Abstract
Background
Japanese encephalitis virus (JEV) is the etiological agent of Japanese encephalitis (JE), one of the most serious viral encephalitis worldwide. Five genotypes have been classified based on phylogenetic analysis of the viral envelope gene or the complete genome. Previous studies based on four genotypes have reported that in evolutionary terms, genotype 1 JEV is the most recent lineage. However, until now, no systematic phylogenetic analysis was reported based on whole genomic sequence of all five JEV genotypes.
Findings
In this study, phylogenetic analysis using Bayesian Markov chain Monte Carlo simulations was conducted on the whole genomic sequences of all five genotypes of JEV. The results showed that the most recent common ancestor (TMRCA) for JEV is estimated to have occurred 3255 years ago (95% highest posterior density [HPD], −978 to−6125 years). Chronologically, this ancestral lineage diverged to produce five recognized virus genotypes in the sequence 5, 4, 3, 2 and 1. Population dynamics analysis indicated that the genetic diversity of the virus peaked during the following two periods: 1930–1960 and 1980–1990, and the population diversity of JEV remained relatively high after 2000.
Conclusions
Genotype 5 is the earliest recognized JEV lineage, and the genetic diversity of JEV has remained high since 2000.
Keywords: Japanese encephalitis virus, Genotype, Genetic diversity
Findings
Japanese encephalitis virus (JEV) is the prototype member of the JEV serogroup within the genus Flavivirus, family Flaviviridae. JEV comprises five genotypes (G1-G5) [1-3]. In previous studies, the phylogenetic characteristics of JEV were analyzed and the most recent common ancestor (TMRCA) was estimated. The TMRCA of JEV was estimated to be 1690 years when calculations were based on the complete sequence of four genotypes (G1-G4) [4], whereas, analysis of JEV using a limited number of whole genomic sequences from five genotypes indicated that TMRCA of JEV appeared approximately 460 years [5]. More recently, however, G5 strain XZ0934 isolated in 2009, which had not been included in earlier analyses, was shown to be significantly different from the G5 Muar isolate [6]. Therefore, in order to improve our understanding of the evolutionary progress and population diversity of JEV, a comprehensive dataset was established for evolutionary analysis of JEV in this study. In the dataset, 100 whole genomic sequences of JEV representing all five genotypes of JEV, isolated from various hosts (humans, pigs and bats) and vectors (mosquitoes and midges) were collected and analyzed.
Two G5 JEV full-length genome sequences (Muar and XZ0934) were downloaded from GenBank (GB No. HM596272 and JF915894, respectively) and added to the database established in a previous report [4], forming a new database for analysis (Table 1). The new JEV sequence database was analyzed using Bayesian Markov chain Monte Carlo (MCMC) method. The General Time Reversible (GTR) model + Invariant (I) + Gamma (G) model was selected using MrModelTest [7]. The nucleotide substitution rates and divergence times of the most recent common ancestor (TMRCA) were estimated using the relaxed (uncorrelated lognormal) molecular clock model in the BEAST software package [8]. Demographic histories of JEV were inferred based on Bayesian skyline reconstruction. The analysis was run through 1,000,000,000 generations to ensure sufficient mixing. Finally, the maximum clade credibility (MCC) tree was built using TreeAnnotator with 10% burn-in (http://beast.bio.ed.ac.uk/).
Table 1.
Information of JEV isolates analyzed in this study
| Strain | Date | Country | Host a | Genotype | GenBank accession no. |
|---|---|---|---|---|---|
| 47 | 1950’s | China:Heilongjiang | CSF | 3 | JF706269 |
| 14178 | 2001 | India | - | 3 | EF623987 |
| 57434 | 2005 | India | - | 3 | EF623988 |
| 04940-4 | 2002 | India | - | 3 | EF623989 |
| B58 | 1989 | China:Yunnan | Bat | 3 | FJ185036 |
| Beijing-1 | 1949 | China | Human brain | 3 | L48961 |
| BL06-50 | 2006 | China:Guangxi | Culex tritaeniorhynchus | 1 | JF706270 |
| BL06-54 | 2006 | China:Guangxi | Culex tritaeniorhynchus | 1 | JF706271 |
| CBH | 1954 | China:Fujian | CSF | 3 | JN381860 |
| CH-13 | 1957 | China:Sichuan | CSF | 3 | JN381870 |
| CH1392 | 1990 | Taiwan | Culex tritaeniorhynchus | 3 | AF254452 |
| CTS | 1955 | China:Fujian | CSF | 3 | GQ429184 |
| CZX | 1954 | China:Fujian | CSF | 3 | JN381865 |
| DH107 | 1989 | China:Yunnan | Aedes lineatopennis | 3 | JN381873 |
| DL04-29 | 2004 | China:Yunnan | Culex theileri | 3 | JF706272 |
| DL04-45 | 2004 | China:Yunnan | Ar. Subalbatus & Mansonia uniform | 3 | JN381854 |
| Fj02-29 | 2002 | China:Fujian | CSF | 3 | JF706273 |
| Fj02-76 | 2002 | China:Fujian | Human blood | 3 | JN381867 |
| FJ03-39 | 2003 | China:Fujian | Human blood | 3 | JN381859 |
| FJ03-94 | 2003 | China:Fujian | Human blood | 3 | JN381858 |
| FU | 1995 | Australia | Human serum | 2 | AF217620 |
| G35 | 1954 | China:Fujian | Mosquito pool | 3 | GQ429185 |
| GB30 | 1997 | China:Yunnan | Murina aurata brain tissue | 3 | FJ185037 |
| GP78 | 1978 | India | Human brain | 3 | AF075723 |
| GS07-TS11 | 2007 | China:Gansu | Culex tritaeniorhynchus | 1 | JN381843 |
| GSBY0801 | 2008 | China:Gansu | Culex tritaeniorhynchus | 1 | JF706274 |
| GSBY0804 | 2008 | China:Gansu | Culex tritaeniorhynchus | 1 | JN381844 |
| GSBY0810 | 2008 | China:Gansu | Culex tritaeniorhynchus | 1 | JN381840 |
| GSBY0816 | 2008 | China:Gansu | Culex tritaeniorhynchus | 1 | JN381842 |
| GSBY0827 | 2008 | China:Gansu | Culex tritaeniorhynchus | 1 | JN381845 |
| GSBY0861 | 2008 | China:Gansu | Culex tritaeniorhynchus | 1 | JN381833 |
| GSS | 1960’s | China:Beijing | CSF | 3 | JF706275 |
| GX0519 | 2005 | China:Guanxi | Culex tritaeniorhynchus | 1 | JN381835 |
| GX0523/44 | 2005 | China:Guanxi | Culex tritaeniorhynchus | 1 | JN381832 |
| GZ04-2 | 2004 | China:Guizhou | Armigeres | 3 | JN381857 |
| GZ56 | 2006 | China:GuiZhou | CSF | 1 | HM366552 |
| Ha-3 | 1960’s | China:Heilongjiang | CSF | 3 | JN381872 |
| HB49 | 1990 | China:Yunnan | Rousettus leschenaulti blood | 3 | JF706284 |
| HB97 | 1990 | China:Yunnan | Rousettus leschenaulti blood | 3 | JF706285 |
| HLJ02-134 | 2002 | China:Heilongjiang | Genus culicoides | 3 | JF706276 |
| HN04-11 | 2004 | China:Henan | Culex | 1 | JN381831 |
| HN04-21 | 2004 | China:Henan | Culex | 1 | JN381841 |
| HN06129 | 2006 | China:Henan | Armigeres | 1 | JF706277 |
| HN0621 | 2006 | China:Henan | Culex | 1 | JN381830 |
| HN0626 | 2006 | China:Henan | Culex | 1 | JN381837 |
| HVI | 1965 | Taiwan | Mosquito | 3 | AF098735 |
| HYZ | 1979 | China:Yunnan | Patient blood | 3 | JN381853 |
| Ishikawa | 1994 | Japan | Culex tritaeniorhynchus | 1 | AB051292 |
| JaGAr 01 | 1959 | Japan | Cluex | 3 | AF069076 |
| JaOArS982 | 1982 | Japan | Mosquito | 3 | M18370 |
| JaOH0566/Japan/1966/human | 1966 | Japan | Human | 3 | AY508813 |
| JEV/sw/Mie/40/2004 | 2004 | Japan | Swine serum | 1 | AB241118 |
| JEV/sw/Mie/41/2002 | 2002 | Japan | Swine serum | 1 | AB241119 |
| JH04-18 | 2004 | China:Yunnan | Whitmorei & Anophelessinensis | 3 | JN381855 |
| JKT6468 | 1981 | Indonesia | Mosquito | 4 | AY184212 |
| K87P39 | 1987 | South Korea | Mosquito | 3 | AY585242 |
| KV1899 | 1999 | Korea | Swine | 1 | AY316157 |
| LFM | 1955 | China:Fujian | Human blood | 3 | JN381863 |
| Ling | 1965 | Taiwan | Human brain | 3 | L78128 |
| LN02-102 | 2002 | China:liaoning | Culex modestus | 1 | JF706278 |
| LN0716 | 2007 | China:Liaoning | Culex tritaeniorhynchus | 1 | JN381849 |
| LYZ | 1957 | China:Fujian | CSF | 3 | JN381869 |
| M28 | 1977 | China:Yunnan | Culex pseudovishnui | 1 | JF706279 |
| Nakayama | 1935 | Japan | Human brain | 3 | EF571853 |
| P3 | 1949 | China:Beijing | Human brain | 3 | U47032 |
| RP-2 ms | 1985 | Taiwan | Mosquito | 3 | AF014160 |
| RP-9 | 1985 | Taiwan | Mosquito | 3 | AF014161 |
| SA14 | 1954 | China | Mosquito | 3 | U14163 |
| SC04-12 | 2004 | China:Sichuan | Culex | 1 | JN381839 |
| SC04-15 | 2004 | China:Sichuan | Culex tritaeniorhynchus | 1 | JN381838 |
| SD0810 | 2008 | China:Shandong | Culex tritaeniorhynchus | 1 | JF706286 |
| SH03-103 | 2003 | China:Shanghai | Culex tritaeniorhynchus | 1 | JN381847 |
| SH03-105 | 2003 | China:Shanghai | Culex tritaeniorhynchus | 1 | JN381846 |
| SH04-10 | 2004 | China:Shanghai | Culex tritaeniorhynchus | 3 | JN381856 |
| SH04-5 | 2004 | China:Shanghai | Culex tritaeniorhynchus | 3 | JN381866 |
| SH17M-07 | 2007 | China | - | 1 | EU429297 |
| SH-3 | 1987 | China:Shanghai | CSF | 3 | JN381864 |
| SH-53 | 2001 | China:Shanghai | Culex tritaeniorhynchus | 1 | JN381850 |
| SH-80 | 2001 | China:Shanghai | Culex tritaeniorhynchus | 1 | JN381848 |
| T1P1 | 1997 | Taiwan | Armigeres subalbatus | 3 | AF254453 |
| TLA | 1971 | China:Liaoning | CSF | 3 | JN381868 |
| Vellore P20778 | 1958 | India | Human brain | 3 | AF080251 |
| XJ69 | 2007 | China | Culex pipiens pallens | 1 | EU880214 |
| XJP613 | 2007 | China | Culex tritaeniorhynchus | 1 | EU693899 |
| XZ0938 | 2009 | China:Xizang | Culex tritaeniorhynchus | 1 | HQ652538 |
| YLG | 1955 | China:Fujian | CSF | 3 | JF706280 |
| YN | 1954 | China:Yunnan | CSF | 3 | JN381871 |
| YN05124 | 2005 | China:Yunnan | Culex tritaeniorhynchus | 1 | JF706281 |
| YN05155 | 2005 | China:Yunnan | Culex tritaeniorhynchus | 1 | JN381852 |
| YN0623 | 2006 | China:Yunnan | Culex tritaeniorhynchus | 1 | JN381836 |
| YN0911 | 2009 | China:Yunnan | Culex tritaeniorhynchus | 1 | JF706267 |
| YN0967 | 2009 | China:Yunnan | Culex tritaeniorhynchus | 1 | JF706268 |
| YN79-Bao83 | 1979 | China:Yunnan | Culex tritaeniorhynchus | 1 | JN381851 |
| YN82-BN8219 | 1982 | China:Yunnan | Mosquito | 1 | JN381834 |
| YN83-Meng83-54 | 1983 | China:Yunnan | Lasiohelea taiwana Shiraki | 1 | JF706282 |
| YN98-A151 | 2003 | China:Yunnan | Mosquitoes | 3 | JN381861 |
| ZMT | 1955 | China:Fujian | CSF | 3 | JF706283 |
| ZSZ | 1955 | China:Fujian | CSF | 3 | JN381862 |
| Muar | 1952 | Malaysia | Human brain | 5 | HM596272 |
| XZ0934 | 2009 | China:Tibet | Culex tritaeniorhynchus | 5 | JF915894 |
a- Information not available.
Based on Bayesian Markov chain Monte Carlo (MCMC) analysis, the maximum clade credibility (MCC) tree for the whole genomic sequences of JEV was established (Figure 1). Representatives of the five distinct lineages were included in the analysis. The posterior probability values for the nodes of each lineage were >0.95, indicating their robustness. JEV was estimated to have emerged 3255 years ago (95% HPD: −978 to −6125 years) and subsequently diverged at least five times to produce the 5 recognized genotypes. In chronological order, they diverged in the order G5, G4, G3, G2 and G1. Thus, G5 represents the most ancestral lineage among genotypes 1–5.
Figure 1.
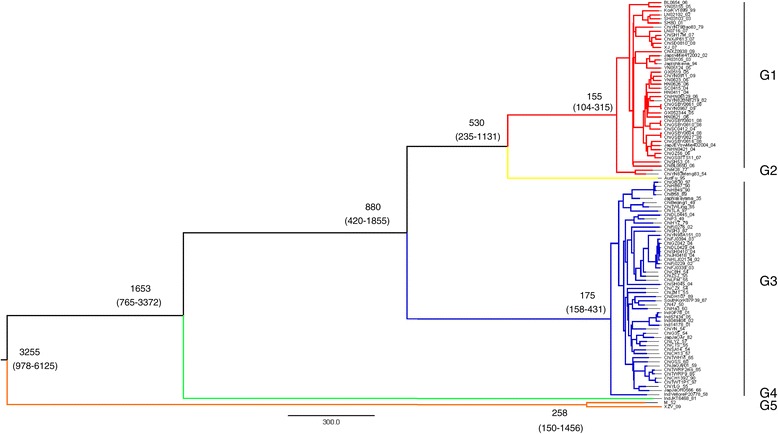
Maximum clade credibility (MCC) tree for 100 whole-genome sequences of JEV. Five distinct lineages were identified: G1 (red), G2 (yellow), G3 (blue), G4 (green) and G5 (orange). Estimated TMRCAs of these lineages (with their 95% HPD values in parentheses) are shown, G1: 155(104–315), G2: 530(235–1131), G3: 880(420–1855), G4: 1653(765–3372), and G5: 3255(978–6125).
The mean rate of nucleotide substitution for the whole genomic sequences of 100 JEV strains isolated from a variety of hosts worldwide, estimated using a Bayesian MCMC approach, was 1.01 × 10−4 nucleotide substitutions per site per year (95% HPD values, 4.37 × 10−5, 1.56 × 10−4). This is similar to previous estimates based on analysis of four JEV genotypes [4].
The population dynamics of JEV are shown in Figure 2. The skyline plot showed that the JEV population had experienced complicated changes during the process of evolution. However, the virus population remained relatively stable during the first 2700 years (Figure 2A), followed by a period of rapid decline from the 1700s, reaching a minimum in the 1900s. It then increased rapidly from the 1930s until the 1960s and formed the first peak. The second peak appeared in the 1980-1990s and subsequently the populations of JEV remained high after 2000 (Figure 2B).
Figure 2.
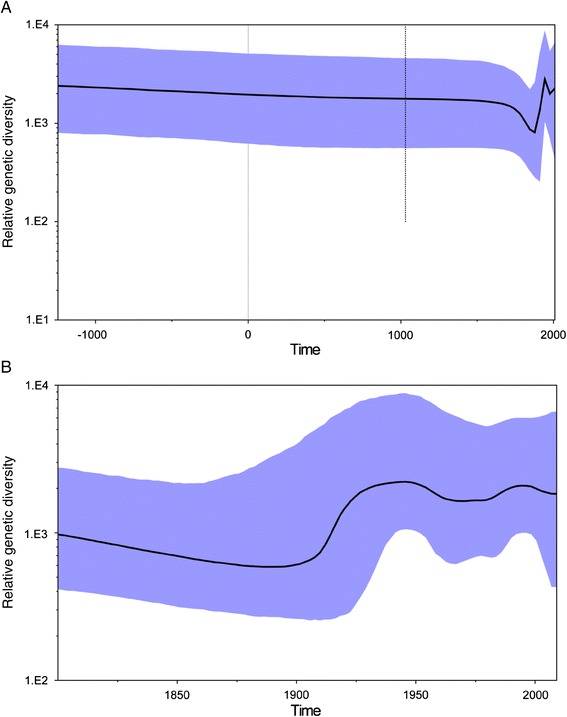
Bayesian skyline plots for JEV. Highlighted areas correspond to 95% HPD intervals. (A) Populations during the whole evolutionary history; (B) Populations during the later evolutionary history since 1800.
The findings in this study have similarities with previous studies [5]. For example, the divergence pattern of the genotypes occurred in the order G5, G4, G3, G2 and G1, and the mean rate of nucleotide substitution was similar to previous estimates. However, the occurrence time of TMRCA determined in this study (~3255 years ago) was quite different compared with that measured in the report (~460 years ago) of Mohammed et al. [5]. The reason for this discrepancy could be attributed to the dataset used for analysis. In Mohammed’s study, only 35 whole genomic sequences were used and the only G5 representative included was the Muar strain. Therefore, since our new dataset includes two G5 representatives with robust sequences, the occurrence time of TMRCA (~3255 years ago) obtained in this study should reflect more precisely the evolutionary patterns and diversity of JEV.
Two main peak periods in population dynamics were identified in this study, 1930–1960 and 1980-1990s, respectively. These fluctuations were reflections of the virus activity in the sylvatic environment. Since the 1930s, JEV strains belonging to G3 emerged and were isolated from Asian countries. G5 was first identified in 1952 [2,4]. This was a good interpretation of the first peak. Subsequently, G2 and G4 strains were isolated during the 1980s. Importantly, the G1 genotype emerged during the 1980s onwards [4]. Therefore, the virus population diversity peaked in the 1980-1990s. Since 2000, G1 JEV has become the dominant genotype in most endemic regions [4], and although a relatively small decrease was observed, the virus remains the most active and G5 reemerged.
Interestingly, although G5 is estimated to be the most ancestral JEV lineage, this virus showed a highly active dispersal capacity following its reemergence. Indeed, this new G5 strain was isolated from mosquitoes collected in southern region of the Asian continent (Tibet, China) in 2009 [2] and northeast region of Asia (South Korea) during the same year [9]. Thus, G5 now appears to be dispersing widely in Asia. A recent study showed that genotype 1 JEV originated in Southeastern Asia and spread to the entire Asian continent [10]. Based on these observations, it seems likely that G5 will follow a dispersal pattern similar to that of G1 JEV, and has dispersed or will disperse over the entire Asian continent. Clearly, G5 should be monitored closely throughout JEV endemic regions.
Finally, the available inactivated and live attenuated JE vaccines are derived from G3 JEVs [11]. Thus, the level of cross protection of the current vaccines against G5 JEV is likely to be sub-optimal and should therefore be analyzed carefully since the reemergence of G5 and its widespread dispersal, and significant genetic variation could impact on its epidemiology. This possibility is emphasized by the fact that Muar (the first G5 JEV) strain was isolated from a patient with severe viral encephalitis [12]. Thus, there is the realistic possibility that the newly isolated G5 viruses could be highly virulent. Thus, the potential disease burden of viral encephalitis caused by G5 JEV requires careful reassessment.
Acknowledgements
We appreciate Dr. Ernest A Gould from Aix Marseille University for his important revisions and comments on the manuscript. This work was supported by grants from the National Natural Science Foundation of China (81290342), the Ministry of Science and Technology, China (2011CB504702), and State Key Laboratory for Infectious Disease Prevention and Control (2014SKLID03).
Abbreviations
- JEV
Japanese encephalitis virus
- JE
Japanese encephalitis
- E
Envelope
- TMRCA
The most recent common ancestor
- HPD
Highest posterior density
- G
Genotype
- MCMC
Markov chain Monte Carlo
- MCC
Maximum clade credibility
- Ser
Serine
- ORF
Open reading frame
Footnotes
Xiaoyan Gao and Hong Liu contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XYG and HL contributed equally to the analysis and interpretation of the results, and involved in drafting the manuscript. MHL and SHF participated in the design and analysis of the dataset. GDL contributed to conception and design of the manuscript, and involved in drafting and revising the manuscript. All authors have read and approved the final version of the manuscript.
Contributor Information
Xiaoyan Gao, Email: g.x.y1018@hotmail.com.
Hong Liu, Email: liuhongseminar@126.com.
Minghua Li, Email: minghua-li@hotmail.com.
Shihong Fu, Email: shihongfu@hotmail.com.
Guodong Liang, Email: gdliang@hotmail.com.
References
- 1.Uchil PD, Satchidanandam V. Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am J Trop Med Hyg. 2001;65(3):242–51. doi: 10.4269/ajtmh.2001.65.242. [DOI] [PubMed] [Google Scholar]
- 2.Li MH, Fu SH, Chen WX, Wang HY, Guo YH, Liu QY, et al. Genotype V Japanese encephalitis virus is emerging. PLoS Negl Trop Dis. 2011;5(7):e1231. doi: 10.1371/journal.pntd.0001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon T, Ni H, Beasley DW, Ekkelenkamp M, Cardosa MJ, Barrett AD. Origin and evolution of Japanese encephalitis virus in southeast Asia. J Virol. 2003;77(5):3091–8. doi: 10.1128/JVI.77.5.3091-3098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan XL, Liu H, Wang HY, Fu SH, Liu HZ, Zhang HL, et al. The Emergence of Genotype I of Japanese Encephalitis Virus as the Dominant Genotype in Asia. J Virol. 2011;85(19):9847–53. doi: 10.1128/JVI.00825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammed MA, Galbraith SE, Radford AD, Dove W, Takasaki T, Kurane I, et al. Molecular phylogenetic and evolutionary analyses of Muar strain of Japanese encephalitis virus reveal it is the missing fifth genotype. Infect Genet Evol. 2011;11(5):855–62. doi: 10.1016/j.meegid.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Li MH, Fu SH, Chen WX, Wang HY, Cao YX, Liang GD. Molecular Characterization of Full-length Genome of Japanese Encephalitis Virus Genotype V Isolated from Tibet. China Biomed Environ Sci. 2014;27(4):231–9. doi: 10.3967/bes2014.046. [DOI] [PubMed] [Google Scholar]
- 7.Posada D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinformatics. 2003. Chapter 6, Unit 6.5 http://www.ncbi.nlm.nih.gov/pubmed/18428705/. [DOI] [PubMed]
- 8.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takhampunya R, Kim HC, Tippayachai B, Kengluecha A, Klein TA, Lee WJ, et al. Emergence of Japanese encephalitis virus genotype V in the Republic of Korea. Virol J. 2011;8:449. doi: 10.1186/1743-422X-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao XY, Liu H, Wang HY, Fu SH, Guo ZY, Liang GD. Southernmost Asia is the source of Japanese encephalitis virus (Genotype 1) diversity from which the viruses disperse and evolve throughout Asia. PLoS Negl Trop Dis. 2013;7(9):e2459. doi: 10.1371/journal.pntd.0002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead SB, Jacobson J. Japanese encephalitis vaccines. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccine. 5. Phildelphia: Elsevier; 2008. pp. 311–52. [Google Scholar]
- 12.Hale JH, Lim KA, Chee PH. Japanese type B encephalitis in Malaya. Ann Trop Med Parasitol. 1952;46(3):220–6. doi: 10.1080/00034983.1952.11685526. [DOI] [PubMed] [Google Scholar]


