated AgNPs were introduced to blood and imaged using darkfield microscopy (Fig. 8, A and B). Primary AgNPs scattered a cyan color associated with rapidly moving particles, while agglomerates appeared as slow-moving yellow spots. Due to either the size of the AgNPs (~75 nm) or the optical properties of Ag, the signal is significantly brighter than from AuNPs. Hyperspectral plots confirm this change in color and intensity with primary AgNPs scattering ~500 nm and agglomerates scattering ~600–700 nm, sometimes displaying multiple peaks. Having demonstrated that agglomeration of AgNPs can be detected, citrate-capped AgNPs were incubated in isolated, heparinized rat blood for 5 min and 18 h (Fig. 8, C and D). After 5 min, the signal returned is primarily between 500–540 nm, with the typical spectrum of the primary particles. There appears to be some background signal from the blood, though it presents less interference than for AuNPs, which is attributable to the increased signal intensity of the AgNPs. After 18 h in blood a mixture of signals was obtained (representative signals shown in Figure 8 E and F) with agglomerated AgNP showing emissions at 550–600 nm (upper two spectra) and primary particles at 500–540 nm (lower three spectra).. Interestingly, the increased brightness of the AgNPs mitigated some of the obscuring effects of the blood on the hyperspectral plot as was seen with AuNP.
Figure 8.
Characterization of Ag nanomaterials in isolated, whole rat blood with typical hyperspectral scattering spectra with inset darkfield image of albumin-stabilized (A) primary Ag nanoparticles and (B) agglomerates. The agglomeration behavior of citrate-capped Ag nanoparticles in isolated whole rat blood after incubation for different time periods: (C) darkfield image and (D) typical hyperspectral scattering spectra at 5 min; (E) darkfield image and (F) typical hyperspectral scattering spectra at 18 h.
4. Conclusions
Three complementary methods, hsDFM, cRM, and spICP-MS, have been demonstrated for the first time for rapid characterization of plasmonic nanomaterials in blood with minimal sample preparation. These methods were capable of distinguishing primary nanoparticles and agglomerates in simple media, which was validated by the standard methods such as TEM, UV-Vis, DLS and NTA. More importantly, hsDFM and spICP-MS demonstrated the unique capability to distinguish primary particles and agglomerates in blood. As a test, AuNPs and AgNPs were incubated in whole rat blood and these methods were used to monitor for the agglomeration of particles. The methods are anticipated to be adaptable to a wide variety of particle morphologies, surface coatings, and biological matrices. Furthermore, these methods could be applicable to blood and tissue extracted from animals following in vivo nanomedicine administration. These methods provide biological and pharmacokinetic information important for the development of nanomedicine, as well as analytical approaches for the assessment of nanomaterial-based products.
Supplementary Material
Acknowledgement
This research was supported in part by an appointment (S. Jenkins) to the Research Participation Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. We thank the pilot project funds from the Arkansas Biosciences Institute, the National Institutes of Health (NIH P30 GM103450), and startup funds from the University of Arkansas, to J. Chen. We appreciate the thorough review of this manuscript by. T. Eustaquio, P. Fu, S. Linder, and A. Patri.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer
This document has been reviewed in accordance with United States Food and Drug Administration (FDA) policy and approved for publication. Approval does not signify that the contents necessarily reflect the position or opinions of the FDA nor does mention of trade names or commercial products constitute endorsement or recommendation for use. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the FDA.
References
- 1.Kim BYS, Rutka JT, Chan WCW. Current Concepts: Nanomedicine. N Eng J Med. 2010;363:2434–2443. doi: 10.1056/NEJMra0912273. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Ferguson SA, Watanabe F, Jones Y, Xu Y, Biris AS, et al. Silver Nanoparticles Decrease Body Weight and Locomotor Activity in Adult Male Rats. Small. 2013;9:1715–1720. doi: 10.1002/smll.201201548. [DOI] [PubMed] [Google Scholar]
- 3.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Ali SF, Dervishi E, Xu Y, Li Z, Casciano D, et al. Cytotoxicity Effects of Graphene and Single-Wall Carbon Nanotubes in Neural Phaeochromocytoma-Derived PC12 Cells. ACS Nano. 2010;4:3181–3186. doi: 10.1021/nn1007176. [DOI] [PubMed] [Google Scholar]
- 5.Karmakar A, Zhang Q, Zhang Y. Neurotoxicity of nanoscale materials. Journal of Food and Drug Analysis. 2014;22:147–160. doi: 10.1016/j.jfda.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamburg MA. FDA's Approach to Regulation of Products of Nanotechnology. Science. 2012;336:299–300. doi: 10.1126/science.1205441. [DOI] [PubMed] [Google Scholar]
- 7.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. The golden age: gold nanoparticles for biomedicine. Chem Soc Rev. 2012;41:2740–2779. doi: 10.1039/c1cs15237h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardhan R, Lal S, Joshi A, Halas NJ. Theranostic Nanoshells: From Probe Design to Imaging and Treatment of Cancer. Acc Chem Res. 2011;44:936–946. doi: 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia Y, Li W, Cobley CM, Chen J, Xia X, Zhang Q, et al. Gold Nanocages: From Synthesis to Theranostic Applications. Acc Chem Res. 2011;44:914–924. doi: 10.1021/ar200061q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins SV, Muldoon TJ, Chen J. Plasmonic Nanostructures for Biomedical and Sensing Applications. In: Xiong Y, Lu X, editors. Metallic Nanostructures. Springer International Publishing; 2015. pp. 133–173. [Google Scholar]
- 11.Libutti SK, Paciotti GF, Byrnes AA, Alexander HR, Jr, Gannon WE, Walker M, et al. Phase I and pharmacokinetic studies of CYT-6091, a novel PEGylated colloidal gold-rhTNF nanomedicine. Clin Cancer Res. 2010;16:6139–6149. doi: 10.1158/1078-0432.CCR-10-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilot Study of AuroLase(tm) Therapy in Refractory and/or Recurrent Tumors of the Head and Neck. 2010 ed. National Institute of Health; 2013. [Google Scholar]
- 13.Rivera-Gil P, Jimenez De Aberasturi D, Wulf V, Pelaz B, Del Pino P, Zhao Y, et al. The Challenge To Relate the Physicochemical Properties of Colloidal Nanoparticles to Their Cytotoxicity. Acc Chem Res. 2012;46:743–749. doi: 10.1021/ar300039j. [DOI] [PubMed] [Google Scholar]
- 14.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 15.Lacerda SHDP, Park JJ, Meuse C, Pristinski D, Becker ML, Karim A, et al. Interaction of Gold Nanoparticles with Common Human Blood Proteins. ACS Nano. 2009;4:365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- 16.Keene AM, Peters D, Rouse R, Stewart S, Rosen ET, Tyner KM. Tissue and cellular distribution of gold nanoparticles varies based on aggregation/agglomeration status. Nanomedicine (London, U K) 2012;7:199–209. doi: 10.2217/nnm.11.125. [DOI] [PubMed] [Google Scholar]
- 17.Chithrani BD, Chan WCW. Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes. Nano Lett. 2007;7:1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 18.Tay CY, Cai P, Setyawati MI, Fang W, Tan LP, Hong CHL, et al. Nanoparticles Strengthen Intracellular Tension and Retard Cellular Migration. Nano Letters. 2013;14:83–88. doi: 10.1021/nl4032549. [DOI] [PubMed] [Google Scholar]
- 19.Setyawati MI, Tay CY, Chia SL, Goh SL, Fang W, Neo MJ, et al. Titanium dioxide nanomaterials cause endothelial cell leakiness by disrupting the homophilic interaction of VE–cadherin. Nat Commun. 2013;4:1673. doi: 10.1038/ncomms2655. [DOI] [PubMed] [Google Scholar]
- 20.Woehl TJ, Park C, Evans JE, Arslan I, Ristenpart WD, Browning ND. Direct Observation of Aggregative Nanoparticle Growth: Kinetic Modeling of the Size Distribution and Growth Rate. Nano Lett. 2013;14:373–378. doi: 10.1021/nl4043328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nativo P, Prior IA, Brust M. Uptake and Intracellular Fate of Surface-Modified Gold Nanoparticles. ACS Nano. 2008;2:1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 22.Dykman LA, Khlebtsov NG. Uptake of Engineered Gold Nanoparticles into Mammalian Cells. Chem Rev. 2013;114:1258–1288. doi: 10.1021/cr300441a. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Y, Liu Y, Wang L, Xu L, Bai R, Ji Y, et al. Surface chemistry and aspect ratio mediated cellular uptake of Au nanorods. Biomaterials. 2010;31:7606–7619. doi: 10.1016/j.biomaterials.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 24.Peckys DB, de Jonge N. Visualizing Gold Nanoparticle Uptake in Live Cells with Liquid Scanning Transmission Electron Microscopy. Nano Lett. 2011;11:1733–1738. doi: 10.1021/nl200285r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu FK. Analysis and applications of nanoparticles in the separation sciences: A case of gold nanoparticles. J Chromatogr A. 2009;1216:9034–9047. doi: 10.1016/j.chroma.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 26.Xie H, Gill-Sharp KL, O'Neal DP. Quantitative estimation of gold nanoshell concentrations in whole blood using dynamic light scattering. Nanomedicine: Nanotechnology, Biology and Medicine. 2007;3:89–94. doi: 10.1016/j.nano.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Hinterwirth H, Wiedmer SK, Moilanen M, Lehner A, Allmaier G, Waitz T, et al. Comparative method evaluation for size and size-distribution analysis of gold nanoparticles. J Sep Sci. 2013;36:2952–2961. doi: 10.1002/jssc.201300460. [DOI] [PubMed] [Google Scholar]
- 28.Shipway AN, Lahav M, Gabai R, Willner I. Investigations into the Electrostatically Induced Aggregation of Au Nanoparticles†. Langmuir. 2000;16:8789–8795. [Google Scholar]
- 29.Wustholz KL, Henry A-I, McMahon JM, Freeman RG, Valley N, Piotti ME, et al. Structure−Activity Relationships in Gold Nanoparticle Dimers and Trimers for Surface-Enhanced Raman Spectroscopy. J Am Chem Soc. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- 30.Wilets KA, Duyne RPV. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh SK, Pal T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: From Theory to Applications. Chem Rev. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- 32.Fairbairn N, Christofidou A, Kanaras AG, Newman TA, Muskens OL. Hyperspectral darkfield microscopy of single hollow gold nanoparticles for biomedical applications. Phys Chem Chem Phys. 2013;15:4163–4168. doi: 10.1039/c2cp43162a. [DOI] [PubMed] [Google Scholar]
- 33.Badireddy AR, Wiesner MR, Liu J. Detection, Characterization, and Abundance of Engineered Nanoparticles in Complex Waters by Hyperspectral Imagery with Enhanced Darkfield Microscopy. Environ Sci Technol. 2012;46:10081–10088. doi: 10.1021/es204140s. [DOI] [PubMed] [Google Scholar]
- 34.Rothen-Rutishauser BM, Schürch S, Haenni B, Kapp N, Gehr P. Interaction of Fine Particles and Nanoparticles with Red Blood Cells Visualized with Advanced Microscopic Techniques†. Environ Sci Technol. 2006;40:4353–4359. doi: 10.1021/es0522635. [DOI] [PubMed] [Google Scholar]
- 35.Aaron J, Travis K, Harrison N, Sokolov K. Dynamic Imaging of Molecular Assemblies in Live Cells Based on Nanoparticle Plasmon Resonance Coupling. Nano Lett. 2009;9:3612–3618. doi: 10.1021/nl9018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X, Wang J, Feizpour A, Reinhard BM. Illuminating the Lateral Organization of Cell-Surface CD24 and CD44 through Plasmon Coupling between Au Nanoparticle Immunolabels. Anal Chem. 2013;85:1290–1294. doi: 10.1021/ac303310j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Wu L, Reinhard BM. Scavenger Receptor Mediated Endocytosis of Silver Nanoparticles into J774A.1 Macrophages Is Heterogeneous. ACS Nano. 2012;6:7122–7132. doi: 10.1021/nn302186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haynes CL, McFarland AD, Duyne RPV. Surface-Enhanced Raman Spectroscopy. Anal Chem. 2005;77:338 A–346 A. [Google Scholar]
- 39.Valley N, Greeneltch N, Van Duyne RP, Schatz GC. A Look at the Origin and Magnitude of the Chemical Contribution to the Enhancement Mechanism of Surface-Enhanced Raman Spectroscopy (SERS): Theory and Experiment. J Phys Chem Lett. 2013;4:2599–2604. [Google Scholar]
- 40.Wang X, Qian X, Beitler JJ, Chen ZG, Khuri FR, Lewis MM, et al. Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles. Cancer Res. 2011;71:1526–1532. doi: 10.1158/0008-5472.CAN-10-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 42.Jans H, Huo Q. Gold nanoparticle-enabled biological and chemical detection and analysis. Chem Soc Rev. 2012;41:2849–2866. doi: 10.1039/c1cs15280g. [DOI] [PubMed] [Google Scholar]
- 43.Shah NB, Dong J, Bischof JC. Cellular Uptake and Nanoscale Localization of Gold Nanoparticles in Cancer Using Label-Free Confocal Raman Microscopy. Mol Pharmaceutics. 2010;8:176–184. doi: 10.1021/mp1002587. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Murphy KE, MacCuspie RI, Winchester MR. Capabilities of Single Particle Inductively Coupled Plasma Mass Spectrometry for the Size Measurement of Nanoparticles: A Case Study on Gold Nanoparticles. Anal Chem. 2014;86:3405–3414. doi: 10.1021/ac403775a. [DOI] [PubMed] [Google Scholar]
- 45.Laborda F, Jimenez-Lamana J, Bolea E, Castillo JR. Selective identification, characterization and determination of dissolved silveRi) and silver nanoparticles based on single particle detection by inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2011;26:1362–1371. [Google Scholar]
- 46.Rakcheev D, Philippe A, Schaumann GE. Hydrodynamic Chromatography Coupled with Single Particle-Inductively Coupled Plasma Mass Spectrometry for Investigating Nanoparticles Agglomerates. Anal Chem. 2013;85:10643–10647. doi: 10.1021/ac4019395. [DOI] [PubMed] [Google Scholar]
- 47.Mitrano DM, Barber A, Bednar A, Westerhoff P, Higgins CP, Ranville JF. Silver nanoparticle characterization using single particle ICP-MS (SP-ICP-MS) and asymmetrical flow field flow fractionation ICP-MS (AF4-ICP-MS) J Anal At Spectrom. 2012;27:1131–1142. [Google Scholar]
- 48.Zook J, Long S, Cleveland D, Geronimo C, MacCuspie R. Measuring silver nanoparticle dissolution in complex biological and environmental matrices using UV–visible absorbance. Anal Bioanal Chem. 2011;401:1993–2002. doi: 10.1007/s00216-011-5266-y. [DOI] [PubMed] [Google Scholar]
- 49.Frens G. Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nature (London), Phys Sci. 1973;241:20–22. [Google Scholar]
- 50.Dominguez-Medina S, Blankenburg J, Olson J, Landes CF, Link S. Adsorption of a Protein Monolayer via Hydrophobic Interactions Prevents Nanoparticle Aggregation under Harsh Environmental Conditions. ACS Sustainable Chem Eng. 2013;1:833–842. doi: 10.1021/sc400042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Draine BT, Flatau PJ. Discrete-dipole approximation for scattering calculations. J Opt Soc Am A. 1994;11:1491–1499. [Google Scholar]
- 52.Johnson PB, Christy RW. Optical Constants of the Noble Metals. Phys Rev B: Condens Matter Mater Phys. 1972;6:4370–4379. [Google Scholar]
- 53.Basu S, Ghosh SK, Kundu S, Panigrahi S, Praharaj S, Pande S, et al. Biomolecule induced nanoparticle aggregation: Effect of particle size on interparticle coupling. J Colloid Interface Sci. 2007;313:724–734. doi: 10.1016/j.jcis.2007.04.069. [DOI] [PubMed] [Google Scholar]
- 54.Halas NJ, Lal S, Chang WS, Link S, Nordlander P. Plasmons in strongly coupled metallic nanostructures. Chem Rev. 2011;111:3913–3961. doi: 10.1021/cr200061k. [DOI] [PubMed] [Google Scholar]
- 55.Chithrani BD, Stewart J, Allen C, Jaffray DA. Intracellular uptake, transport, and processing of nanostructures in cancer cells. Nanomedicine: Nanotechnology, Biology and Medicine. 2009;5:118–127. doi: 10.1016/j.nano.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 56.Dominguez-Medina S, McDonough S, Swanglap P, Landes CF, Link S. In Situ Measurement of Bovine Serum Albumin Interaction with Gold Nanospheres. Langmuir. 2012;28:9131–9139. doi: 10.1021/la3005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eustaquio T, Leary JF. Nanobarcoding: detecting nanoparticles in biological samples using in situ polymerase chain reaction. Int J Nanomedicine. 2012;7:5625–5639. doi: 10.2147/IJN.S37433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith BR, Ghosn EEB, Rallapalli H, Prescher JA, Larson T, Herzenberg LA, et al. Selective uptake of single-walled carbon nanotubes by circulating monocytes for enhanced tumour delivery. Nat Nanotechnol. 2014 doi: 10.1038/nnano.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K, Cui Y, Lee LP, Irudayaraj J. Quantitative imaging of single mRNA splice variants in living cells. Nat Nanotechnol. 2014 doi: 10.1038/nnano.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Xu Y, Li Z, Chen T, Lantz SM, Howard PC, et al. Mechanistic toxicity evaluation of uncoated and PEGylated single-walled carbon nanotubes in neuronal PC12 cells. ACS Nano. 2011;5:7020–7033. doi: 10.1021/nn2016259. [DOI] [PubMed] [Google Scholar]
- 61.Qian X, Zhou X, Nie S. Surface-Enhanced Raman Nanoparticle Beacons Based on Bioconjugated Gold Nanocrystals and Long Range Plasmonic Coupling. J Am Chem Soc. 2008;130:14934–14935. doi: 10.1021/ja8062502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Z, Zhu T, Liu Z. Raman scattering enhancement contributed from individual gold nanoparticles and interparticle coupling. Nanotechnology. 2004;15:3574–3564. [Google Scholar]
- 63.Barker EF, Sleator WW. The Infrared Spectrum of Heavy Water. J Chem Phys. 1935;3:660–663. [Google Scholar]
- 64.Jang NH. The Coordination Chemistry of DNA Nucleosides on Gold Nanoparticles as a Probe by SERS. Bull Korean Chem Soc. 2002;23:1790–1800. [Google Scholar]
- 65.Tuma R. Raman spectroscopy of proteins: from peptides to large assemblies. J Raman Spectrosc. 2005;36:307–319. [Google Scholar]
- 66.Margueritat J, Gehan H, Grand J, Lévi G, Aubard J, Félidj N, et al. Influence of the Number of Nanoparticles on the Enhancement Properties of Surface-Enhanced Raman Scattering Active Area: Sensitivity versus Repeatability. ACS Nano. 2011;5:1630–1638. doi: 10.1021/nn103256t. [DOI] [PubMed] [Google Scholar]
- 67.Yan B, Thubagere A, Premasiri WR, Ziegler LD, Dal Negro L, Reinhard BM. Engineered SERS Substrates with Multiscale Signal Enhancement: Nanoparticle Cluster Arrays. ACS Nano. 2009;3:1190–1202. doi: 10.1021/nn800836f. [DOI] [PubMed] [Google Scholar]
- 68.Fraire JC, Pérez LA, Coronado EA. Cluster Size Effects in the Surface-Enhanced Raman Scattering Response of Ag and Au Nanoparticle Aggregates: Experimental and Theoretical Insight. J Phys Chem C. 2013;117:23090–23107. [Google Scholar]
- 69.Shah Neha B, Bischof John C. Blood protein and blood cell interactions with gold nanoparticles: the need for in vivo studies. Bionanomaterials. 2013;14:65. [Google Scholar]
- 70.Rivera Gil P, Oberdörster G, Elder A, Puntes V, Parak WJ. Correlating Physico-Chemical with Toxicological Properties of Nanoparticles: The Present and the Future. ACS Nano. 2010;4:5527–5531. doi: 10.1021/nn1025687. [DOI] [PubMed] [Google Scholar]
- 71.Dreaden EC, Austin LA, Mackey MA, El-Sayed MA. Size matters: gold nanoparticles in targeted cancer drug delivery. Ther Delivery. 2012;3:457–478. doi: 10.4155/tde.12.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arvizo RR, Bhattacharyya S, Kudgus RA, Giri K, Bhattacharya R, Mukherjee P. Intrinsic therapeutic applications of noble metal nanoparticles: past, present and future. Chem Soc Rev. 2012;41:2943–2970. doi: 10.1039/c2cs15355f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.