Abstract
Cleidocranial dysplasia (CCD), an autosomal dominant skeletal dysplasia characterized by hypoplastic clavicles and delayed closure of the cranial sutures, is caused by mutations of the runt-related transcription factor 2 (RUNX2) gene. The RUNX2 gene consists of a glutamine and alanine repeat domain (Q/A domain, 23Q/17A), a DNA-binding Runt domain and a proline/serine/threonine-rich domain. We report on a familial case of CCD with a novel mutation within the Q/A domain of the RUNX2 gene, which is an insertion in exon 1 (p.Q71_E72insQQQQ) representing the Q-repeat variant (27Q/17A). Functional analysis of the 27Q variant revealed abolished transactivation capacity of the mutated RUNX2 protein. This is the first case report that demonstrated a glutamine repeat variant of the RUNX2 gene causes CCD.
Key Words: Cleidocranial dysplasia, Glutamine repeat variant, Q/A domain, RUNX2
Cleidocranial dysplasia (CCD) is an autosomal dominant skeletal dysplasia characterized by hypoplastic or aplastic clavicles, delayed closure of the fontanelles and cranial sutures, delayed ossification of the pelvis, dental abnormalities such as late eruption of permanent teeth and multiple supernumerary teeth, and moderately short stature [Cooper et al., 2001]. CCD is caused by hypomorphic or haploinsufficiency of the runt-related transcription factor 2 (RUNX2) gene [Lee et al., 1997; Mundlos et al., 1997].
To date, the mutations occur throughout the RUNX2 gene, but are clustered in the Runt domain in CCD. Most of the mutations within the Runt domain are missense mutations. On the other hand, nonsense mutations, insertions or deletions are predominant within the Q/A domain or the proline/serine/threonine-rich domain [Kim et al., 2006]. The Q/A domain has the capacity to mutate via strand slippage during DNA replication [Yoshida et al., 2002]. Glutamine repeat sequence expansion has been the cause of some diseases that show genetic anticipation, where severity increases in subsequent generations as the repeat length increases due to errors in replication [McMurray, 2010]. Wild-type human RUNX2 contains a 23Q/17A repeat: 23 consecutive glutamine residues followed by 17 alanine residues. An insertion of the polyalanine tract (23Q/27A) was previously observed in only one CCD patient [Mundlos et al., 1997].
Here, we describe a familial case of CCD with a novel mutation within the Q/A domain, which is an insertion of the polyglutamine tract (27Q/17A). In vitro functional analysis was performed to assay the transactivation capacity of the mutant RUNX2 protein.
Case Report
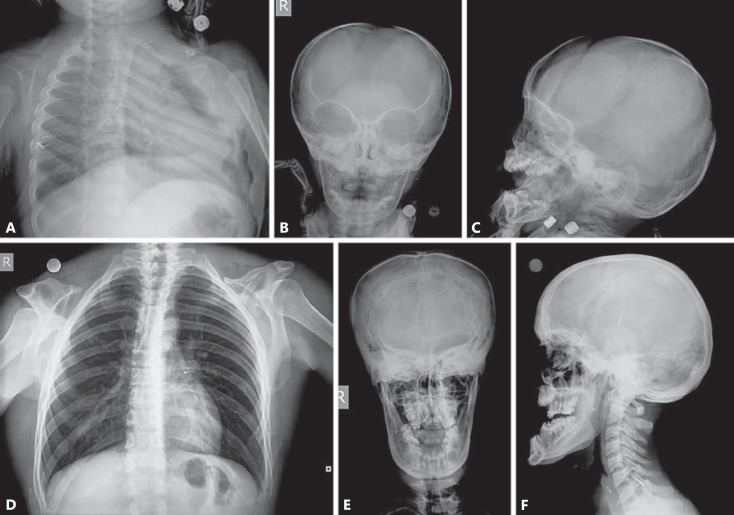
A family with the clinical diagnosis of CCD from the Erciyes University, Turkey, was examined in this study. The proband, a 2-year-old boy, is the only child of an affected father (27 years old) and a healthy mother (23 years old). Radiographs of the proband showed a large defect of the parietal and occipital bones, supernumerary teeth, sclerosis of the cranial base, multiple wormian bones, and bilateral absence of clavicles (fig. 1A–C). The last 2 radiographic manifestations (multiple wormian bones and absent clavicles) were also observed in the boy's father (fig. 1D, E), although he had a relatively thick skull and prognathism (fig. 1F). The craniofacial manifestations, including frontal bossing, midface hypoplasia and a small face, were shared in both the proband and his father.
Fig. 1.
Radiographs of the proband's chest and skull demonstrating the complete absence of bilateral clavicles, open large fontanelles, multiple wormian bones, supernumerary teeth, and mandibular protrusion (A-C). Anteroposterior radiograph of the father's chest demonstrating bilateral absence of clavicles (D) and radiographs of his skull demonstrating multiple wormian bones, a relatively thick skull and prognathism (E, F).
Methods
After informed consent was obtained from all family members, genomic DNA was extracted from peripheral blood leukocytes. The exons (0-7) and their flanking intronic regions of the RUNX2 gene were amplified by PCR using sets of primers. Direct sequence analysis of the affected patients' DNA from this family demonstrated a novel heterozygous mutation within the Q/A domain, c.213_214insCAGCAGCAGCAG (p.Q71_E72insQQQQ).
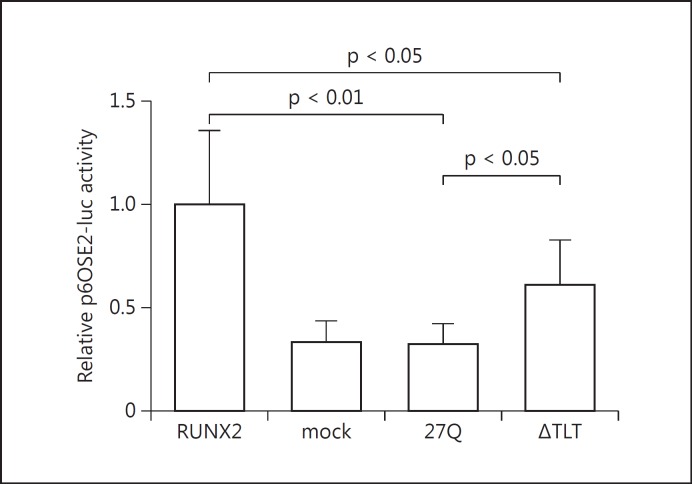
For in vitro functional studies of the mutant RUNX2 protein identified in this family, the entire cDNA of p.Q71_E72insQQQQ (27Q) was constructed as follows. We confirmed that the mutation was located between 2 PstI sites (181 bp) in exon 1 and obtained the oligonucleotide duplex containing the mutation (Integrated DNA Technologies MBL, Japan). PCR fragments of the oligonucleotide duplex were double-digested with PstI. This insert was cloned into the human full-length RUNX2 cDNA (Ori-Gene Technologies, Rockville, Md., USA) at the PstI sites. On the other hand, p.T198_T200del (ΔTLT), which was previously identified in a patient with CCD, was constructed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif., USA) [Matsushita et al., 2014]. Transient transfection experiments in COS7 cells were performed using FuGENE 6 (Roche, Indianapolis, Ind., USA). Aliquots of 400 ng expression plasmid containing either wild-type or mutagenized RUNX2 were cotransfected with 400 ng of a reporter plasmid p6OSE2-luc (kindly provided by T. Komori, Nagasaki University, Japan) [Harada et al., 1999]. All transfection experiments were done 8 times. The transactivation study showed that the 27Q variant and ΔTLT mutants had significantly lower transcription activities (32 and 61% of the wild type, respectively) (fig. 2).
Fig. 2.
Transactivation ability of the wild-type and mutant RUNX2 proteins. COS7 cells were transfected with p6OSE2-luc as a reporter plasmid, full-length, wild-type or mutant RUNX2 as effector plasmids, and phRL-TK as an internal control of transfection efficiency. Data are presented as fold activation relative to the activity obtained with wild-type RUNX2 vector plasmid. Bars represent the average ratios of luciferase to Renilla activity. Standard deviations are represented by error bars. Both the 27Q variant and ΔTLT198_200 mutants showed significantly reduced transactivation ability compared to the wild type. Moreover, transactivation of the 27Q variant was significantly lower than that of the ΔTLT198_200 mutant.
Discussion
Clinical and radiographic manifestations of the present cases seemed to be typical for CCD, including complete absence of bilateral clavicles, multiple wormian bones and supernumerary teeth. Mutation analysis of this family showed a Q-repeat variant within the Q/A domain, which resulted in a significant reduction of transactivation of the RUNX2 protein.
Q-repeat variants within the RUNX2 gene were identified in an Australian fracture cohort (15Q, 16Q, 24Q, and 30Q) [Vaughan et al., 2002], a randomly selected population from Aberdeen (16Q) [Vaughan et al., 2004], and a Spanish population study (16Q, 18Q and 30Q) [Pineda et al., 2010]. A 30Q variant of the RUNX2 gene has never been reported to be associated with CCD phenotypes. On the other hand, a novel 27Q variant caused CCD by downregulating the transactivation activity of the RUNX2 protein. Generally, triplet repeat expansion disorders accelerate their phenotypes according to the repeat length. Huntington's disease, for example, is one of the polyQ-repeat disorders, and its severity is usually associated with the length of the polyQ tracts. It has been suggested that aggregation of the polyQ fibers is pathogenic of the disease. Perutz [1996] reported that Huntington's disease has not been observed in individuals with <37 repeats, and absence of disease has never been found in those with >41 repeats. This indicated that polyQ expansion beyond the pathological threshold of 36-40 repeats leads to a clinical manifestation. According to the model of Perutz et al. [2002], polyQ fibers are composed of nanotubes with 20 residues per turn, and a minimum of 2 turns (40 repeats) is necessary for pathogenic polyQ aggregates. It is possible that the 27Q variant is pathogenic, while the 30Q variant is benign, since the repeat length is not necessarily related to the severity of the disease when it is <40 repeats.
Sears et al. [2007] showed that Q/A tandem repeat ratio correlated to RUNX2 transcriptional activity. Morrison et al. [2012] demonstrated that transactivation activity was reduced by the RUNX2 Q-repeat variants, but rescued by PEBP2β, which is the partner subunit for heterodimerization with the Runt domain. In a study on dogs, Fondon and Garner [2004] demonstrated that the length of the Q repeat is significantly associated with midface length and nose curvature. We previously reported a CCD patient with the in-frame deletion (ΔTLT) who showed a milder phenotype than the present cases, including mild short stature (-1.75 SD), delayed fontanelle, midface hypoplasia, pseudoarthrosis of the right clavicle, and hypoplasia of the left clavicle [Matsushita et al., 2014]. ΔTLT mutation decreased the transactivation activity of the RUNX2 protein by abolishing the heterodimerization of the RUNX2 protein with the PEBP2β. Significantly lower transactivation activity of the 27Q variant than that of the ΔTLT mutant may reflect the phenotypic severity of the disease.
References
- 1.Cooper SC, Flaitz CM, Johnston DA, Lee B, Hecht JT. A natural history of cleidocranial dysplasia. Am J Med Genet. 2001;104:1–6. doi: 10.1002/ajmg.10024. [DOI] [PubMed] [Google Scholar]
- 2.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, et al. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem. 1999;274:6972–6978. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ, Nam SH, Kim HJ, Park HS, Ryoo HM, et al. Four novel RUNX2 mutations including a splice donor site result in the cleidocranial dysplasia phenotype. J Cell Physiol. 2006;207:114–122. doi: 10.1002/jcp.20552. [DOI] [PubMed] [Google Scholar]
- 5.Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, et al. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 6.Matsushita M, Kitoh H, Kaneko H, Mishima K, Itoh Y, et al. A novel in-frame deletion of the RUNX2 gene causes a classic form of cleidocranial dysplasia. J Bone Miner Metab. 2014;32:96–99. doi: 10.1007/s00774-013-0456-7. [DOI] [PubMed] [Google Scholar]
- 7.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison NA, Stephens AA, Osato M, Polly P, Tan TC, et al. Glutamine repeat variants in human RUNX2 associated with decreased femoral neck BMD broadband ultrasound attenuation and target gene transactivation. PLoS One. 2012;7:e42617. doi: 10.1371/journal.pone.0042617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 10.Perutz MF. Glutamine repeats and inherited neurodegenerative diseases: molecular aspects. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 11.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pineda B, Hermenegildo C, Laporta P, Tarín JJ, Cano A, García-Pérez MÁ. Common polymorphisms rather than rare genetic variants of the Runx2 gene are associated with femoral neck BMD in Spanish women. J Bone Miner Metab. 2010;28:696–705. doi: 10.1007/s00774-010-0183-2. [DOI] [PubMed] [Google Scholar]
- 13.Sears KE, Goswami A, Flynn JJ, Niswander LA. The correlated evolution of Runx2 tandem repeats, transcriptional activity, and facial length in carnivora. Evol Dev. 2007;9:555–565. doi: 10.1111/j.1525-142X.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 14.Vaughan T, Pasco JA, Kotowicz MA, Nicholson GC, Morrison NA. Alleles of RUNX2/CBFA1 gene are associated with differences in bone mineral density and risk of fracture. J Bone Miner Res. 2002;17:1527–1534. doi: 10.1359/jbmr.2002.17.8.1527. [DOI] [PubMed] [Google Scholar]
- 15.Vaughan T, Reid DM, Morrison NA, Ralston SH. RUNX2 alleles associated with BMD in Scottish women; interaction of RUNX2 alleles with menopausal status and body mass index. Bone. 2004;34:1029–1036. doi: 10.1016/j.bone.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, et al. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–638. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]