Abstract
Whole-exome sequencing (WES) is being used clinically to diagnose rare Mendelian disorders, especially when standard tests have failed. The diagnostic yield from WES is reported to be ∼15-30%; however, data regarding the clinical utility and interpretative challenges from the clinician's perspective are lacking. Here, we present a series of the first 6 unselected consecutive cases seen over a period of 6 months where WES was employed in clinical labs via trio-based testing (proband and parents). While we do not discount the value of WES in the clinical setting, our cases and experience illustrate the significant clinical challenges of WES, even when a diagnosis may be achieved.
Key Words: Diagnostic odyssey, Schinzel-Giedion syndrome, Whole-exome sequencing
The exome, or protein-coding portion of the genome, makes up ∼1-2% of the entire genome. Despite this, variations in the DNA sequence of the exome are much more likely to be associated with a particular phenotype of a Mendelian disorder. Whole-exome sequencing (WES) is a technique used to analyze the DNA sequence of the exome and has begun to show promise, specifically in the genetic work-up of patients who present with a challenging constellation of phenotypic features that has sent both clinicians and patients on a ‘diagnostic odyssey’. There are many challenges in the clinical application of WES as a diagnostic tool including, by definition, lack of coverage of non-coding regions of the genome, variable depth of coverage of coding regions, the burden of dealing with incidental findings, and a high likelihood of finding DNA sequence variants of unclear significance (VUSs) [Bamshad et al., 2011; Goh and Choi, 2012]. In addition, even though the name WES implies ‘whole-exome sequencing’, it is clear that in any given run of the test, only 80-90% of the exome is covered, thus raising concerns about false negative results. Moreover, WES may or may not pick up significant copy number variations, depending upon the calling method used (each of which currently available will not guarantee the detection of all), and further, may miss these important variations altogether with the inability to detect nucleotide repeat expansion/contraction [Tan et al., 2014]. Despite these challenges, WES continues to be used in increasing frequency as a clinical diagnostic tool. Current opinion suggests that when standard genetic testing has failed to establish a diagnosis of a suspected Mendelian disorder, it is reasonable to consider WES. On the other hand, it can be tempting to consider WES early on in the genetic diagnostic assessment, prior to more specific testing based on clinical clues, as WES may lead to an earlier diagnosis. Because WES is now much easier to access as a clinical test, it is being sent by clinicians of all backgrounds [Yang et al., 2013], and it remains unclear whether appropriate pre-test counseling regarding the pros and cons of such testing is performed. An American College of Medical Genetics (ACMG) policy statement [ACMG Board of Directors, 2012] reviews indications for the use of WES, and there remains variability in the clinical threshold for using this test. Various studies have reported a diagnostic yield with the use of WES ranging from 16-50% [de Ligt et al., 2012; Need et al., 2012; Yang et al., 2013; Shashi et al., 2014]. Most of these studies report results from a laboratory perspective. The clinician's perspective regarding whether a ‘positive result’ truly explains the phenotype, the interpretive challenges of WES, and whether WES is needed to achieve a diagnosis, are very important when assessing the clinical performance of WES for diagnostic purposes. In addition, the clinical burden on families and the medical system created by incidental findings and VUSs needs to be well-thought-out before WES is considered ‘successful’ in each individual case. We present our experience of the first 6 consecutive cases where WES was used as a clinical diagnostic test, providing further insight into the challenges of using WES in the clinic, even when a diagnosis is achieved. We provide a clinician's perspective to this discussion, highlighting opportunities for pre- and post-test counseling prior to its application in future cases.
Materials and Methods
This is a retrospective case-series of patients seen in the Department of Medical Genetics at Mayo Clinic, Rochester, on whom WES was performed. The first 6 unselected consecutive cases for whom results were available were included in the study. The patients were evaluated in the Medical Genetics clinic at our institution over a period of 6 months. All were ‘diagnostic odyssey’ cases where a previous extensive evaluation had failed to reveal a diagnosis. All tests were sent to 1 of 2 clinical labs offering WES, and were ordered prior to the ACMG policy statement on incidental findings in WES [Green et al., 2013]. Samples for cases 1-4 and 6 were sent to a lab that estimated coverage of >95% of target bases at greater than 20× depth, with >85% of target bases covered at greater than 40× with mean coverage of greater than 100×, therefore leading to an estimate that 5% of coding regions were not well covered. This laboratory reported the use of the ACMG guidelines for standards for interpretation and reporting of sequence variations as their policy for reviewing and interpreting variants and incidental findings [Richards et al., 2008]. For instance, the focused report would contain genes unrelated to disease phenotype, unclassified variants and deleterious mutations in genes with no known association with disease in humans, while findings such as carrier status for autosomal recessive disorders would only be reported per the recommendations of those listed by professional societies such as ACMG or ACOG. Samples for case 5 were sent to a lab that reported 90% coverage of target bases at greater than 10× depth, leading to an estimate that 10% of the coding regions were not well covered [Yang et al., 2013; Genetics, unpubl. data]. This laboratory reported a policy of using HGMD, OMIM or medical literature to interpret variants and incidental findings. For instance, this laboratory stated that variants which were noted in clinically novel genes would not be reported, nor analyzed. In addition, this laboratory would not release incidental findings, such as carrier status, to individuals undergoing WES if they were under the age of 18. In all cases, trio-based testing was performed (proband and both biological parents). WES was only performed on the probands and data obtained from each proband was subsequently filtered down to a candidate list. The candidate list was generated based upon a laboratory-derived algorithm, developed and defined by the laboratory [Yang et al., 2013; Genetics, unpubl. data], and further refined by the clinical information provided by the ordering clinician. Variants ascertained from the candidate list were further interrogated using parental samples to determine the likelihood of association to the reported phenotype. In case 5, where the proband's brother had a similar phenotype, variants were also interrogated using the brother's sample. Each patient's chart was reviewed with respect to the clinical presentation, previous diagnostic studies and WES results.
Results
Findings are detailed below and summarized in table 1.
Table 1.
Summary of cases, findings and diagnoses
| Case | Sex, age | Clinical indication | Findings |
Diagnosis | |
|---|---|---|---|---|---|
| primary | incidental | ||||
| 1 | female, 3 years | craniofacial dysmorphism, developmental delay | de novo, heterozygous SETBP1 mutation; mutation in CLCN1; VUSs in CLCN1, SCN9A, KIF1A, ACACA, RNASEH2A, PNKP, TOP1MT, ERLIN2 | yes (Schinzel-Giedion syndrome) | |
| 2 | female, 2 years | arthrogryposis, seizures, microcephaly, progressive cerebellar atrophy and axonal neuropathy | de novo VUS in KIF1A; compound heterozygous VUSs in FASN; VUS in DNAJC5 | unclear | |
| 3 | male, 3 years | microcephaly, developmental delay and hypotonia | de novo, heterozygous VUS in SYNGAP1 | VUS in FBN1 | unclear |
| 4 | male, 3 years | failure to thrive, short stature and known A1AT deficiency | heterozygous GAMT mutation; VUS in ACAN; homozygous SERPINA1 mutations | no | |
| 5 | male, 10 years | global developmental delay and craniofacial dysmorphism | 3 VUSs: homozygous VUS in SACS, hemizygous VUSs in SMS and MED12 | none reported | no |
| 6 | male, 2 years | infantile spasms, developmental delay and retinal dystrophy | 4 VUSs: hemizygous VUS in CACNA1F, heterozygous VUSs in ROGDI, PEPD, ANKRD11 | none reported | no |
In all cases, WES was performed after initial genetic studies were negative.
Case 1
This patient has recently been reported in a series by a laboratory performing WES [Yang et al., 2013]. The patient presented to us at 3 years of age. She was born to a 29-year-old primigravida mother after natural conception without any reported prenatal exposures. Prenatal ultrasound showed bilateral hydronephrosis and cystic fluid collections in the brain. Amniocentesis showed a normal 46,XX karyotype, and postnatally, a chromosome microarray using a 180K Agilent platform was normal. Melnick-Needles syndrome was excluded by FLNA sequencing, and biochemical testing for congenital disorders of glycosylation was normal. The patient was born via induced vaginal delivery at 39 weeks gestation, had immediate respiratory distress, but only required blow-by oxygen. Due to poor feeding and multiple ureteral stent placements to relieve urinary obstruction, she was hospitalized for the first month of life. Prior to discharge from the hospital, a head MRI revealed increased extra-axial fluid-filled spaces over the frontal and temporal bones, widened Sylvian fissures, cave of septum pellucidum, but otherwise normal brain tissue.
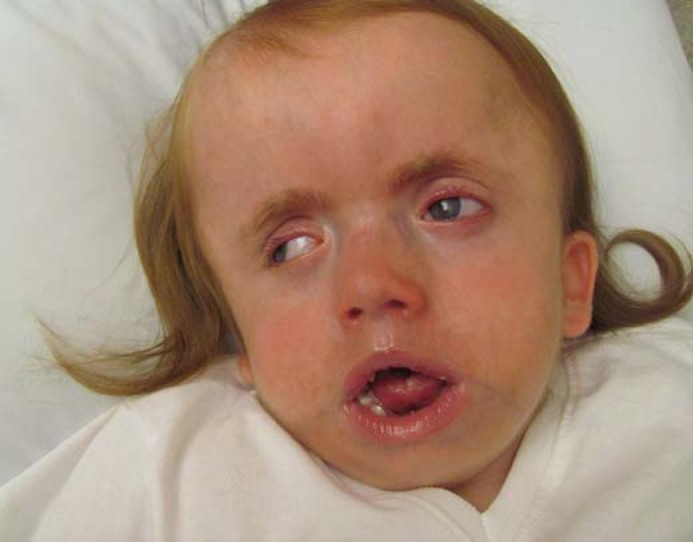
At 2 months of age, the patient developed seizures that remained intractable despite multiple anti-epileptic medications. She demonstrated severe global developmental delay, significant hypotonia, and bilateral sensorineural hearing loss. More specifically, at age 3, she was able to coo, but not to babble, and had no discernable words; she was able to move all 4 extremities, without purpose, and was neither able to roll over nor grasp purposefully. Additionally, she had significant craniofacial dysmorphism including brachyturricephaly, enophthalmos, down-slanting palpebral fissures, depressed nasal bridge, prominent forehead, deep-set eyes with bilateral ptosis and corneal scarring, small, simplified ears, short and upturned nose, retrognathia, and a narrow, crowded palate. The overall facial appearance was significant for midface retraction and malar hypoplasia (fig. 1). Family history revealed that her father had bilateral congenital retinoblastoma with a known mutation in RB1. There was no other significant paternal family history. Maternal family history revealed a paternal first cousin with developmental delay and dysmorphic features including a high forehead without a known diagnosis.
Fig. 1.
Case 1. Image at 3 years of age.
When she presented to our clinic at age 3 for a second opinion regarding the diagnosis, WES had already been ordered by the referring neurologist, after evaluation by both the neurologist and an outside geneticist; initial genetic studies were negative.
After evaluation, we had a strong suspicion for Schinzel-Giedion syndrome which was subsequently confirmed when a de novo pathogenic mutation in SETBP1 (heterozygous c.2602G>A, p.D868N) was found via WES. In addition, the lab reported a primary finding of a mutation (c.2680C>T, p.R894X) and a VUS (c.2545G>A, p.A849T) in CLCN1, the gene responsible for both the autosomal dominant and autosomal recessive forms of myotonia congenita. The mother was heterozygous for the R894X mutation, and the father was heterozygous for the A849T VUS. The patient had no clinically discernible features of myotonia congenita. The patient and the father were found to be heterozygous for a primary VUS in SCN9A (c.4612T>C, p.W1538R), the gene associated with various phenotypes including Dravet syndrome, febrile seizures and erythromelalgia. Neither parent had any abnormal neurologic findings. In addition, 6 other missense VUSs were reported as primary findings in various genes (table 1).
Case 2
The patient presented to us at 13 months of age with intractable epilepsy, microcephaly, arthrogryposis, severe axonal neuropathy, and progressive cerebellar atrophy. It was the mother's third pregnancy and she had gastroenteritis lasting 24 h approximately 3 months prior to delivery. Three weeks following this illness, the mother reported that an ultrasound showed decreased growth parameters, a low amniotic fluid index, and confirmed that the fetus was small for gestational age. The girl was born at 39 weeks gestation by caesarian section for breech presentation and weighed 2,240 g. At birth she had closed fontanelles, microcephaly, optic nerve hypoplasia, hypotonia, and lack of movement of lower extremities. At 13 months of age she rarely cried, had some vocalization, but no cooing or babbling, and did not laugh. Her weakness was progressive with less movement noted at the hips with axial and facial hypotonia; proximal upper limb-girdle movement was relatively preserved. Electromyography showed severe axonal neuropathy. Onset of infantile spasms began at 9 months; she has medically intractable epilepsy with spasms and tonic seizures. MRI at birth and again at 9 months of age showed dysplastic hippocampi, a small dysplastic corpus callosum, cerebellar atrophy, and delayed myelination. There was no significant family history.
Initial genetic studies including a chromosome microarray and karyotype were normal. EGR2 sequencing was sent because EGR2 mutations are associated with congenital hypomyelinating neuropathies, and this was also normal. Results from WES revealed a heterozygous de novo variant (c.757G>A, p.E253K) in KIF1A, a gene encoding a kinesin motor protein that functions as an axonal transporter of synaptic vesicles [Riviere et al., 2011]. The patient was also found to be compound heterozygous for paternally and maternally inherited primary VUSs in FASN (c.3988A>T, p.S1330C and c.3128A>T, p.K1043M), which encodes a fatty acid synthase; however, mutations in this gene have not yet been implicated in human disease. A paternally-inherited primary VUS was also noted in DNAJC5 (c.188C>T, p.A63V), a gene associated with a late-onset autosomal dominant form of neuronal ceroid lipofuscinosis (CLN4B). No neurological symptoms were described in the father or other paternal relatives.
Case 3
The patient is a 29-month-old male who presented with hypotonia and developmental delay. Pregnancy was uncomplicated and ultrasounds were reported normal with TORCH titers within normal limits. The patient was born at term, weighed 2,920 g, and had hypotonia and feeding difficulties. Feeding issues at the time of evaluation included oral aversion to both solids and liquids, and he was only able to tolerate pureed feeds, therefore requiring a G-tube for nutrition supplementation. He was noted to have global developmental delay with the greatest delays in fine motor and language skills. The patient began walking at 21 months. At the time of examination, he was unable to verbalize any recognizable syllables, could not feed himself with a spoon, but could use his thumb and forefinger to pick up objects. Additionally, he had infantile strabismus and difficulty with spatial perception. Family history did not reveal any similar issues; however, his mother had a history of neuroblastoma in infancy and epilepsy as a child. Initial genetic studies including extensive metabolic testing, muscle biopsy with histology and respiratory chain enzyme analysis, Angelman/Prader-Willi methylation studies, and chromosome microarray were all negative. Sequencing and deletion/duplication analysis of SLC9A6, CDKL5, FOXG1, UBE3A, MECP2, ZEB2, and TCF4 were also normal. After all of this testing, WES was ordered. Results revealed a primary finding of a de novo SYNGAP1 heterozygous mutation (c.2212_2213del, p.S738X). Hamdan et al. [2011] recently reported 3 other SYNGAP1 protein-truncating mutations (c.412A>T, p.Lys138X; c.1735C>T, p.Arg579X; and c.2438delT, p.Leu813ArgfsX23) in patients with moderate to severe intellectual disability (ID). Additionally, for our patient, there was a secondary finding of a maternally inherited FBN1 VUS (c.G3509A, p.R1170H) that was reported to be potentially pathogenic. An echocardiogram of the patient revealed an aortic root Z score of +4. Neither the patient nor the mother had any other phenotypic features of the Marfan syndrome, and the mother's echocardiogram was reported to be normal.
Case 4
The patient presented to us at age 3 years with failure to thrive, short stature and mild developmental delay. The patient was born to a 26-year-old primigravida mother who discontinued metformin use after knowledge of pregnancy and required an emergent caesarian section at term, due to fetal distress after induction. Birth weight was 2,550 g. He remained in the special care nursery for the first week of life for temperature regulation and oxygen supplementation. He also became significantly jaundiced during the first few days of life and, as a result, was treated with ursodeoxycholic acid. Developmentally, he was delayed with gross and fine motor skills, but did not have any language delays. At 3 years of age, he was noted to suffer from poor appetite, hypotonia, abdominal pain with hepatosplenomegaly, musculoskeletal pain and joint hypermobility. Previous studies for cholestatic liver disease with elevated liver function studies had revealed alpha-1-antitrypsin (A1AT) deficiency with a ZZ phenotype, and the family history was significant for a maternal second cousin with A1AT deficiency diagnosed in her fourth decade of life.
Standard genetic studies were done including a normal karyotype, and a chromosomal microarray that revealed a maternally inherited duplication of a 0.1-Mb region at Xp11.21, partially involving KLF8, but this was thought to be of doubtful clinical significance. Given that this did not reveal an underlying cause for his other problems, WES was ordered. WES results showed that both the patient and mother were heterozygous carriers of a GAMT (guanidinoacetate methyltransferase) splicing mutation (c.327G>A, p.K109K). GAMT deficiency is an autosomal recessive disorder characterized by developmental delay or regression, mental retardation, speech disorders, seizures, and movement disturbances. Additionally, a likely benign, maternally inherited, primary finding of a heterozygous VUS was detected in ACAN (c.5318T>C, p.L1773P), which has been associated with both an autosomal dominant skeletal dysplasia (osteochondritis dissecans, short stature and early-onset arthritis) and an autosomal recessive skeletal dysplasia (spondyloepimetaphyseal dysplasia, aggrecan type). Both the mother and child had no such skeletal features, and a skeletal survey on the child was normal. Homozygous SERPINA1 mutations (c.1096G>A, p.E366K) responsible for the patient's A1AT deficiency were not initially reported as a secondary finding in the report, but were only included in a revised report after the lab was alerted that the patient also had a diagnosis of A1AT deficiency.
Case 5
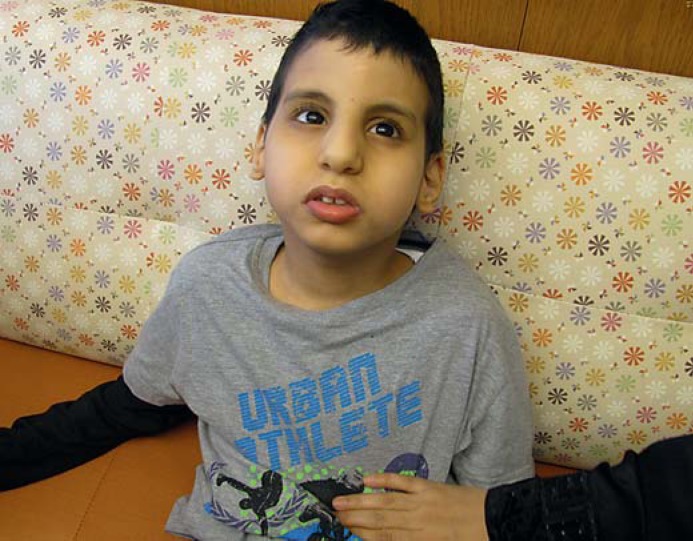
The patient presented to us at age 10 years with global developmental delay and craniofacial dysmorphism (fig. 2); he came from a highly consanguineous Arab family. After extensive testing and evaluations in Saudi Arabia, the 10-year-old patient presented to the United States for further evaluation. He was born at term to a 23-year-old gravida 2 para 2 mother. He was the product of natural conception, pregnancy was uncomplicated, and he weighed 3,200 g. At birth he had some respiratory distress that required blow-by oxygen for 4 days, and he had a cleft palate that was repaired at age 2 years. He was developmentally delayed and, at presentation, walked with assistance with an unstable wide-based gait. He was non-verbal, but was able to recognize parents and express discomfort. Subjectively, he had no visual issues and seemed to respond to sounds. He had a normal head MRI at the age of 3. The patient's older brother was 12 years old, was born with a cleft palate and an unspecified congenital heart defect, but had similar microcephaly, facial dysmorphology and comparable developmental delays to that of the patient. Interestingly, a chromosomal analysis on the 12-year-old brother had revealed a 47,XXY karyotype, consistent with a diagnosis of Klinefelter syndrome. Chromosome analysis performed on the younger brother (our patient) revealed a normal 46,XY karyotype. The genetic workup done prior to WES was all normal including biochemical testing, Smith-Lemli-Opitz screening, head MRI, chromosome microarray, fragile X methylation testing, and Prader-Willi/Angelman syndrome methylation analysis. WES was done with a strong suspicion for an autosomal recessive or X-linked recessive disorder, given the fact that an older brother had a similar phenotype. Results revealed 3 primary VUSs in disease-causing candidate genes. However, based on inheritance, clinical phenotype and segregation pattern, all 3 VUSs were thought to be likely benign (table 2). Both parents were heterozygous, and the patient and brother homozygous for a VUS in SACS (c.1562G>C, p.S521T), a gene associated with Charlevoix-Saguenay spastic ataxia. The mother was heterozygous, the patient hemizygous, and the brother negative for a VUS in SMS, a gene associated with X-linked Snyder-Robinson syndrome. Finally, the mother was heterozygous, and the father and both brothers were hemizygous for a MED12 VUS (c.3797G>A, p.R1266H), a gene associated with Lujan or Opitz-Kaveggia syndrome. Additionally, the laboratory was originally made aware of the brother's diagnosis of Klinefelter syndrome, confirmed by karyotype; however, the segregation pattern of X-linked variants in the brother did not make this clear.
Fig. 2.
Case 5. Image at 10 years of age.
Table 2.
Familial cosegregation table for case 5
| SACS | SMS | MED12 | |
|---|---|---|---|
| Variant filtering model | AR | XLR | XLR |
| Location of alteration | exon 8 | exon 6 | exon 27 |
| Alteration | c.1562G>C | c.535C>T | c.3797G>A |
| (p.S521T) | (p.R179W) | (p.R1266H) | |
| Affected | |||
| Proband | +/+ | + | + |
| Brother | +/+ | − | + |
| Unaffected | |||
| Father | +/− | − | + |
| Mother | +/− | +/− | +/− |
AR = Autosomal recessive; XLR = X-linked recessive.
Case 6
The patient presented to us at 12 months of age with global delay, infantile spasms and retinal dystrophy. He was born to a 26-year-old primigravida mother at 39 weeks gestation with a birth weight of 3,400 g. He reportedly had normal development up until the age of 5 months, and had onset of seizures at this time. By 7 months, he had developmental plateauing along with regression of some skills. A head MRI and MR spectroscopy at 9 months of age was not informative. Family history was only significant for distant cousins on both sides of the family with seizures. At age 2, the patient was evaluated by an ophthalmologist and diagnosed with retinal dystrophy. After the initial genetic studies were negative, WES was ordered. No disease-causing mutations were reported, only primary VUSs. The mother was heterozygous and the patient hemizygous for a VUS (c.1903G>A, p.V635I) in CACNA1F, a gene associated with X-linked congenital stationary blindness; however, this disorder is not associated with seizures or global delays. The patient and his unaffected father were both found to be heterozygous for a VUS in ANKRD11 (c.6125A>G, p.D2042G), a gene associated with autosomal dominant KBG syndrome (craniofacial dysmorphism, short stature, skeletal anomalies, developmental delay, seizures, mental retardation, and macrodontia). Two additional heterozygous novel variants were found in genes typically associated with an autosomal recessive pattern of inheritance: ROGD1 (c.127C>G, p.R43G) and PEPD (c.796A>G, p.T266A). Based on inheritance, clinical phenotype and segregation pattern, all VUSs were thought to be likely benign.
Discussion
WES can help identify inherited and de novo disease-causing variants in the protein-coding portion of the genome. WES is a powerful tool that has the potential to impact and improve patient care, but it also has limitations [Goh and Choi, 2012], which were highlighted by our experience reported in this case series. Before considering WES a success or failure in each case, we suggest that more careful consideration is needed before WES is widely applied in the clinical setting.
The first issue to consider is whether WES was needed to achieve a diagnosis, and this juxtaposition is highlighted by case 1. Schinzel-Giedion syndrome was suspected for this patient by the geneticist based on the clinical studies, but WES had already been ordered prior to this evaluation. Focused genetic testing for SETBP1 mutations would have been sufficient to establish the diagnosis, and raises the question of whether WES was the appropriate test to order in this situation. We do recognize that not all clinicians are skilled at recognizing dysmorphic features, and while WES may facilitate diagnoses in ‘atypical’ cases [Yang et al., 2013], we suggest that WES should not replace the need for a thorough clinical exam by a trained dysmorphologist. Thus, to simply consider WES in this case a ‘success’, as has been previously reported from the lab perspective [Yang et al., 2013], is in question. From our perspective, we believe that the diagnosis could have been achieved with focused, hypothesis-based testing, without the need for WES, and saved valuable time and resources. If we take our 6 cases and only deem case 1 as a success, then the reported success rate in our small case series is 1/6 or 16.6%. However, the question of whether WES was necessary remains paramount and if this case is removed, we do not have a single clearly successful case in our small series.
The second issue to consider is whether the finding of a de novo variant that is not clearly pathogenic constitutes a success or not. This is highlighted by cases 2 and 3, where no definite disease-causing mutations in known genes were found, although de novo variants in genes possibly associated with the phenotype were reported. In case 2, a de novo KIF1A variant was detected. Homozygous nonsense mutations in KIF1A have recently been implicated in autosomal recessive forms of hereditary autonomic and sensory neuropathy [Erlich et al., 2011], and missense homozygous mutations have been associated with autosomal recessive spastic paraparesis (SPG30) [Riviere et al., 2011]. Hamdan et al. [2011] reported a different de novo heterozygous missense mutation in KIF1A (c.296C>T, p.Thr99Met) in a female with moderate ID as well as axial hypotonia and peripheral spasticity. However, the child in that report did not have seizures, an important part of the clinical picture in our case. Further, in case 3, the variant in SYNGAP1 is an interesting potential explanation for the patient's ID. At the time this WES report was issued, there was very limited information on the SYNGAP1 mutation and the phenotype associated with mutations in this gene. Therefore, it was difficult to say with confidence that this is indeed the disease-causing mutation accounting for the patient's phenotype. Thus, we would have to follow the literature over time to see if this suspicion is indeed true. Therefore, we feel that it may be premature to assign these variants as disease-causing mutations, and the literature will need to be followed over time as more functional information and clinical correlations are made with reference to mutations in these genes. Results may be reported as successes from a lab perspective, yet clinically, while suspicious, additional follow-up over time is needed to determine whether the diagnosis is truly accurate.
The third issue our cases highlight relates to findings and interpretation of multiple VUSs, in genes associated with the phenotype (primary findings), and in genes unrelated to the phenotype (incidental findings). In case 1, in addition to confirming the diagnosis of Schinzel-Gideon syndrome, WES results showed an additional primary finding of a maternally-inherited mutation and a paternally-inherited VUS in CLCN1, the gene associated with the autosomal dominant and recessive forms of myotonia congenita. Neither the mother who carries the mutation nor the father who carries the VUS had any findings consistent with myotonia congenita. In addition, the patient who was examined by 2 pediatric neurologists did not exhibit any features of myotonia congenita. Thus, the significance of this secondary finding is unclear, and even though this case has been used as an example where WES found a dual molecular diagnosis [Yang et al., 2013], we feel that this diagnosis is in contention, and implications for the care of this patient or this family remain unclear.
In case 3, the incidental finding of the FBN1 VUS could also be clinically significant, given the fact that the child did demonstrate aortic root dilatation with a Z score of +4. It is also possible though that this is a benign variant since the mother has the same VUS, normal aortic dimensions and no phenotypic features of Marfan syndrome. The child's aortic dimensions will have to be followed over time to determine whether he continues to have aortic root dilatation or not. Only long-term clinical follow-up will show whether this information led to appropriate surveillance or wasteful imaging and unnecessary anxiety for the family. Thus, in the 2 above-mentioned cases, the incidental findings are both unclear, and may bear no clinical significance for the patients or their families.
In the remainder of cases, the VUSs reported are likely not clinically significant. In case 2, given the lack of clinical correlation with the patient's symptoms and family history, the reported primary VUSs in FASN and DNAJC5 are unlikely to be clinically significant. In case 4, in spite of all primary findings, none of the variants explained the phenotype of the patient, and the diagnosis remained unclear. Even though mutations in GAMT are associated with GAMT deficiency, the patient's phenotype did not match with this disorder. Given the autosomal recessive nature of this disorder, and the fact that both mother and child were heterozygous for the change, we believe they are both asymptomatic carriers. WES did not lead to a diagnosis in case 5. Multiple variants in a number of genes were found, but they neither fully explained the clinical findings, nor segregated with the phenotype. Lastly, in case 6, there were no disease-causing mutations reported that were related to the patient's phenotype, and although a number of VUSs were found, none appears to be clinically relevant to the underlying clinical diagnosis.
While we understand that with WES the reporting of VUSs is expected, further supporting the importance of pre- and post-test counseling, this also shows the burden WES may place on the patient and the healthcare system. It is not clear that as VUSs are noted, to what extent each one does necessitate further exploration, and what is the manner of that investigation. It is not feasible to perform functional studies to clarify the significance of most VUSs, and in silico analyses are not refined enough to rely on for clinical decision-making [Thusberg et al., 2011]. While we do not have objective data for patient dissatisfaction or harm done due to these VUSs, and the retrospective nature of our study did not allow us to track the amount of time and resources spent in trying to clarify variants, we felt dissatisfied due to the lack of clear information we were providing to the patients and their families. We believe that as our understanding of genomics increases, the burden of VUSs will decrease, but it currently remains an issue that must be considered prior to ordering WES.
Finally, as noted in cases 4 and 5, the importance of provided information and communication with the laboratory used, is highlighted. It was concerning that despite notation to the testing laboratory that the patient in case 4 had known A1AT deficiency, the initial report did not report mutations in SERPINA1; even it was well documented by the laboratory that secondary findings unrelated to the phenotype would be reported. It was only after repeated prompting that an amended report noted this finding. This should alert the clinician that the ‘filters’ used by testing laboratories to narrow down the genes of interest for any given phenotype are not perfect, and that even though certain ‘actionable’ findings unrelated to the phenotype are supposed to be reported, this does not always happen. This should hopefully change after the ACMG's recommendations on incidental findings in WES [Green et al., 2013].
Additionally, in case 5, where WES was conducted by a different lab than in the case above, the older brother of the patient was suspected to have the same disorder based on a very similar phenotype and was also reported to have a 47,XXY karyotype (Klinefelter syndrome). Upon confirmation of this finding in our lab, the information was supplied to the testing laboratory that performed WES. The segregation pattern of the X-linked variants in the brother did not clarify this situation, and it cannot be taken for granted that sex chromosome aneuploidy will become apparent just by simply looking at the segregation pattern of variants on the X chromosome. These cases highlight the importance of supplying the laboratory with as much clinical information as possible for them to appropriately focus their filtering when looking for disease causing variants. Importantly, detailed and complete clinical information provided to the laboratory is essential for decreasing turnaround time, narrowing searches and revealing clinically significant mutations. While we understand this technology is in its infancy, the importance of cooperation and communication between clinicians and laboratories is paramount as WES develops at a rapid pace and increases in use.
Beyond making a clear diagnosis for the patient, in some cases, the primary motivation to proceed with WES may be for a couple to expand their family with the ability to perform pre-implantation genetic diagnosis or prenatal testing. However, in the majority of our cases, this is not a possibility. As such, patients and their families need to be appropriately counseled and cautioned on how WES results may be used for future reproductive and medical decision making.
Our findings highlight the importance of careful consideration prior to sending WES as part of clinical genetic testing. Although WES may seem more attractive as a cost-effective option compared to standard genetic testing, there still remain significant challenges. For example, unclear or unexpected results may lead to confusion, necessitating additional testing and follow-up. Importantly, this all may lead to anxiety for patients and families. Therefore, first and foremost an emphasis must be put on pre-test counseling by appropriately trained care providers, as this is essential to ensure families have realistic expectations from WES. We recommend that WES be offered only after a thorough evaluation by a clinical geneticist and with appropriate pre-test counseling by trained genetic counselors. We also recognize that some of the limitations we have highlighted may reflect institution-specific issues related to bioinformatics or sequencing technology. These are likely to improve with experience. Long-term follow-up will be needed to assess whether incidental findings will improve outcomes or lead to wasteful use of medical resources. WES is certainly a powerful tool that has the potential to impact and improve patient care, but risks and benefits must be carefully weighed in each case. It will take time to develop a clear consensus on which clinical scenarios necessitate the use of WES, but until then, more consideration is needed regarding the clinical threshold for using WES in ‘diagnostic odyssey’ cases.
References
- 1.ACMG Board of Directors. Points to consider in the clinical application of genomic sequencing. Genet Med. 2012;14:759–761. doi: 10.1038/gim.2012.74. [DOI] [PubMed] [Google Scholar]
- 2.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 3.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 4.Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, et al. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res. 2011;21:658–664. doi: 10.1101/gr.117143.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goh G, Choi M. Application of whole exome sequencing to identify disease-causing variants in inherited human diseases. Genomics Inform. 2012;10:214–219. doi: 10.5808/GI.2012.10.4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 10.Riviere JB, Ramalingam S, Lavastre V, Shekarabi M, Holbert S, et al. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet. 2011;89:219–230. doi: 10.1016/j.ajhg.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 12.Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, et al. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat. 2014;35:899–907. doi: 10.1002/humu.22537. [DOI] [PubMed] [Google Scholar]
- 13.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]