Abstract
The Paramyxoviridae family includes many viruses that are pathogenic in humans, including parainfluenza viruses, measles virus, respiratory syncytial virus and the emerging zoonotic Henipaviruses. No effective treatments are currently available for these viruses, and there is a need for efficient antiviral therapies. Paramyxoviruses enter the target cell by binding to a cell surface receptor and then fusing the viral envelope with the target cell membrane, allowing the release of the viral genome into the cytoplasm. Blockage of these crucial steps prevents infection and disease. Binding and fusion are driven by two virus encoded glycoproteins, the receptor-binding protein and the fusion protein, that together form the viral “fusion machinery”. The development of efficient antiviral drugs requires a deeper understanding of the mechanism of action of the Paramyxoviridae fusion machinery, which is still controversial. Here we review recent structural and functional data on these proteins and the current understanding of the mechanism of the paramyxovirus cell entry process.
Keywords: Paramyxoviridae, fusion machinery, viral entry, receptor-binding protein, fusion protein
1. Introduction to paramyxoviruses
1.1 Classification and medical significance
The Paramyxoviridae family, among the Mononegavirales order, is composed of enveloped viruses containing non-segmented negative strand RNA (reviewed in Refs. 1–3). Its members are found worldwide (Fig.1) and infect a broad range of host species including humans, pigs, horses and birds. Several paramyxoviruses such as measles virus (MeV), mumps virus (MuV), human parainfluenza viruses (HPIV) and respiratory syncytial virus (RSV) continue to have a major impact on global health. These viruses cause severe infections mainly affecting the respiratory tract of children and immunocompromised patients (Table 1).
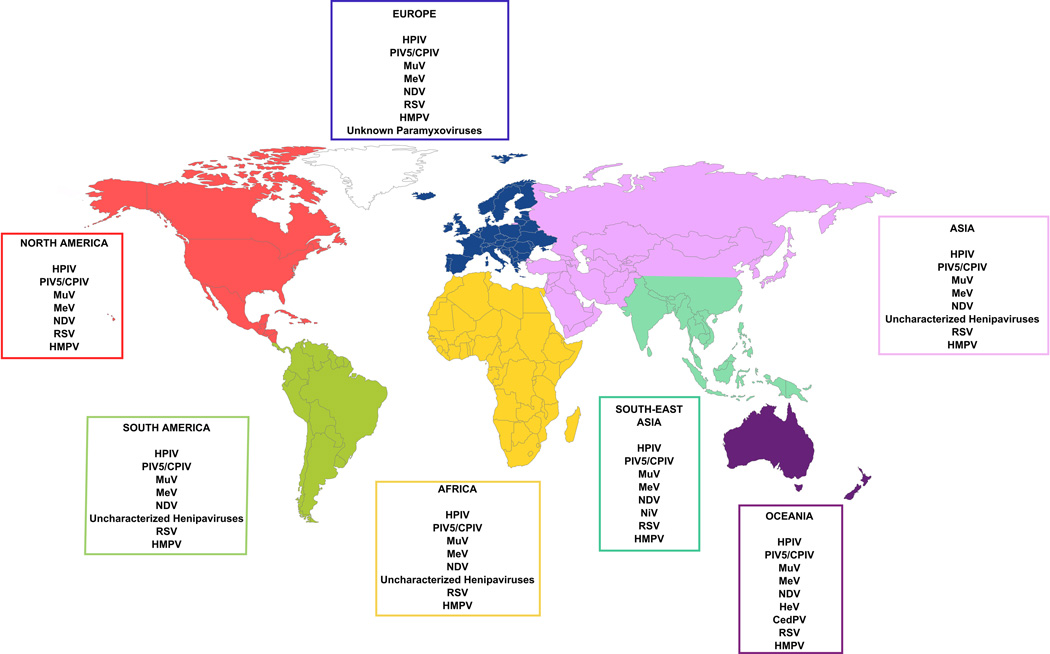
Figure 1. World distribution of major paramyxoviruses.
Paramyxoviruses are found on every continent. Henipa- and Henipa-like viruses have been found in Oceania, Asia, Africa and South America, but human infections have only been reported in Oceania and South-East Asia. Data gathered from Enders et al.2, Ganar et al.12, Croser et al.13 the World Health Organization, the World Organization for Animal Health, and recent studies8. Abbreviations are as in Table 1.
Table 1.
Paramyxoviruses classification and associated pathologies.
| Order | Family | Sub-family | Genus | Species | Associated diseases |
|---|---|---|---|---|---|
| Respirovirus | HPIV1, HPIV3 | Rhinitis, pharyngitis, pneumonia | |||
| Mononegavirales | Paramyxoviridae | Paramyxovirinae | Rubulavirus | MuV, HPIV2, HPIV4, PIV5/CPIV | Mumps, orchitis, meningo-encephalitis, tracheobronchitis |
| Morbillivirus | MeV | Measles, encephalitis | |||
| Avulavirus | NDV | Fatal respiratory tract infection of poultry | |||
| Aquaparamyxovirus | ASPV | Salmonid gill disease? | |||
| Ferlavirus | FDLV | Unknown | |||
| Henipavirus | HeV, NiV | Fatal encephalitis | |||
| Pneumovirus | RSV | Rhinitis, pneumonia | |||
| Pneumovirinae | |||||
| Metapneumovirus | HMPV | Rhinitis, fever, bronchiolitis | |||
The Paramyxoviridae family is divided into two sub-families and seven genera. Most paramyxoviruses cause a wide range of pathology. Data gathered from ICTV, Enders et al.2 and recent articles4,5. PIV: parainfluenza virus. HPIV: human parainfluenza virus. CPIV: canine parainfluenza virus. MuV: mumps virus. MeV: measles virus. NDV: Newcastle disease virus. ASPV: atlantic salmon paramyxovirus. FDLV: Fer-de-Lance virus. HeV: Hendra virus. NiV: Nipah virus. RSV: respiratory syncytial virus. HMPV: human metapneumovirus.
The Paramyxoviridae family is divided into two sub-families: Paramyxovirinae and Pneumovirinae. The Paramyxovirinae sub-family consists of seven genera, Respirovirus (which includes human parainfluenza virus type 3; HPIV3), Rubulavirus (which includes MuV); Morbillivirus (which includes MeV), Avulavirus (which includes Newcastle disease virus; NDV), Aquaparamyxovirus (which only includes atlantic salmon paramyxovirus; ASPV4), Ferlavirus (which only includes Fer-de-Lance virus; FDLV5), and Henipavirus (which includes Nipah virus [NiV] and Hendra virus [HeV] as well as Cedar virus [CedPV] recently discovered in bats in Australia6); Table 1. Other paramyxoviruses such as J-virus (JPV) and Beilong virus (BeiPV), as well as some recently discovered bat paramyxoviruses, are closely related, but remain unassigned to any sub-family7,8. The Pneumovirinae sub-family consists of two genera: Pneumovirus (which includes RSV) and Metapneumovirus (which includes human metapneumovirus; HMPV); Table 1.
Epidemiological studies have shown that HPIV is responsible for around 7% of hospitalizations for fever and/or respiratory diseases in children under five9. RSV alone is responsible for at least 3–9% (66,000 – 199,000) of deaths caused by acute lower respiratory tract infection worldwide, mainly in children under the age of five10. Human metapneumovirus (HMPV) also causes acute respiratory infections. Studies led on hospitalized patients in Virginia revealed that HMPV is involved in 90% of wheezing cases requiring hospitalization11. In terms of the pathogens that do not infect humans but cause problems to society, NDV infects poultry and is associated with a high mortality rate due to respiratory tract infections, generally occurring in developing countries where this disease has a negative economic impact (reviewed in Ref. 12).
The emerging henipaviruses NiV and HeV is associated with high mortality and/or lethal outbreaks. In the first outbreak of NiV in Malaysia in 1999, 265 people were infected, and 105 patients died of fatal encephalitis. HeV first emerged in Australia in 1994 primarily affecting horses, however, seven people have been infected resulting in four deaths. All Henipaviruses are classified as Biosafety level 4 agents, due to the high lethality of infection and the lack of established treatment. The main reservoir for Henipaviruses is fruit bats, notably the Pteropus genus. These bats are mainly present in Africa, South-East Asia and Oceania (reviewed in Refs.13,14). Recent studies have identified new paramyxoviruses in European insectivorous bats8 which produce symptoms resembling HeV infection. While these viruses remain unassigned to any genus, they are more closely related to Paramyxovirinae than to Pneumovirinae8.
The only preventive vaccines currently available for members of the Paramyxoviridae family are those against MeV, MuV, and NDV (for poultry). Even these viruses are still a major health concern. According to the World Health Organization (WHO), each year, around 200,000 deaths are associated with MeV infection mainly in developing countries. The Centers for Disease Control and Prevention (CDC) declares that more people have been infected with measles in the United States during the first four months of 2014 than have been infected in the first four months of the past 18 years.
In 2012, 687,000 cases of MuV infection were reported across the world. (WHO). Usually, MuV infection is not lethal but it can lead to complications such as meningitis, encephalitis, and orchitis, with possible permanent sequelae (WHO). Furthermore, there are currently no therapies to treat patients infected by any paramyxovirus (reviewed in Ref. 2), making these viruses a significant public health issue.
1.2 Structure
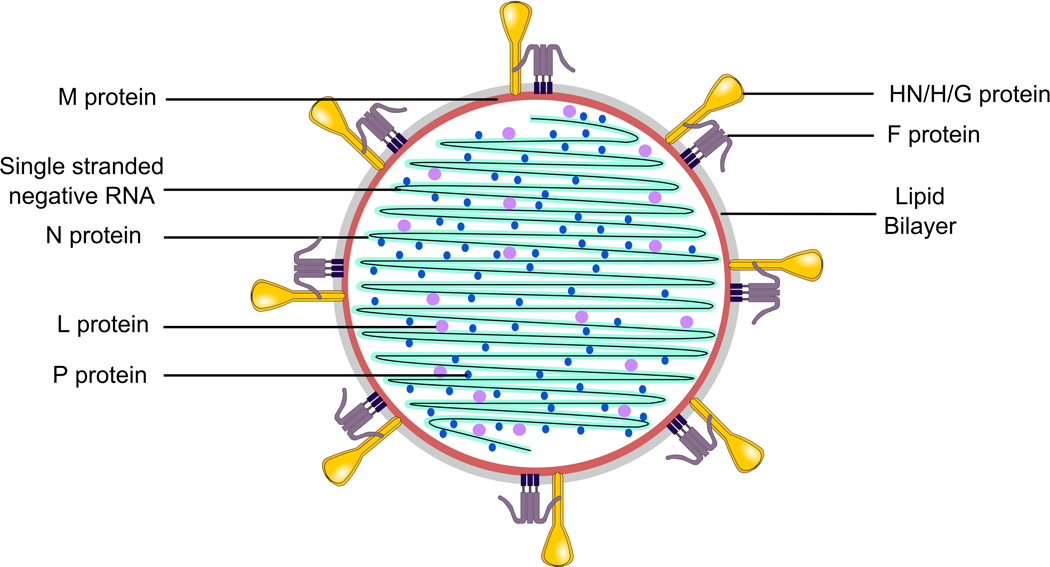
Paramyxoviruses are 150 to 300nm in diameter with envelopes composed of host cell lipids and viral glycoproteins (reviewed in Refs. 1,2). The genome is a non-segmented RNA strand of negative polarity, between 15,210 (RSV type 2, GU591759.1, Kumaria et al.15) and 15,894 nucleotides (MeV, NC_001498.1, Takeuchi et al.16) with the exception of Henipaviruses which contain a longer genome (NC_001906, Wang et al.17, Yu et al.18, NC_007454.1, Jack et al.19, NC_007803.1, Li et al.20). The length of the genome of each paramyxovirus is always a multiple of six nucleotides, an organization required for efficient replication by the viral polymerase15,16. The genomic RNA strand is encapsidated by the helical nucleocapsid protein, N or NP (Fig. 2). The Large protein (L) and the phospho protein (P) constitute the viral RNA-dependent transcriptase / replicase complex. In the virion, L and P are associated with the RNA-nucleocapsid complex (Fig. 2). The N or NP protein also interacts with the matrix protein (M), a non-structural protein that lines the envelope of the viral particle (Fig. 2). The lipid bilayer envelope of the virus is derived from the host cell membrane, formed when the virus buds from a region of membrane expressing the viral receptor-binding protein (HN/H/G) and the fusion protein (F). As for many other enveloped viruses, the virions are labile, and can be easily inactivated ex vivo by heat, organic solvents such as ethanol, or detergents.
Figure 2. Schematic representation of the common structure of Paramyxoviruses.
Paramyxoviridae are enveloped viruses. They contain single-stranded negative RNA coated with nucleocapsid (N) protein as well as a large (L) protein and a phosphoprotein (P) that carries out polymerase activity. The matrix (M) protein lines the viral lipid bilayer. The two viral glycoproteins -- hemagglutinin-neuraminidase (HN)/ hemagglutinin (H)/ glycoprotein (G) and fusion (F) -- protude from the viral membrane.
The six proteins N/NP, L, P, M, HN/H/G and F are conserved among the Paramyxoviridae family. In addition, some structural proteins are restricted to specific viruses and their roles may be less clear, for example the small hydrophobic proteins (SH) and the transmembrane (TM) proteins7,17–20. Paramyxoviridae also encode for non-structural proteins that are involved in the inhibition of the interferon response21. In addition, the alternative splicing of the P-gene leads to the expression of C, V and W proteins, whose role is to counteract host innate immunity (reviewed in Ref. 22).
1.3 Viral entry and life cycle
Paramyxovirus fusion is mediated by two different viral proteins that in most cases must work in concert to accomplish viral entry. The receptor-binding protein first engages the cellular receptor, then in most cases activates the fusion protein, and the fusion protein inserts itself into the target cellular membrane, allowing the viral envelope and target cellular membrane to merge. Upon fusion of the viral envelope with the target cell membrane, the genetic material is released into the cytoplasm. The negative sense RNA, which is present in the form of a nucleocapsid or RNA/protein complex, is converted into positive sense message-length RNAs by the RNA-dependent RNA polymerase that is provided by the virus. This step allows for translation of virally encoded proteins. Replication of the viral genome occurs via transcription of a full length positive sense strand which is then copied into a full-length negative sense new genome, and encapsidated by the viral nucleocapsid protein. The matrix protein binds to the nucleocapsid and interacts with the cytosolic tails of the membrane bound HN/H/G and F proteins, facilitating the process of budding of progeny virions. Release of new viral particles from the cell surface, in some paramyxoviruses requiring a receptor-cleaving enzymatic function carried out by the receptor binding protein23, permits infection of new target cells and spread of infection.
Most paramyxovirus fusion events occur in a pH-independent manner, at the cell surface, however some viruses enter the cell via endocytosis (reviewed in Refs. 1,3). How the receptor-binding protein and the fusion protein (together called “fusion machinery’) work together to promote fusion has been an area of active investigation since it was first shown that the paramyxovirus receptor binding protein plays an active role in the fusion process during entry24–26. Several models have been proposed, and the molecular details of the fusion process mediated by the paramyxovirus fusion machinery remain controversial. Previous models postulate a duality amongst Paramyxoviridae (reviewed in Refs. 1,3). It has been proposed that for paramyxoviruses that bind a proteinaceous receptor, the role of the receptor binding protein is mainly a repressive one27–32 (reviewed in Refs. 33–35) and that upon receptor binding, the fusion protein is released and proceeds to fusion ; on the other hand, the receptor binding proteins of sialic-acid binding viruses have been thought to interact with F only upon receptor engagement27,28,36,37 (reviewed in Refs. 33,34,38,39). Our data suggest that a common mechanism applies to all paramyxoviruses that use a receptor-binding protein to activate a fusion protein, including those that bind a proteinaceous receptor40,41.The debate will be detailed in the sections below. In this chapter we review recent advances in the field of paramyxovirus entry. We first summarize structural data about Paramyxoviridae virions and specifically HN/H/G and F. The focus then turns to receptor engagement and its effects on HN/H/G. We detail the interaction between these two surface glycoproteins before, during, and after receptor engagement, as well as the membrane fusion process mediated by F, and propose a potential unifying model for Paramyxoviridae fusion.
2. Structure and function of the paramyxovirus glycoproteins
2.1 The receptor binding protein
The paramyxovirus receptor binding proteins present on different members of the virus family are known as HN, H, or G. These proteins are distinguished by the type of receptor they engage, their ability to cleave sialic acid (neuraminidase activity), and their ability to agglutinate red blood cells (reviewed in Ref. 1). The HN protein carried by the Respirovirus, Rubulavirus and Avulavirus genera (Table 1) possesses both sialic acid binding (hemagglutinating) and sialic acid cleaving (neuraminidase) activities. Sialic acid binding is active during viral entry while neuraminidase activity is involved in viral budding and prevents the virus from self-aggregating. The H protein carried by the Morbillivirus genus (Table 1) does not bind to sialic acid during viral entry. Both HN and H proteins have the ability to agglutinate red blood cells, but the H binds proteinaceous receptors during MV entry. The H protein lacks neuraminidase activity, suggesting that following viral release, self-aggregation mediated by sialic acid binding does not occur. The G protein carried by the Pneumovirinae and Henipaviruses genera (Table 1) does not bind sialic acid and does not possess neuraminidase activity. Like H, G proteins bind proteinaceous receptors.
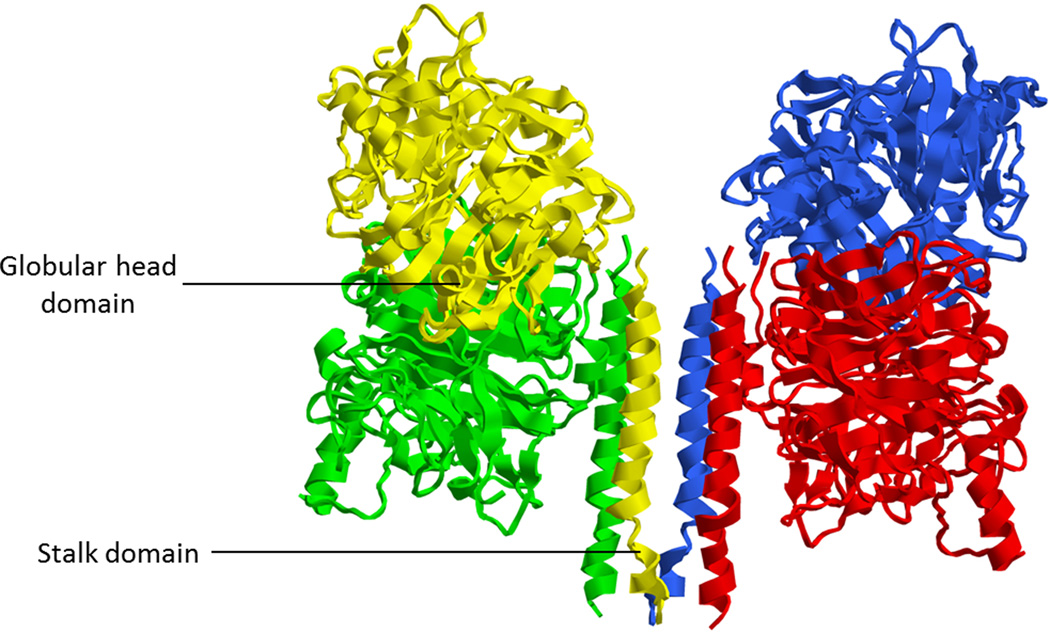
The three types of receptor-binding proteins differ in the type of receptor they bind, but share the same general architecture. HN, H, and G are type II transmembrane proteins, with N- termini inside the viral particle (Fig. 3). Each is present on the viral membrane as a tetramer composed of two dimers, an arrangement known as a dimer of dimers. A dimer consists of an association of two monomers (Fig. 3), each of which monomer contains a cytoplasmic tail domain, a transmembrane domain, a stalk domain, and a globular head domain (reviewed in Refs. 1,3).
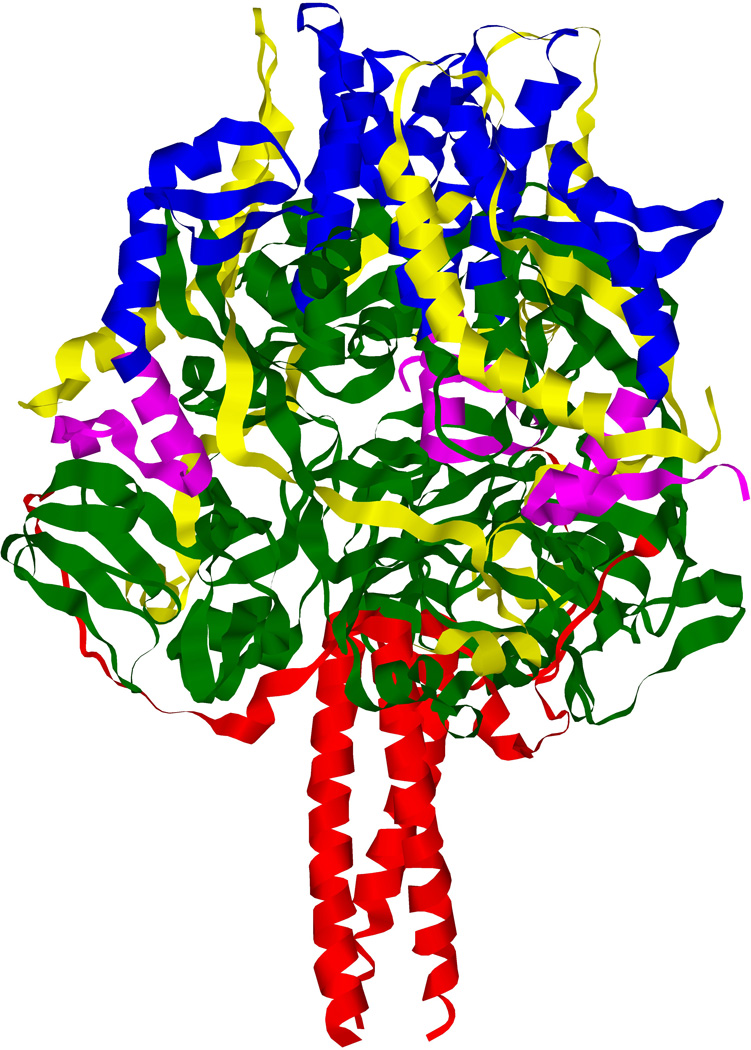
Figure 3. Structure of the Newcastle disease virus hemagglutinin-neuraminidase protein.
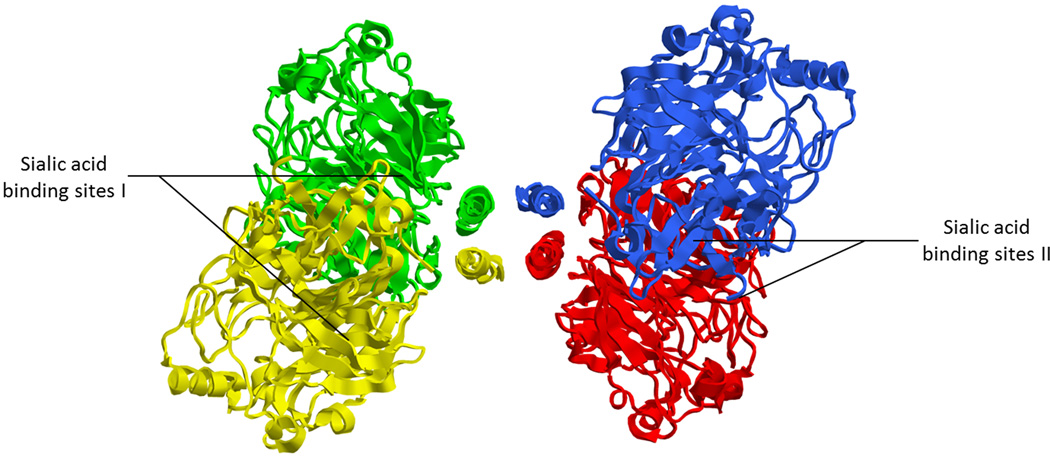
3.1 Side view of the crystal structure of the tetramerized NDV-HN ectodomain showing the stalk and the globular domains of each monomer. Each color represents one monomer of the receptor-binding protein. One dimer is composed of green and yellow monomers, the other of red and blue monomers. (PDB ID: 3T1E; Yuan et al.53). 3.2 Top view of the crystal structure of the tetramerized NDV-HN ectodomain, showing sialic acid binding sites I and II. Each monomer bears a site I and a site II. (PDB ID: 3T1E; Yuan et al.53) 3.3 Schematic representation of the domains of NDV HN.
Dimers of the receptor binding proteins are formed by disulfide bridges between the stalk domains of two monomers42–53 and are also linked via the stalk domain of the proteins, as described for PIV5-HN42, although the transmembrane domain may stabilize the tetramer, as described for NDV-HN53. These inter-dimer links mainly involve non-covalent bonds which are weaker than the intra-dimer disulfide linkages. Tetramers of HN/H/G are more suitable for crystallization than dimers, as described for HeV-G52. Alteration of disulfide bridges via in vitro mutagenesis alters dimer and tetramer stability50,51. Interestingly, a recent study from Navaratnarajah et al.51 reported that the complete stalk domain of MeV-H is not directly involved in the tetrameric structure, and the extent of involvement of the stalk in formation of the tetramer for other members of the Paramyxoviridae family is unclear.
Crystallographic studies of the tetrameric globular heads show each monomer carriying an N-terminal six-blade β-propeller, characteristic of neuraminidase enzymes43–55. Interestingly, HN, H and G share this structure, although only HN possesses neuraminidase activity. H and G carry a structural vestigial neuraminidase site43–47, consistent with the hypothesis of a common evolutionary origin for these three receptor-binding proteins.
In the case of HN, the sialic acid binding site of each monomer, known as sialic acid binding site I, is located at the top of the globular head domain, in the center of the β-propeller. In addition, a second sialic acid binding site (known as site II) was identified crystallographically at the dimer interface of NDV-HN.56 This site II is involved in receptor binding57,58. Functional analysis have suggested a second sialic acid site on HPIV159,60 and have identified a second sialic acid binding site on HPIV3 that is also important for activating F61, although these sites have not been demonstrated crystallographically. The receptor binding site of G shares the same location as site I46,47 whereas the H binding sites are located on the side of the β-propeller45,55, 138. The structure of the tetrameric receptor-binding protein ectodomain, comprised of the head and stalk domains, have been solved for PIV548 and NDV53. The stalk domain adopts a four helix bundle conformation (Fig. 3) with a hydrophobic core located at the upper part of the stalk domain48,53.
2.2 The fusion protein
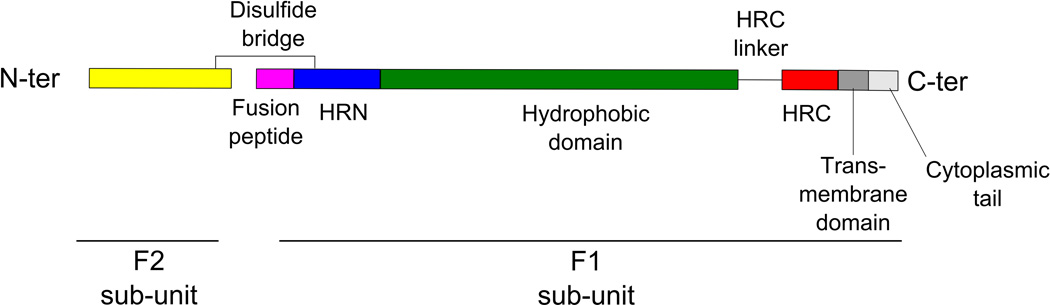
The paramyxovirus fusion protein, F, is a type I transmembrane protein, with its N-terminus outside the viral particle. It is synthesized as an inactive F0 precursor (reviewed in Refs.1,3). F0 is then cleaved into its active form, F, which is composed of two sub-units, F1 and F2. The two sub-units are linked by a disulfide bridge between the HRN of F1 and F2 (Fig. 4). The cleavage creates the hydrophobic fusion peptide, which is inserted into the target membrane during the fusion process, once an activation step exposes the peptide at the surface of the molecule.
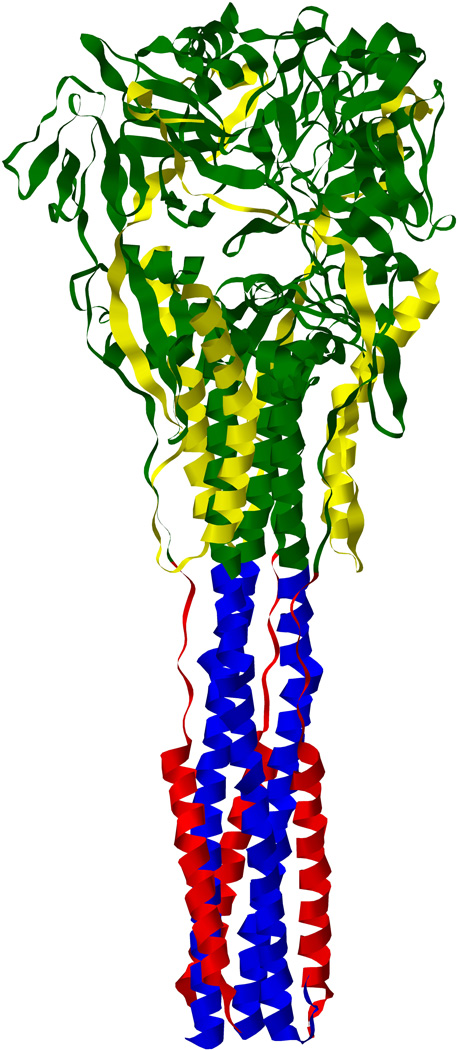
Figure 4. Structure of the paramyxovirus fusion protein.
4.1 Crystal structure of the pre-fusion state of the trimeric fusion protein of PIV5 showing the fusion peptide (purple) in the hydrophobic pocket formed by ae hydrophobic domain (deep green), the HRN domain (deep blue) and the F2 sub-unit (yellow). (PDB ID: 4GIP; Welch et al.70). 4.2 Crystal structure of the post-fusion state of the fusion protein of HPIV3 showing the 6-helix bundle structure formed by the HRN (deep blue) and HRC (red) domains interacting together. (PBDB ID: 1ZTM; Yin et al73). 4.3 Schematic representation of the main domains of a monomer of the cleaved paramyxovirus fusion protein. HRC/HRN: Heptad Repeat C-/N-terminal domain.
F acts as a homotrimer in which each monomer is linked to each other via a transmembrane domain62,63 and contains a cytoplasmic tail, a transmembrane domain, a heptad repeat C-terminal domain (HRC), a heptad repeat N-terminal domain (HRN), and the fusion peptide (Fig. 4). The heptad repeat domains are regions of 7-mer repeats in which every seventh residue is either a Leucine, Isoleucine or Valine, and the whole structure is an amphipatic α-helix.
The F cleavage step is crucial for the viruses, as uncleaved F proteins are unable to promote fusion. For most Paramyxoviridae (with the exception of Henipaviruses) furin proteases within the trans-Golgi network cleave F0 at an R-X-K/R-R consensus motif (reviewed in Ref. 1). Unlike most Paramyxoviridae, RSV F possesses two cleavage sites which are required for efficient fusion64. For Henipaviruses, F0 is cleaved in the endosomal compartment by cathepsins L and B at a VGDVR/K consensus motif65–68. Henipavirus F0 is expressed at the plasma membrane, re-internalized and then cleaved before associating with the rest of the viral particle. The cleavage also seems highly dependent on the valine content of the fusion peptide, as reported for HeV69.
The structure of cleaved PIV5-F in its pre-fusion state has been solved70. The fusion peptide is initially buried in a hydrophobic pocket, preventing premature exposure70. This pocket is composed of the HRN (Fig. 4). In this pre-fusion state, HRC forms an α-helix close to the viral membrane71. The crystallographic data suggested that few conformational changes occurred after cleavage, when compared to uncleaved forms of PIV5 F and HPIV3 F72,73. However, uncleaved F is fusion incompetent. Established models describe the state of pre-fusion F as being metastable, and destabilized following activation by the receptor-binding protein.
In the post-fusion state, the fusion peptide is exposed in an open α-helical domain, and the heptad repeat domains associate, forming a highly stable 6-helix bundle. The formation of this stable structure is a significant driver of the process of membrane fusion74,75 (reviewed in Ref. 76) (Fig. 4). Like the receptor binding proteins, F is highly glycosylated. For NiV-F, it seems that some of these glycosylation sites decrease the fusogenicity of the virus77. In vivo these additional carbohydrates may protect the virus from recognition by the host immune system77.
3. Proposed mechanisms of receptor binding protein and fusion protein interactions
3.1 The globular heads of the receptor-binding protein selectively engage specific cellular receptors
The fusion process begins when the receptor-binding protein engages its receptor. HN recognizes sialic acid bearing membrane proteins, whereas H and G bind proteinaceous receptors. H binds different proteinaceous receptors for each virus. For example, MeV H engages CD46, CD150/SLAM (signaling lymphocyte-activation molecule), and Nectin 445,55,78–82. CD46 binding seems to be unique to laboratory-adapted strains. CD150 is expressed on the cell surface of macrophages and dendritic cells, and MeV engages this receptor to infect the host immune system83. Nectin-4 is expressed on the basal surface of the epithelium cells, allowing MeV to be spread from macrophages to epithelium and then into the lung lumen.81,82 The neurotropic Henipavirus G engages Ephrin B2 and B3 on cell surfaces6,46,47,84; these molecules are expressed in neurons in the brain. Ephrin B2 and B3 are also found in other cell types, and are conserved among many species, allowing Henipaviruses to infect a range of species including humans, pigs, horses and bats. Henipaviruses can spread within the host by binding lymphocytes and using them as transporters85. The G protein of Pneumovirinae binds heparan sulfate proteoglycans86–89. RSV-G has been shown to interact with the chemokine receptor CXC3CR1, through a CX3C motif90. While it is unlikely that this interaction would promote fusion, this interaction strongly inhibits the host immune response91. The diversity in receptor usage confers paramyxoviruses the ability to adapt, gain access and infect new tissues and new hosts.
3.2 The stalk domain the receptor-binding protein interacts with and activates F
HN/H/G is the driving force for fusion initiation and then for sustaining F’s role in mediating viral entry40 (reviewed in Refs.1,92). Under a variety of in vitro experimental situations, F can fuse alone40,58,93,94, or a “headless” HN/H/G may be sufficient to mediate F activation41,94,99 ; see specific examples below. However, as discussed in section 3.6 the function of specific residues in the globular head of HN is essential for infection in the host, and any subtle change at the dimer interface of the globular domain can affect HN dimer association, impact the HN/F fusion machinery, and markedly alter host infection.
The globular heads of the HN/H/G proteins bind the cellular receptor. The stalk domains of HN/H/G proteins are responsible for specific interaction with the homologous F proteins and are critical for F activation once they receive the signal from the receptor bound globular head27,37,42,49–51,53,95–98. After initial identification of the importance of the stalk of the receptor binding protein for activating F, this stalk function has been assessed using a variety of approaches including the use of the “headless” receptor binding proteins mentioned above. A construct consisting of the PIV5 HN stalk domain (residues 1–117) lacking the globular binding domain was sufficient to activate F. This activation seemed to be specific; the PIV5 HN stalk could not activate heterotypic Fs, and required direct interaction with F94. This set of experiments was used to postulate that for PIV5 HN, activation of F requires that the stalk domain be “freed”. Receptor engagement would drive the movement of the heads that would free the stalk. Similar experiments have been performed using different “headless” stalks proteins with varied results. However, only very specific MeV H, NiV G, and PIV5 stalk lengths can activate the F protein94,96,99 and, for MeV H, the stalk must be partially stabilized in order to be functional96. Only one out of several different headless stalk constructs of mumps HN100, NDV HN100, and NiV G99 can activate F suggesting that the specific sequence of the receptor binding protein stalk and the F protein are crucial for this activity. For HPIV3, a headless HN does not seem to be capable of activating HPIV3 F. Thus how the stalk domain of paramyxovirus HN/H/G activates F remains to be further characterized, and as described below, we contend that the interaction of the globular head of the receptor binding protein with its receptor provides a critical signal to the stalk in the process of F activation.
Chimeric proteins bearing the globular domain from NDV and the stalk domain of either HPIV3-HN, NiV-G, or MeV-H revealed that receptor engagement by the NDV-HN globular head is sufficient for transmitting the activating signal through the stalk domain of these other paramyxoviruses and trigger the homologous fusion protein58,101. These chimeric receptor-binding proteins are only capable of triggering an F protein that is homologous to the stalk domain of the chimeric protein. Thus a chimeric protein with an NDV-HN globular head and an HPIV3-HN stalk can only activate HPIV3-F58. The only exceptions are Henipaviruses NiV-G and HeV-G whose stalks demonstrate enough sequence similarity to activate both F proteins102. Closely related Henipaviruses, such as the recently discovered Cedar virus6, may share the same property. The chimeric receptor-binding proteins reveal one of the ways in which HN, H, and G protein function is conserved at least amongst the Paramyxovirinae sub-family, and support the hypothesis of a unified model for the paramyxovirus fusion machinery in which the globular head domain of the receptor-binding protein acts as a receiving unit that is independent of the rest of the protein. The receptor-binding protein engages its receptor and transmits a signal to the stalk domain. The stalk domain likely undergoes conformational changes allowing it to activate its homologous fusion protein50,103 (reviewed in Ref.39).
3.3 The role of the receptor-binding protein before receptor engagement
Several distinct models describing the interaction between the HN/H/G protein and the F protein have been proposed (reviewed in Refs. 1,3). One model, the dissociation or clamp model, postulates that the HN/H/G and F proteins interact prior to receptor engagement and that receptor engagement abrogates this interaction. Another model, the association or provocateur model, suggests that HN/H/G only interact with the homologous F protein following receptor engagement. Recent studies from our group have uncovered elements in support of a unified model among paramyxoviruses41,104. We used a Bimolecular Fluorescence Complementation (BiFC) strategy where HPIV3 HN and HPIV3 F were respectively fused with the N-terminus of YFP and C- terminus of CFP. Only if HN and F proteins interact, the fluorescent protein is reconstitiuted and fluorescence is emitted upon excitation. We observed that HPIV3 HN interacts with F in the absence of receptor engagement. Upon receptor engagement, HN and F continue to interact and cluster at the point where the fusion pore will form41. Whether clustering occurs before or after activation of the F protein has not been firmly established, but recent data are more consistent with clustering occurring first, and F activation occurring in the cluster (unpublished). HN and F continue to interact throughout the fusion process40 and dissociate only once fusion is complete (unpublished).
For HPIV3, it appears that non-receptor engaged HN protein stabilizes F, maintaining it in the pre-fusion state. When HPIV3 F alone is exposed to high temperatures, it enters the post fusion state (as assessed by acquisition of sensitivity to proteinase K digestion); however, in the presence of non-receptor engaged HN, F remains in its pre-fusion state, resistant to proteinase digestion104. These data support the idea that HPIV3 HN serves a “protective” role for the fusion protein104. Prior to receptor engagement the receptor-binding protein stabilizes HPIV3 F and prevents it from premature activation. Ader et al.103 recently showed that the F protein of some morbilliviruses is highly stable and suggested that it is unlikely that the H stabilizes F, however it cannot be excluded that in vivo, stabilization may be required since many parameters could prematurely trigger fusion.
An intriguing role of pH in the NDV cell entry process has recently emerged105. NDV entry is reduced when caveolin-associated traffic is inhibited. Cholesterol seems to be important in the process since the drug methyl-β-cyclodextrin, which inhibits cholesterol trafficking, also diminishes NDV-HN binding. Moreover, NDV particles were shown to colocalize with EEA1, a marker of early endosome formation suggesting that NDV could enter the cell through caveolin-mediated endocytosis. Past work showed that HMPV, NiV, and RSV can use the endosomal pathway to enter cells106–108. Low pH exposure increases NDV fusion and subsequent syncytia formation while, reciprocally, fusion decreases in the presence of pH-acidification inhibitors109. Consistent with these results, the stability of NDV F (assessed through fusion assays) decreases after exposure to low pH; F is more easily activated under these conditions109. There may be an accessory pH-dependent pathway through the caveolin-mediated endocytosis for NDV.
For some paramyxoviruses, the F protein can mediate fusion in the absence of the receptor-binding protein, in some cases permitting viral infection110–116. However very few viruses are infectious when lacking their receptor-binding protein. Infectious virions that lack a receptor-binding protein have been studied for RSV, HMPV, and laboratory adapted strains of NDV, however in these cases, infectivity is enhanced when the receptor-binding protein is present89,111,117,118. In the case of RSV, the F protein itself can bind nucleolin119, the HMPV F can bind integrins120, and both can engage heparan sulfate121–123, potentially permitting G-independent entry. It will be important to determine whether in these cases the receptor binding protein serves a role in vivo in stabilization of F prior to receptor engagement; an F-stabilizing role for HN/H/G protein may be a conserved feature at least among the Paramyxovirinae sub-family.
3.4 The receptor-binding protein transmits a triggering signal to the fusion protein upon receptor engagement
Upon receptor engagement, the HN/H/G protein activates the F protein to undergo its final fusion-readiness structural changes. The mechanism whereby this activation occurs – where the signal for activation originates, and how it is transmitted from the receptor binding protein to the fusion protein – is a topic of significance. The stalk domain of HN/H/G protein is critical to this activation process51,95–97,100,124,125. Recent studies from our group indicate that for NiV the domain that connects the globular head to the stalk domain is required for transmission of the triggering signal to F124. Chimeric receptor-binding proteins containing the globular heads of NDV and the stalk domain of NiV can efficiently activate NiV-F only if specific residues are present in the head-stalk junction124.
A proposed model deriving from PIV5 postulated that the heads of HN/H/G could change their position upon receptor engagement, shifting from a “heads-down” conformation, in which the stalk domain is masked, to a “heads-up” conformation exposing the stalk domain. A masked stalk domain would prevent HN/H/G and F interaction, and upon receptor engagement the heads would move aside, allowing the stalk to activate the F protein42,48,53,94. However, recent analyses of the MV fusion process do not support this model, suggesting that at least this mechanism may not apply to other paramyxoviruses (reviewed in Ref. 39). MeV H and MeV F interact prior to receptor engagement126, indicating that an H head must be up to allow H/F interaction during transit to the cell membrane. However F is not triggered prematurely (i.e., before receptor engagement during infection), suggesting that for MeV the exposure of the stalk is not the crucial requirement for F activation. It has been suggested that MeV receptor engagement with a “pulling” of the H molecule could induce a conformational change in the stalk that triggers F27. Brindley et al. noted that MeV H proteins with truncated globular heads promoted fusion in vitro96, implying that the globular heads may mask a portion of the stalk domain responsible for activating the F protein, consistent with a “heads-down, heads-up” model. However, the truncated H proteins that were studied are highly specific, with truncation at precise sites on H being required for fusion complementation. Headless Hs, to promote F-mediated fusion, require stabilization at the C-terminus, e.g. by a yeast-derived GCN4 motif96. Thus it seems likely that the specific truncated H proteins do not represent a general mechanism, except when they are stabilized or otherwise modified to adopt a structure similar to the receptor-engaged conformation that activates F. Finally, key residues on the MeV H that are involved in F protein activation are located at the membrane-proximal part of the stalk domain, where the globular heads of MeV H do not reach even when the protein is in a “heads-down” conformation51,95,100,124.
How does receptor engagement affect the receptor binding protein and modulate the F activation step? The first studies to analyze how receptor engagement modifies the receptor binding protein structure showed that receptor engagement does not appear to affect the monomer-monomer interface127. Stabilization of NDV HN monomers by disulfide bridges restricted monomer movement but enhanced HN’s fusion promotion activity, suggesting that large movements between monomers are not needed for successful fusion promotion. Crystallographic data of MeV H bound to SLAM receptors revealed H present in two tetrameric conformations. In the pre-F triggering conformation H is in a planar form and upon receptor engagement, a “sliding” movement between the dimers occurs that gives rise to the post F-triggering form45. One recently proposed model suggests that upon receptor engagement, after exposure of the stalk of the receptor binding protein, a specific conformational change occurs in the complex formed by HN/H/G and F, to reach an “induced-fit” state that leads to F activation100. In this case, the nature of the signal activating F is a modulation of its interaction with HN/H/G rather than a loss or a gain of interaction (except perhaps for PIV5).
Even after F activation and during the fusion process itself, the receptor-binding protein continues to regulate the fusion process. For HPIV3, the presence of HN is crucial until merger of the viral and cellular membranes40. Using a mutant HN that lacks neuraminidase activity and thus constitutively engages its sialic acid receptor, we specifically disrupted receptor engagement at precise times using the small molecule zanamivir. Only when HN continuously engages its receptor can F proceed through the fusion process. Even after insertion of the fusion peptide into the target membrane, F still requires the activating signal from HN/H/G to complete fusion40.
3.5 The fusion protein inserts its hydrophobic fusion peptide into the target membrane leading to the formation of the fusion pore
After activation, F undergoes several conformational changes (Fig. 4). Fusion can be prevented by introducing disulfide bridges to stabilize F128. F acts as an indivisible unit. The cytoplasmic tail of F protein must bear conserved Ser/Thr residues for fusion to properly occur, and the cytoplasmic tail is likely to be virus-specific; it cannot be interchanged with that of another virus129. The current model for F action is based on several available F protein structures (Fig. 4), and predicts that F inserts its fusion peptide into the target membrane by extending its α-helical domains. The amphipathic HRC and HRN domain then interact with each other, driving fusion. Supporting this hypothesis, Donald et al found that the α-helical domains on PIV5-F interact avidly with each other130. The model postulates that once the fusion peptide is inserted, the F protein is in a pre-hairpin intermediate state (Fig. 5). This transient intermediate was recently observed for PIV571. Kim et al. used nanobeads bearing a lipid bilayer as targets for viral particles, and HRC-derived peptides that interact with the HRN domain to prevent F from refolding after fusion peptide insertion. They then measured the distance between the viral bilayer and the nanobead bilayer using EM, and compared it to in silico predictions based on the hypothesis of the pre-hairpin fusion intermediate. The experimental results fit the computational model, supporting the existence of the intermediate state69. F proteins blocked in the transient state were identified by electronic microscopy by gold coupling of HRC-derived peptides, and were observed to be present in clusters. This observation is consistent with our model for HPIV3, where receptor-binding and fusion protein clustering is required in order for fusion to occur41. Attaining this unstable fusion intermediate is likely to be the event that is prevented by the non-receptor-engaged HN/H/G protein104.
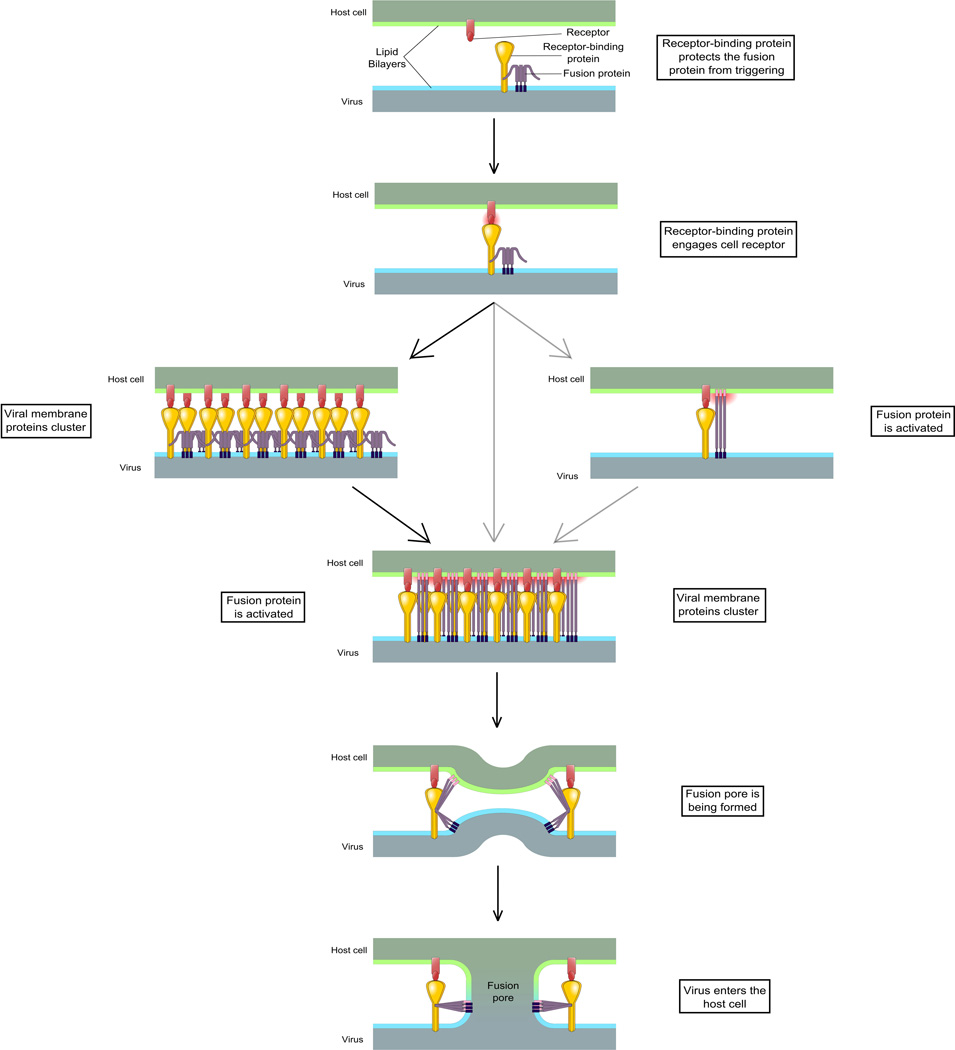
Figure 5. Unified model for Paramyxoviridae fusion process.
See text for description. Pink: HRN domain and fusion peptide. HRC/HRN: Heptad Repeat C-/N-terminal domain.
After the unstable intermediate state, F adopts a hairpin intermediate state (Fig. 5). In this form, the two heptad repeat regions (HRN and HRC) interact with each other, leading to a 6 Helix-Bundle conformation, which mechanically forces the two membranes together (Fig. 5). In this process, the length of the HRC-linking region separating the HRC domain from the membrane (Fig. 4) is crucial. Linkers that are too short or too long were associated with fusion-incompetent F proteins, as described for NDV75. This is consistent with the hypothesis that the HRC-linker contributes to the stabilization of the HRC domain and thus can modulate its function, as previously described for HeV-F63.
A critical element of this refolding is that the HRN and HRC domains form a close interaction. Inhibitors of this interaction an attractive strategy for blocking fusion and viral entry, and peptides derived from the HRC domain of paramyxovirus F molecules have recently been shown to inhibit the fusion process in vivo131,132. The peptide inhibitors are far more efficient when conjugated with lipids. This conjugation allows for peptide insertion into target cell membranes, placing the anti-fusion peptides in close proximity to the viral glycoproteins. These conjugated peptides can reach the brain and may be efficient against neurotropic viruses such as NiV, or neurotropic variants of MeV131–133.
At the end of the F refolding process, F is in its stable post-fusion state. Experimentally expressed, soluble F protein naturally adopts this post-fusion state73,134. The stability of this final state implies that the fusion process is irreversible, and supports the importance of the “protective role”104 of the receptor binding protein in preventing premature initiation of the activation process. Indeed, virions whose F proteins have been prematurely activated cannot fuse and are non-infectious135,136.
3.6 The interaction between HN/H/G and F modulates infection in the natural host
A series of recent experiments showed that the communication between the HPIV3 HN and F directly impacts infection in the natural host101,135. An HPIV3 virus bearing HN molecules that trigger F rapidly or interact with F avidly are effective at fusing in cell culture, but fare poorly in natural tissues or in vivo26,41,61,135. Enhanced F-triggering by HN, while advantageous in vitro, results in non-infectious viral particles in airway epithelium137, in which the F protein may have been prematurely activated before contacting target cells135,136. Recent structural analysis of the HN molecules with specific biological effects of revealed properties that are critical for infection in vivo. The second sialic acid binding site that was identified by functional assays as a key operative site on HPIV3 HN (site II)61 has a clear structural correlate and modulates viral growth in vivo101. Specific structural changes at the HN dimer interface, where site II exists, modulate the interaction between HN and F, impact fusion triggering, and directly impact viral infection. For example, the HN from a virus that is well adapted to growth in lung tissue has a wider separation across the dimer interface near the regions of site II than a variant that is restricted in growth in the lung. These conformational changes are propagated to nearby loops at the dimer interface and reduce dimer association in the lung-adapted virus’s HN, as measured by buried interface areas, indicating that the dimer interface (and its modulation of HN/F interaction) is critical to infection in the host101. The viruses that grow well in lung tissue and in vivo bear a less active HN/F fusion machinery, with lower receptor avidity and less efficient fusion triggering137. One may speculate that during natural infection in the host, receptors may be widely available, and an overly fusogenic virus may prematurely trigger its F before reaching the target membrane, a notion consistent with the evolution of laboratory-adapted HPIV3 variants in vivo to become less fusogenic than reference strains137. In support of this concept, several highly fusogenic glycosylation mutants from NiV have not been identified in vivo77. It seems apparent that most paramyxoviruses require a specific balance between the various properties of the fusion machinery for viability in vivo.
Conclusions
A variety of models that reflect the diversity of the paramyxoviruses have been proposed for the steps in entry. However, recent data may be taken together to support a common fusion mechanism. Reviewing recent advances in the field of Paramyxoviridae entry, we propose a unified model for the fusion process (Fig. 5). Before receptor engagement, the receptor-binding protein interacts with the fusion protein and, if necessary, prevents its untimely activation. After the globular heads of the receptor binding protein engage the target receptor, an activating signal is transmitted via the stalk domain to the fusion protein. This signal appears likely to be in the form of a conformational change in the stalk domain and -- in an “induced-fit” model -- induces a structural change in the fusion protein. The fusion protein is destabilized and inserts its hydrophobic fusion peptide into the target membrane. The two heptad repeat domains of the fusion protein interact with each other as the molecule progresses to its stable post-fusion state. This process drives the formation of the fusion pore (Fig. 5).
Many open questions remain about the process of fusion activation. While the importance of the HN/H/G stalk domain is well-established42,95,96 the mechanism by which the signal is transmitted to the homologous F protein is just beginning to be understood. Structural analysis of the complete HN/H/G molecule both engaged to its receptor and free, has been challenging in the face of the important hydrophobic domains42,48,52,53. Measuring the precise structural changes that trigger F protein activation will require novel approaches including cryo-electron microscopic analysis of specific stages in the fusion activation process.
Understanding the fusion and entry processs of paramyxoviruses is key for the design of new therapies. The unifying model we suggest for the Paramyxoviridae fusion machinery can support the development of new anti-viral strategies that may have broad-spectrum potential for inhibiting entry. Potentially promising therapies that target the entry step include small molecules that prematurely trigger the fusion protein136, or lipid conjugated HRC-derived peptides131,132. Together with other antiviral approaches that target common mechanisms, these strategies hold promise for wide applicability for preventing and treating this important group of pathogens.
Acknowledgments
We are grateful to Ashton Kutcher for his support for microscopy, to Dan and Nancy Paduano for their essential support of innovative research projects, and to the Friedman Family Foundation for our laboratories at Weill Cornell Medical College. MP is a Friedman Family Research Scholar in Pediatric Infectious Diseases. The work was supported by NIH R01 AI31971 and NIH 3R01 AI031971-19S1 to AM, NIH Region II Center of Excellence for Bio-Defense and Emerging Infectious Disease Research U54AI057158 Research Grant to AM, NIH R21 AI100292, NIH R21 EB011707, and NIH R21 NS073781 to MP.
References
- 1.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4(4):613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enders G. Paramyxoviruses. In: Baron S, editor. Medical Microbiology. 4th ed. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. [Accessed April 11, 2014]. Available at: http://www.ncbi.nlm.nih.gov/books/NBK8461/ [Google Scholar]
- 3.Bossart KN, Fusco DL, Broder CC. Paramyxovirus entry. Adv Exp Med Biol. 2013;790:95–127. doi: 10.1007/978-1-4614-7651-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falk K, Batts WN, Kvellestad A, Kurath G, Wiik-Nielsen J, Winton JR. Molecular characterisation of Atlantic salmon paramyxovirus (ASPV): a novel paramyxovirus associated with proliferative gill inflammation. Virus Res. 2008;133(2):218–227. doi: 10.1016/j.virusres.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Kurath G, Batts WN, Ahne W, Winton JR. Complete genome sequence of Fer-de-Lance virus reveals a novel gene in reptilian paramyxoviruses. J Virol. 2004;78(4):2045–2056. doi: 10.1128/JVI.78.4.2045-2056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh GA, de Jong C, Barr JA, et al. Cedar virus: a novel Henipavirus isolated from Australian bats. Plos Pathog. 2012;8(8):e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magoffin DE, Mackenzie JS, Wang L-F. Genetic analysis of J-virus and Beilong virus using minireplicons. Virology. 2007;364(1):103–111. doi: 10.1016/j.virol.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 8.Kurth A, Kohl C, Brinkmann A, et al. Novel paramyxoviruses in free-ranging European bats. Plos One. 2012;7(6):e38688. doi: 10.1371/journal.pone.0038688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg GA, Hall CB, Iwane MK, et al. Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. J Pediatr. 2009;154(5):694–699. doi: 10.1016/j.jpeds.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 10.Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JV, Tollefson SJ, Heymann PW, Carper HT, Patrie J, Crowe JE. Human metapneumovirus infection in children hospitalized for wheezing. J Allergy Clin Immunol. 2005;115(6):1311–1312. doi: 10.1016/j.jaci.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganar K, Das M, Sinha S, Kumar S. Newcastle disease virus: Current status and our understanding. Virus Res. 2014;184C:71–81. doi: 10.1016/j.virusres.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croser EL, Marsh GA. The changing face of the henipaviruses. Vet Microbiol. 2013 doi: 10.1016/j.vetmic.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Clayton BA, Wang LF, Marsh GA. Henipaviruses: an updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Public Heal. 2013;60(1):69–83. doi: 10.1111/j.1863-2378.2012.01501.x. [DOI] [PubMed] [Google Scholar]
- 15.Hausmann S, Jacques JP, Kolakofsky D. Paramyxovirus RNA editing and the requirement for hexamer genome length. Rna New York N. 1996;2(10):1033–1045. [PMC free article] [PubMed] [Google Scholar]
- 16.Halpin K, Bankamp B, Harcourt BH, Bellini WJ, Rota PA. Nipah virus conforms to the rule of six in a minigenome replication assay. J Gen Virol. 2004;85(Pt 3):701–707. doi: 10.1099/vir.0.19685-0. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology. 2000;273(1):210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi K, Tanabayashi K, Hishiyama M, Yamada A. The mumps virus SH protein is a membrane protein and not essential for virus growth. Virology. 1996;225(1):156–162. doi: 10.1006/viro.1996.0583. [DOI] [PubMed] [Google Scholar]
- 19.He B, Leser GP, Paterson RG, Lamb RA. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250(1):30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 20.Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J Virol. 2005;79(19):12608–12613. doi: 10.1128/JVI.79.19.12608-12613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atreya PL, Peeples ME, Collins PL. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72(2):1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Audsley MD, Moseley GW. Paramyxovirus evasion of innate immunity: Diverse strategies for common targets. World J Virol. 2013;2(2):57–70. doi: 10.5501/wjv.v2.i2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huberman K, Peluso RW, Moscona A. Hemagglutinin-neuraminidase of human parainfluenza 3: role of the neuraminidase in the viral life cycle. Virology. 1995;214(1):294–300. doi: 10.1006/viro.1995.9925. [DOI] [PubMed] [Google Scholar]
- 24.Moscona A, Peluso RW. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J Virol. 1991;65(6):2773–2777. doi: 10.1128/jvi.65.6.2773-2777.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison T, McQuain C, McGinnes L. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: requirements for membrane fusion. J Virol. 1991;65(2):813–822. doi: 10.1128/jvi.65.2.813-822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moscona A, Peluso RW. Relative affinity of the human parainfluenza virus type 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J Virol. 1993;67(11):6463–6468. doi: 10.1128/jvi.67.11.6463-6468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navaratnarajah CK, Oezguen N, Rupp L, et al. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol. 2011;18(2):128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirza AM, Aguilar HC, Zhu Q, et al. Triggering of the newcastle disease virus fusion protein by a chimeric attachment protein that binds to Nipah virus receptors. J Biol Chem. 2011;286(20):17851–17860. doi: 10.1074/jbc.M111.233965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81(18):9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguilar HC, Matreyek KA, Filone CM, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80(10):4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishop KA, Hickey AC, Khetawat D, et al. Residues in the stalk domain of the hendra virus g glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82(22):11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol. 2002;76(10):5051–5061. doi: 10.1128/JVI.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iorio RM, Melanson VR, Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 2009;4(4):335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee B, Ataman ZA. Modes of paramyxovirus fusion: a Henipavirus perspective. Trends Microbiol. 2011;19(8):389–399. doi: 10.1016/j.tim.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorio RM, Mahon PJ. Paramyxoviruses: different receptors - different mechanisms of fusion. Trends Microbiol. 2008;16(4):135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porotto M, Murrell M, Greengard O, Doctor L, Moscona A. Influence of the human parainfluenza virus 3 attachment protein’s neuraminidase activity on its capacity to activate the fusion protein. J Virol. 2005;79(4):2383–2392. doi: 10.1128/JVI.79.4.2383-2392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porotto M, Murrell M, Greengard O, Moscona A. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J Virol. 2003;77(6):3647–3654. doi: 10.1128/JVI.77.6.3647-3654.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dutch RE. Entry and fusion of emerging paramyxoviruses. Plos Pathog. 2010;6(6):e1000881. doi: 10.1371/journal.ppat.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plattet P, Plemper RK. Envelope protein dynamics in paramyxovirus entry. MBio. 2013;4(4) doi: 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porotto M, Devito I, Palmer SG, et al. Spring-loaded model revisited: paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J Virol. 2011;85(24):12867–12880. doi: 10.1128/JVI.05873-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porotto M, Palmer SG, Palermo LM, Moscona A. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J Biol Chem. 2012;287(1):778–793. doi: 10.1074/jbc.M111.298059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 2011;85(24):12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14(12):1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 44.Hashiguchi T, Kajikawa M, Maita N, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A. 2007;104(49):19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashiguchi T, Ose T, Kubota M, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18(2):135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 46.Bowden TA, Aricescu AR, Gilbert RJC, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15(6):567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 47.Xu K, Rajashankar KR, Chan Y-P, Himanen JP, Broder CC, Nikolov DB. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A. 2008;105(29):9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. Plos Pathog. 2013;9(8):e1003534. doi: 10.1371/journal.ppat.1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence MC, Borg NA, Streltsov VA, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335(5):1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 50.Navaratnarajah CK, Negi S, Braun W, Cattaneo R. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J Biol Chem. 2012;287(46):38543–38551. doi: 10.1074/jbc.M112.410563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navaratnarajah CK, Kumar S, Generous A, Apte-Sengupta S, Mateo M, Cattaneo R. The measles virus hemagglutinin stalk: structure and function of the central fusion-activation and membrane-proximal segments. J Virol. 2014 doi: 10.1128/JVI.02846-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowden TA, Crispin M, Harvey DJ, Jones EY, Stuart DI. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84(12):6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A. 2011;108(36):14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7(11):1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 55.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17(1):124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 56.Zaitsev V, von Itzstein M, Groves D, et al. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J Virol. 2004;78(7):3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bousse TL, Taylor G, Krishnamurthy S, Portner A, Samal SK, Takimoto T. Biological significance of the second receptor binding site of Newcastle disease virus hemagglutinin-neuraminidase protein. J Virol. 2004;78(23):13351–13355. doi: 10.1128/JVI.78.23.13351-13355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porotto M, Salah Z, DeVito I, et al. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J Virol. 2012;86(10):5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bousse T, Takimoto T. Mutation at residue 523 creates a second receptor binding site on human parainfluenza virus type 1 hemagglutinin-neuraminidase protein. J Virol. 2006;80(18):9009–9016. doi: 10.1128/JVI.00969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alymova IV, Portner A, Mishin VP, McCullers JA, Freiden P, Taylor GL. Receptor-binding specificity of the human parainfluenza virus type 1 hemagglutinin-neuraminidase glycoprotein. Glycobiology. 2012;22(2):174–180. doi: 10.1093/glycob/cwr112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porotto M, Fornabaio M, Kellogg GE, Moscona A. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J Virol. 2007;81(7):3216–3228. doi: 10.1128/JVI.02617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith EC, Smith SE, Carter JR, et al. Trimeric transmembrane domain interactions in paramyxovirus fusion proteins: roles in protein folding, stability, and function. J Biol Chem. 2013;288(50):35726–35735. doi: 10.1074/jbc.M113.514554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith EC, Culler MR, Hellman LM, Fried MG, Creamer TP, Dutch RE. Beyond anchoring: the expanding role of the hendra virus fusion protein transmembrane domain in protein folding, stability, and function. J Virol. 2012;86(6):3003–3013. doi: 10.1128/JVI.05762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.González-Reyes L, Ruiz-Argüello MB, García-Barreno B, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98(17):9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diederich S, Moll M, Klenk H-D, Maisner A. The nipah virus fusion protein is cleaved within the endosomal compartment. J Biol Chem. 2005;280(33):29899–29903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- 66.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79(20):12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pager CT, Craft WW, Jr, Patch J, Dutch RE. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346(2):251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diederich S, Sauerhering L, Weis M, et al. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J Virol. 2012;86(7):3736–3745. doi: 10.1128/JVI.06628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith EC, Gregory SM, Tamm LK, Creamer TP, Dutch RE. Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion. J Biol Chem. 2012;287(35):30035–30048. doi: 10.1074/jbc.M112.367862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012;109(41):16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim YH, Donald JE, Grigoryan G, et al. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc Natl Acad Sci U S A. 2011;108(52):20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin H-S, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin H-S, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102(26):9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85(15):7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swanson K, Wen X, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402(2):372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biering SB, Huang A, Vu AT, et al. N-Glycans on the Nipah virus attachment glycoprotein modulate fusion and viral entry as they protect against antibody neutralization. J Virol. 2012;86(22):11991–12002. doi: 10.1128/JVI.01304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dörig RE, Marcil A, Chopra A, Richardson CD. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75(2):295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 79.Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406(6798):893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 80.Erlenhoefer C, Wurzer WJ, Löffler S, Schneider-Schaulies S, ter Meulen V, Schneider-Schaulies J. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J Virol. 2001;75(10):4499–4505. doi: 10.1128/JVI.75.10.4499-4505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noyce RS, Bondre DG, Ha MN, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. Plos Pathog. 2011;7(8):e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mühlebach MD, Mateo M, Sinn PL, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480(7378):530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kato S-I, Nagata K, Takeuchi K. Cell tropism and pathogenesis of measles virus in monkeys. Front Microbiol. 2012;3:14. doi: 10.3389/fmicb.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonaparte MI, Dimitrov AS, Bossart KN, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102(30):10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mathieu C, Pohl C, Szecsi J, et al. Nipah virus uses leukocytes for efficient dissemination within a host. J Virol. 2011;85(15):7863–7871. doi: 10.1128/JVI.00549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krusat T, Streckert HJ. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142(6):1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- 87.Bourgeois C, Bour JB, Lidholt K, Gauthray C, Pothier P. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J Virol. 1998;72(9):7221–7227. doi: 10.1128/jvi.72.9.7221-7227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feldman SA, Hendry RM, Beeler JA. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol. 1999;73(8):6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thammawat S, Sadlon TA, Hallsworth PG, Gordon DL. Role of cellular glycosaminoglycans and charged regions of viral G protein in human metapneumovirus infection. J Virol. 2008;82(23):11767–11774. doi: 10.1128/JVI.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harcourt JL, Karron RA, Tripp RA. Anti-G protein antibody responses to respiratory syncytial virus infection or vaccination are associated with inhibition of G protein CX3C-CX3CR1 binding and leukocyte chemotaxis. J Infect Dis. 2004;190(11):1936–1940. doi: 10.1086/425516. [DOI] [PubMed] [Google Scholar]
- 91.Chirkova T, Boyoglu-Barnum S, Gaston KA, et al. Respiratory syncytial virus G protein CX3C motif impairs human airway epithelial and immune cell responses. J Virol. 2013;87(24):13466–13479. doi: 10.1128/JVI.01741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steffen DL, Xu K, Nikolov DB, Broder CC. Henipavirus mediated membrane fusion, virus entry and targeted therapeutics. Viruses. 2012;4(2):280–308. doi: 10.3390/v4020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. Embo J. 2001;20(15):4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc Natl Acad Sci U S A. 2012;109(39):E2625–E2634. doi: 10.1073/pnas.1213813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ader N, Brindley MA, Avila M, et al. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem. 2012;287(20):16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. A Stabilized Headless Measles Virus Attachment Protein Stalk Efficiently Triggers Membrane Fusion. J Virol. 2013 doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Talekar A, Moscona A, Porotto M. Measles fusion machinery activated by sialic acid binding globular domain. J Virol. 2013 doi: 10.1128/JVI.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78(23):13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Q, Stone JA, Bradel-Tretheway B, et al. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. Plos Pathog. 2013;9(11):e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bose S, Song AS, Jardetzky TS, Lamb RA. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol. 2014 doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu R, Palmer SG, Porotto M, et al. Interaction between the Hemagglutinin-Neuraminidase and Fusion Glycoproteins of Human Parainfluenza Virus Type III Regulates Viral Growth In Vivo. MBio. 2013;4(5) doi: 10.1128/mBio.00803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bossart KN, Wang L-F, Flora MN, et al. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J Virol. 2002;76(22):11186–11198. doi: 10.1128/JVI.76.22.11186-11198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ader N, Brindley M, Avila M, et al. Mechanism for active membrane fusion triggering by morbillivirus attachment protein. J Virol. 2013;87(1):314–326. doi: 10.1128/JVI.01826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Porotto M, Salah ZW, Gui L, et al. Regulation of paramyxovirus fusion activation: the hemagglutinin-neuraminidase protein stabilizes the fusion protein in a pretriggered state. J Virol. 2012;86(23):12838–12848. doi: 10.1128/JVI.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cantín C, Holguera J, Ferreira L, Villar E, Muñoz-Barroso I. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. J Gen Virol. 2007;88(Pt 2):559–569. doi: 10.1099/vir.0.82150-0. [DOI] [PubMed] [Google Scholar]
- 106.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J Virol. 2009;83(3):1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kolokoltsov AA, Deniger D, Fleming EH, Roberts NJ, Jr, Karpilow JM, Davey RA. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J Virol. 2007;81(14):7786–7800. doi: 10.1128/JVI.02780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Diederich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375(2):391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sánchez-Felipe L, Villar E, Muñoz-Barroso I. Entry of Newcastle Disease Virus into the host cell: Role of acidic pH and endocytosis. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamem.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kahn JS, Schnell MJ, Buonocore L, Rose JK. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology. 1999;254(1):81–91. doi: 10.1006/viro.1998.9535. [DOI] [PubMed] [Google Scholar]
- 111.Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001;75(15):6825–6834. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schowalter RM, Smith SE, Dutch RE. Characterization of Human Metapneumovirus F Protein-Promoted Membrane Fusion: Critical Roles for Proteolytic Processing and Low pH. J Virol. 2006;80(22):10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sergel TA, McGinnes LW, Morrison TG. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol. 2000;74(11):5101–5107. doi: 10.1128/jvi.74.11.5101-5107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seth S, Vincent A, Compans RW. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J Virol. 2003;77(1):167–178. doi: 10.1128/JVI.77.1.167-178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ito M, Nishio M, Kawano M, Komada H, Ito Y, Tsurudome M. Effects of multiple amino acids of the parainfluenza virus 5 fusion protein on its haemagglutinin-neuraminidase-independent fusion activity. J Gen Virol. 2009;90(Pt 2):405–413. doi: 10.1099/vir.0.006437-0. [DOI] [PubMed] [Google Scholar]
- 116.Karron RA, Buonagurio DA, Georgiu AF, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997;94(25):13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dutch RE, Joshi SB, Lamb RA. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, influenza virus HA. J Virol. 1998;72(10):7745–7753. doi: 10.1128/jvi.72.10.7745-7753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Herfst S, Mas V, Ver LS, et al. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J Virol. 2008;82(17):8891–8895. doi: 10.1128/JVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17(9):1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 120.Wei Y, Zhang Y, Cai H, et al. Roles of the putative integrin-binding motif of the human metapneumovirus fusion (f) protein in cell-cell fusion, viral infectivity, and pathogenesis. J Virol. 2014;88(8):4338–4352. doi: 10.1128/JVI.03491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74(14):6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barretto N, Hallak LK, Peeples ME. Neuraminidase treatment of respiratory syncytial virus-infected cells or virions, but not target cells, enhances cell-cell fusion and infection. Virology. 2003;313(1):33–43. doi: 10.1016/s0042-6822(03)00288-5. [DOI] [PubMed] [Google Scholar]
- 123.Chang A, Masante C, Buchholz UJ, Dutch RE. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol. 2012;86(6):3230–3243. doi: 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Talekar A, DeVito I, Salah Z, et al. Identification of a region in the stalk domain of the nipah virus receptor binding protein that is critical for fusion activation. J Virol. 2013;87(20):10980–10996. doi: 10.1128/JVI.01646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bose S, Heath CM, Shah PA, Alayyoubi M, Jardetzky TS, Lamb RA. Mutations in the parainfluenza virus 5 fusion (F) protein reveal domains important for fusion triggering and metastability. J Virol. 2013 doi: 10.1128/JVI.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brindley MA, Takeda M, Plattet P, Plemper RK. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci U S A. 2012;109(44):E3018–E3027. doi: 10.1073/pnas.1210925109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mahon PJ, Mirza AM, Musich TA, Iorio RM. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J Virol. 2008;82(21):10386–10396. doi: 10.1128/JVI.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zokarkar A, Connolly SA, Jardetzky TS, Lamb RA. Reversible inhibition of fusion activity of a paramyxovirus fusion protein by an engineered disulfide bond in the membrane-proximal external region. J Virol. 2012;86(22):12397–12401. doi: 10.1128/JVI.02006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zokarkar A, Lamb RA. The paramyxovirus fusion protein C-terminal region: mutagenesis indicates an indivisible protein unit. J Virol. 2012;86(5):2600–2609. doi: 10.1128/JVI.06546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Donald JE, Zhang Y, Fiorin G, et al. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc Natl Acad Sci U S A. 2011;108(10):3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Porotto M, Rockx B, Yokoyama CC, et al. Inhibition of Nipah virus infection in vivo: targeting an early stage of paramyxovirus fusion activation during viral entry. Plos Pathog. 2010;6(10):e1001168. doi: 10.1371/journal.ppat.1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Welsch JC, Talekar A, Mathieu C, et al. Fatal Measles Infection Prevented by Brain-Penetrant Fusion Inhibitors. J Virol. 2013 doi: 10.1128/JVI.02436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pessi A, Langella A, Capitò E, et al. A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. Plos One. 2012;7(5):e36833. doi: 10.1371/journal.pone.0036833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen L, Gorman JJ, McKimm-Breschkin J, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Struct Lond Engl 1993. 2001;9(3):255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 135.Palermo LM, Porotto M, Yokoyama CC, et al. Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J Virol. 2009;83(13):6900–6908. doi: 10.1128/JVI.00475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Farzan SF, Palermo LM, Yokoyama CC, et al. Premature activation of the paramyxovirus fusion protein before target cell attachment with corruption of the viral fusion machinery. J Biol Chem. 2011;286(44):37945–37954. doi: 10.1074/jbc.M111.256248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Palmer SG, Porotto M, Palermo LM, Cunha LF, Greengard O, Moscona A. Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue. MBio. 2012;3(3) doi: 10.1128/mBio.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leonard VH, et al. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 2008;118:2448–2458. doi: 10.1172/JCI35454. [DOI] [PMC free article] [PubMed] [Google Scholar]