Abstract
Cancer cells can divert metabolites into anabolic pathways to support their rapid proliferation and to accumulate the cellular building blocks required for tumor growth. However, the specific bioenergetic profile of invasive and metastatic cancer cells is unknown. Here we report that migratory/invasive cancer cells specifically favor mitochondrial respiration and increased ATP production. Invasive cancer cells use transcription co-activator, PGC-1α to enhance oxidative phosphorylation, mitochondrial biogenesis and oxygen consumption rate. Clinical analysis of human invasive breast cancers revealed a strong correlation between PGC-1α expression in invasive cancer cells and formation of distant metastases. Silencing of PGC-1α in cancer cells suspended their invasive potential and attenuated metastasis without affecting proliferation, primary tumor growth or epithelial-to-mesenchymal (EMT) program. While inherent genetics of cancer cells determine the transcriptome framework required for invasion and metastasis, mitochondrial biogenesis and respiration induced by PGC-1α is also essential for functional motility of cancer cells and metastasis.
Introduction
It is generally well established that dividing cells, including cancer cells, meet their metabolic demands through the process of aerobic glycolysis(1, 2). The energy generated through aerobic glycolysis is thought to be sufficient to offset the energy demands associated with rapid cancer cell division, while simultaneously allowing accumulation of biosynthetic precursors needed for anabolic reactions(1, 2). Despite enhanced glycolysis, cancer cells also operate mitochondrial respiration to derive a significant fraction of their ATP(3). In a growing tumor, adaptive metabolic reprogramming, precipitated in part by oncogenic transformation(4), gives cancer cells a proliferative advantage(5, 6). The autonomous metabolic reprogramming of rapidly proliferating cancer cells promotes self-sustaining signal transduction mechanisms to foster growth and survival(5). However, the metabolic requirements of invasive and metastatic cancer cells that suspend their proliferative program to acquire a migratory phenotype are unknown. An improved understanding of the energetic demands of invading cancer cells may inform therapeutic strategies to impair metastasis, the primary reason for death in cancer patients. We set out to conduct experiments to study the specific energy requirements of invasive and metastatic cancer cells, with a hope of unraveling additional mechanisms of metastasis.
Results
Circulating cancer cells exhibit enhanced mitochondria biogenesis and respiration
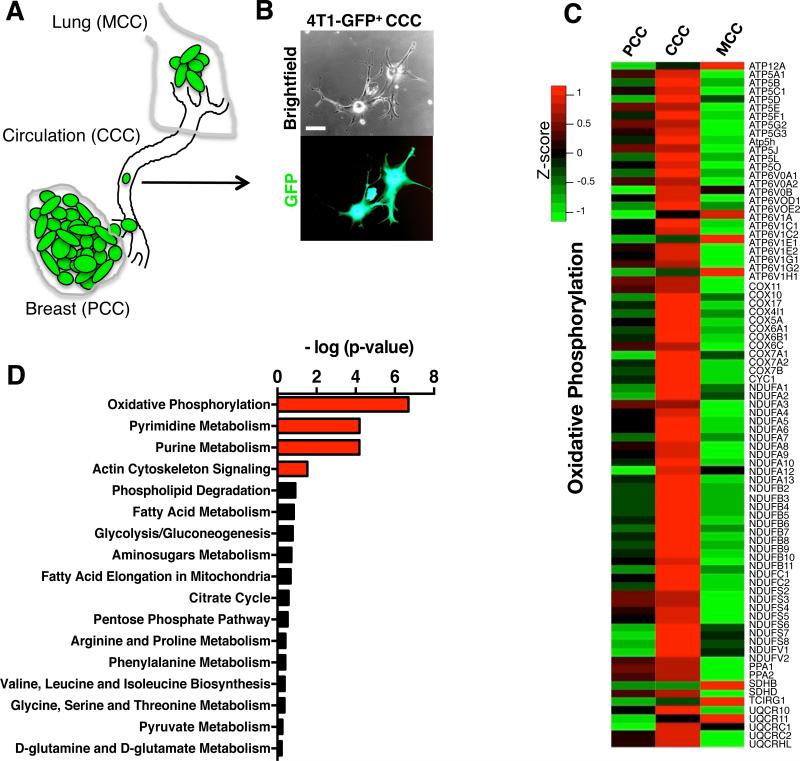
GFP-labeled 4T1 mammary epithelial cancer cells were orthotopically implanted in the mammary fat pads of mice (Fig. 1A-B). Primary tumors emerge following implantation of cancer cells into the mammary fat pads of female mice and lung metastases develop with 100% penetrance(7). Circulating cancer cells (CCC, also referred to as circulating tumor cells or CTC) and cancer cells from the primary tumors (PCC) and metastatic lungs (MCC) were FACS purified and their transcriptome assayed by gene expression microarray. Gene expression profiling coupled with bioinformatic analyses revealed that the oxidative phosphorylation was the most differentially modulated canonical pathway in CCC when compared to PCC, with a significant increase in transcript levels associated with oxidative phosphorylation in CCC (Fig. 1C-D). Actin cytoskeleton signaling pathway was also differentially regulated in CCC compared to PCC (Fig. 1D & Supplementary Fig. 1). We did not observe a significant deregulation in glycolysis/gluconeogenesis, pyruvate metabolism, TCA cycle, pentose phosphate pathway (PPP), amino-sugar metabolism, fatty acid metabolism, fatty acid elongation in the mitochondria, phospholipids degradation, glycine/serine/threonine metabolism, arginine/proline metabolism, phenylalanine metabolism, and valine/leucine/isoleucine metabolism in CCC compared with PCC (Fig. 1D & Supplementary Fig. 1).
Figure 1. Circulating cancer cells (CCC) exhibit enhanced oxidative phosphorylation.
A. 4T1-GFP+ cells were injected orthotopically in the mammary fat pad of mice and primary tumor cancer cells (PCC), circulating cancer cells (CCC) and cancer cells from lung metastases (MCC) were purified by FACS sorting for gene expression profiling assay. B. Representative image of CCC isolated from 4T1 orthotopic tumor model based on their GFP expression. Scale bar: 10 μm. C. Heat map of differentially regulated genes in the oxidative phosphorylation gene set in PCC, CCC and MCC. D. Pathway analyses of transcriptomes of CCC compared to PCC identify oxidative phosphorylation as the most differentially regulated gene set. Actin cytoskeleton signaling, pyrimidine and purine metabolism pathways were also significantly differentially regulated in CCC compared to PCC, while all other metabolic pathways were only minimally changed.
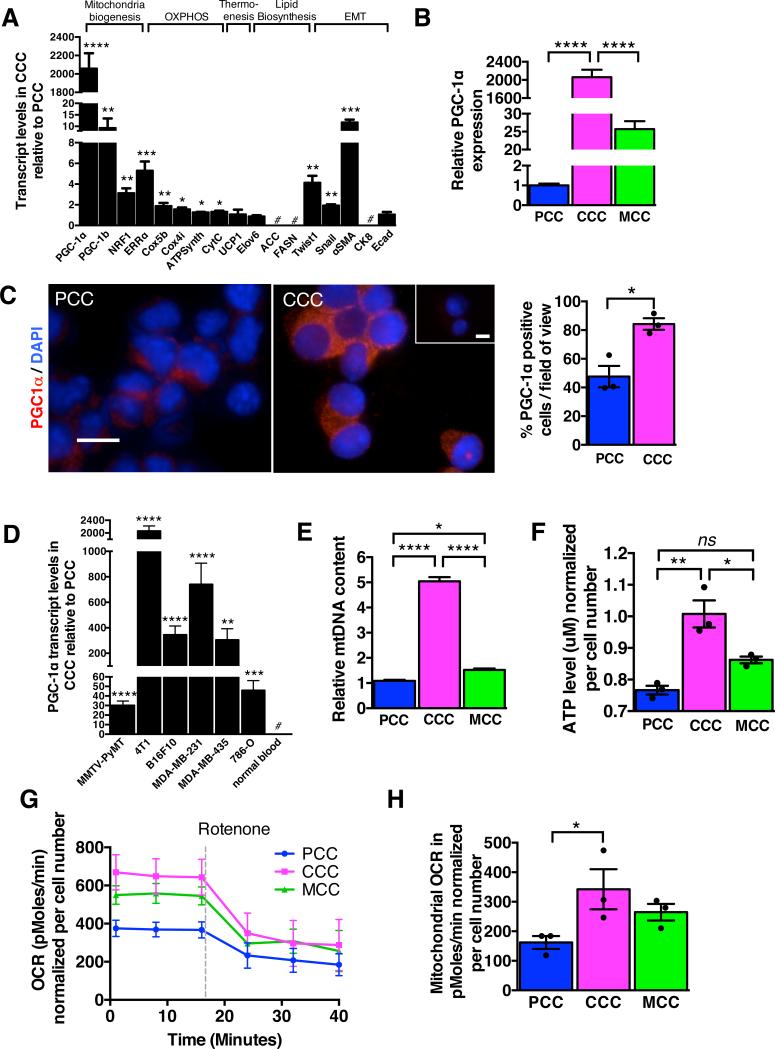
Quantitative PCR analyses showed specific up-regulation of genes associated with mitochondrial biogenesis (PGC-1α, PGC-1β, NRF1, and ERRα) and oxidative phosphorylation (Cox5b, Cox4i, ATPsynth, CytC) in CCC compared to PCC (Fig. 2A). MCC and PCC showed similar gene expression levels associated with mitochondria biogenesis and oxidative phosphorylation (Supplementary Fig. 2A), suggestive of a reversible expression of these genes when CCC are retained in their preferred site of metastasis. The expression levels of some MCC genes were only partially restored to values obtained in PCC, and this may be due to collective mixture of MCC at different stages of metastasis (arrest, extravasation, migration, proliferation). The reversible shift in patterns of metabolic gene expression pattern paralleled that of genes frequently associated with epithelial-to-mesenchymal (EMT) program (Fig. 2A, Supplementary Fig. 2A). Activation of EMT program is a characteristic feature of invading epithelial cancer cells(8, 9). Mesenchymal genes (Twist, Snail, αSMA) were significantly up-regulated in CCC, while epithelial genes (CK8, Ecad) were down-regulated in CCC when compared to PCC and MCC (Fig. 2A, Supplementary Fig. 2A). Genes associated with thermogenesis, uncoupled respiration (UCP1) and lipid biosynthesis (ACC, Elovl6, FASN) were unchanged (Fig. 2A) in CCC compared to PCC. Collectively, these results suggest that CCC assume a characteristic EMT phenotype and transition to a bioenergetic program that utilizes mitochondrial biogenesis and oxidative phosphorylation.
Figure 2. CCC display increased oxygen consumption rate associated with PGC-1α expression and mitochondria biogenesis.
A. Quantitative PCR analyses of relative expression of indicated genes in CCC normalized to PCC. Genes were grouped based on known association with mitochondria biogenesis, oxidative phosphorylation, thermogenesis, lipid biosynthesis, and epithelial to mesenchymal transition (EMT). #: no transcript was detected (n=5 RNA samples from 5 mice, unpaired two-tailed Student's t-test). B. Relative PGC-1α expression by quantitative PCR analysis in CCC compared to PCC in mice with 4T1 orthotopic tumors (n=5 RNA samples from 5 mice, one-way ANOVA). C. Immunostaining for PGC-1α of cytospin of PCC and CCC and quantification of relative percent of PGC-1α positive cells. Scale bars: 10 μm. Nuclear staining (DAPI, blue) (n=3 average percent positive cells from 3 mice, unpaired two-tailed Student's t-test). D. Relative PGC-1α expression by quantitative PCR analysis in CCC compared to PCC in indicated orthotopic tumor models. # (normal blood): no PGC-1α expression detected (n=5 RNA samples from 5 mice, unpaired two-tailed Student's t-test). E. mitochondrial DNA (mtDNA) content (n=5 DNA samples from 5 mice, one-way ANOVA), F. intracellular ATP levels (n=3 lysates from 3 mice, one-way ANOVA), G. oxygen consumption rate (OCR), and H. mitochondrial OCR (delta OCR pre and post rotenone treatment) in PCC, CCC and MCC from 4T1 orthotopic tumor model (n=3 wells of cells from 3 mice, one-way ANOVA). Data is represented as mean +/− SEM. ns: not significant. * p<0.05, ** p<0.01, ** p< 0.001, ****p < 0.0001.
Peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) is enriched in CCCs
The enhanced mitochondrial oxidative phosphorylation in CCC compared to PCC and MCC was associated with a marked up-regulation PGC-1αan inducer of mitochondrial biogenesis (Fig. 2A-B). PGC-1α functions as a master integrator of cellular signals that regulate mitochondrial biogenesis, oxidative phosphorylation, adaptive thermogenesis and fatty acid biosynthesis/degradation(10, 11). PGC-1α promotes ATP production and energy homeostasis during bioenergetic crises, which renders cells resistant to necrosis and apoptosis(12). While PGC-1α has been implicated in tumorigenesis(13, 14), its function in metastasis remains unknown. PGC-1α expression in CCC was highest among all other genes known to promote mitochondria biogenesis, including ERRα, NRF1 and PGC-1β (Fig. 2A). Immunostaining for PGC-1α in PCC and CCC shows in increase in the percentage of PGC-1α+ CCC compared to PCC (Fig. 2C). PGC-1α expression was not detected in peripheral blood cells that were isolated from non-tumor bearing mice (Fig. 2D). We measured PGC-1α expression levels in CCC isolated from the MMTV-PyMT spontaneous mouse breast cancer model (these mice spontaneously develop primary mammary tumors that metastasize primarily to the lung(15, 16)), as well as in CCC from mice harboring a variety of orthotopically implanted tumors, including B16F10 mouse melanoma, MDA-MB-231 (triple-negative human breast cancer cells), MDA-MB-435 human melanoma(17) and 786-O human renal cell carcinoma. In all of the models studied, PGC-1α expression was markedly up-regulated in CCC compared to PCC (Fig. 2D, Supplementary Fig. 2B-C, E).
To determine whether increased expression of genes associated with mitochondria biogenesis and oxidative phosphorylation in CCC correlated with increased mitochondria number and mitochondrial respiration, we first measured oxygen consumption rate, ATP production and mitochondria DNA content in purified CCC (Fig. 2E-H). When compared to PCC, CCC exhibited increased mitochondrial DNA content (Fig. 2E), elevated intracellular ATP levels (Fig. 2F), higher basal respiration rate (Fig. 2G) and enhanced mitochondrial oxygen consumption rate (OCR) (Fig. 2H). The mitochondrial DNA content of CCC from mice with B16F10 and MDA-MB-231 tumors was also increased (Supplementary Fig. 2D, F). These results suggested that the enhanced oxidative phosphorylation was associated with an increase in the number of mitochondria per cell and that enhanced PGC-1α expression and mitochondrial biogenesis are features of CCC.
PGC-1α expression facilitates mitochondria biogenesis and invasion of cancer cells
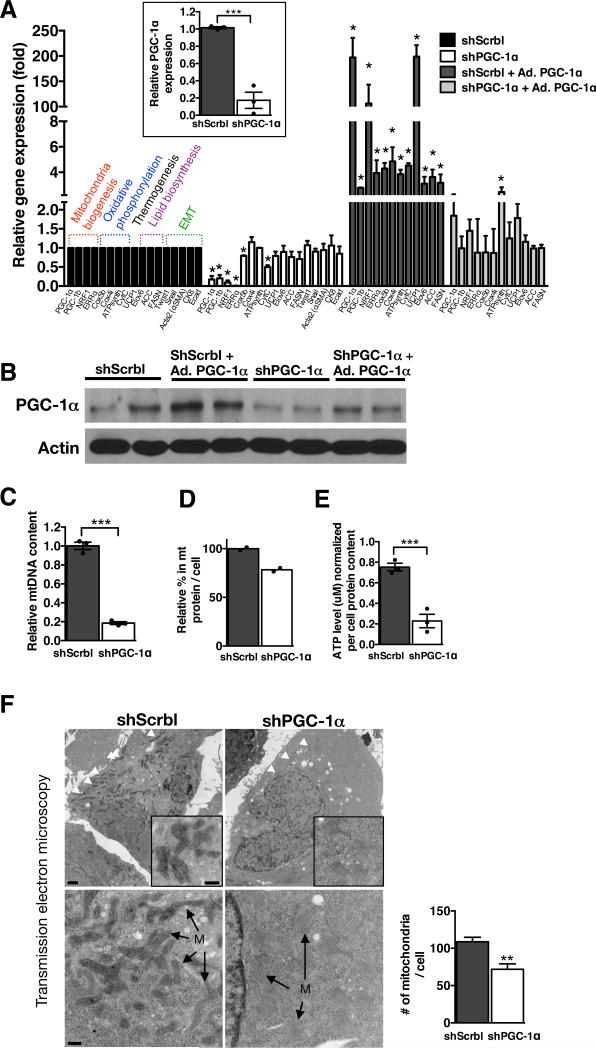
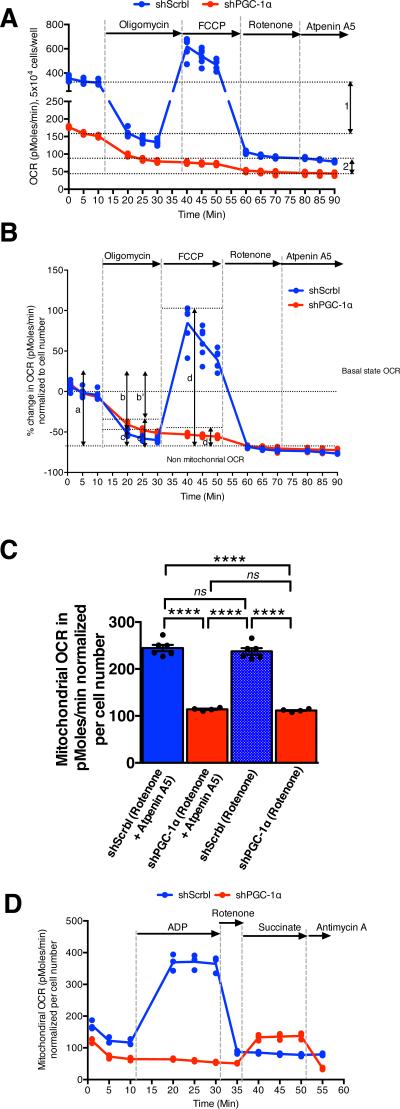
PGC-1α was silenced (gene expression knockdown, shPGC-1α)or overexpressed (adenoviral induced over-expression, Ad. PGC-1α) in 4T1, B16F10 and MDA-MB-231 cells (Fig. 3A-B and Supplementary Fig. 4A-B, 5A-B). Quantitative PCR analyses revealed significant down-regulation of genes associated with mitochondrial biogenesis (PGC-1α, NRF1, ERRα) and oxidative phosphorylation (CytC, Cox5b) in 4T1shPGC-1α cells compared to 4T1shScrbl control cells. Genes associated with lipid biosynthesis (ACC, Elovl6, FASN) and EMT program (CK8, Ecad, Twist, Snail and αSMA) were unchanged (Fig. 3A). Induction of PGC-1α expression reversed the suppression of genes associated with mitochondrial biogenesis and oxidative phosphorylation in 4T1shPGC-1α cells, while genes associated with lipid biosynthesis remained unchanged (Fig. 3A). A significant reduction in PGC-1α transcript and protein levels in 4T1, B16F10 and MDA-MB-231 cells resulted in suppressed mitochondrial biogenesis, as assessed by reduced mitochondrial DNA (Fig. 3C, Supplementary Fig. 4C, 5C) and reduced amount of mitochondrial protein per cell (Fig. 3D, Supplementary Fig. 4D, 5D). PGC-1α depleted cells displayed decreased cellular ATP levels (Fig. 3E, Supplementary Fig. 4E, 5E). Transmission electron microscopy analyses reveal reduced numbers of mitochondria and swollen mitochondria with disorganized cristae, suggesting impaired mitochondria respiration (Fig. 3F, Supplementary Fig. 3A-D). We verified this by measuring the oxygen consumption rate (OCR) in a series of mitochondria stress tests performed on cultured cells. These experiments indicated that mitochondria respiration capacity was diminished when PGC-1α was suppressed (Fig. 4A-C, Supplementary Fig. 4F, 5F). Specifically, basal respiration rate was decreased in shPGC-1α cells compared to shScrbl control cells (‘1’ in Fig. 4A). We noted an elevated non-mitochondrial OCR in shScrbl cells compared to shPGC-1α cells (‘2’ in Fig. 4A) following inhibition of electron transport chain (ETC) Complex I+II (rotenone + atpenin A5 treatments). Targeted mass spectrometry and metabolomics analyses(18) of 4T1shPGC-1α cells compared to 4T1shScrbl cells revealed lower levels of some of the metabolites levels associated with glycolysis, TCA cycle, amino-sugar metabolism, pyruvate metabolism, phospho-lipid metabolism, pyrimidine and purine metabolism, while some of these associated metabolites showed elevated levels (Supplementary Fig. 6A). Metabolomics analyses of 4T1 shPGC-1α cells cultured using labeled 13C-labeled glucose also showed insignificant differences in accumulation rate of glycolytic/gluconeogenic metabolites when compared to shScrbl control cells (Supplementary Fig. 7A). OCR measurements also revealed a decreased capacity for ATP-coupled OCR in shPGC-1α cells compared to shScrbl control cells (b’<b in Fig. 4B) and an enhanced proton leak (c’>c in Fig. 4B). shPGC-1α cells respire at their maximal capacity (d’<a in shPGC-1α cells vs. d>a in shScrbl cells in Fig. 4B), possibly to compensate for the loss in ATP-coupled respiration subsequent to the loss of mitochondria lowering the basal respiration rate. Mitochondrial OCR was decreased in shPGC-1α cells compared to shScrbl cells, which also occurred when either complex I or complex I+II of the ETC was inhibited (Fig. 4C). Compensation via complex II activity is not observed when mitochondrial respiration is compromised by suppression of PGC-1αin live cells, however OCR measurements of permeabilized cells suggest a possible differential regulation of complex II linked OCR activity (increased OCR following succinate injection) in shPGC-1α cells compared to shScrbl cells (Fig. 4D). These results indicate that PGC-1α modulates mitochondrial biogenesis in cancer cells without significantly affecting glycolysis. These results also suggest that PGC-1α suppression impacts oxidative phosphorylation capacity of cancer cells by suppressing both mitochondria biogenesis and directly impacting ATP-coupled respiration and efficacy of electron transport chain (ETC) within the remaining mitochondrial pool. The transition from PCC to CCC appears to be associated with enhanced reliance on mitochondrial respiration, which is likely mediated, in part, by PGC-1α. PGC-1β, ERRα and NRF1 were up-regulated in CCC (Fig. 2A) and down-regulated when PGC-1α was suppressed (Fig. 3A), highlighting an upstream action of PGC-1α in this setting(19).
Figure. 3. PGC-1α expression is associated with mitochondria respiration and biogenesis in cancer cells.
A. Real-time PCR analyses of relative expression of indicated genes in 4T1shPGC-1α normalized to 4T1shScrbl cells, and 4T1shPGC-1α and 4T1shScrbl cells with adenoviral over-expression of PGC-1α(Ad. PGC1α), also normalized to 4T1shScrbl cells (arbitrarily set to 1). Insert shows PGC-1α expression specifically in 4T1shScrbl and 4T1shPGC-1α cells (n=3 RNA samples from cells, unpaired two-tailed Student's t-test, * p<0.05). B. Western blot for PGC-1α in indicated cells/treatment. See also Supplementary Figure 9. C. Mitochondrial DNA (mtDNA) content (n=3 DNA samples from cells, unpaired two-tailed Student's t-test, see also Supplementary Figure 9) and D. mitochondrial protein content relative to total cell protein content in 4T1shPGC1α normalized to 4T1shScrbl cells (n=2 extracted cell lysates). E. Intracellular ATP levels in 4T1shPGC1α normalized to 4T1shScrbl cells (n=3, unpaired two-tailed Student's t-test). F. Transmission electron microscopy of 4T1 shScrbl and shPG1α, white arrowheads and ‘M’ identify mitochondria. Scale bar upper panel: 2 μm, insert and lower panel: 500 nm. Quantitation of the number of mitochondria per cell (shScrbl, n=9 cells; shPGC-1α, n=6 cells, unpaired two-tailed Student's t-test). Data is represented as mean +/− SEM. * p<0.05, *** p< 0.001.
Figure 4. PGC-1α expression modulates complex I-driven oxidative phosphorylation in 4T1 cancer cells.
A. Relative oxygen consumption rate (OCR) normalized to cell number over time in 4T1shPGC-1α (n=4 wells) and 4T1shScrbl (n=6 wells) cells. 1: differential basal state OCR; 2: differential non-mitochondrial OCR. B. Percent change in OCR normalized per cell number in in 4T1shPGC-1α (n=4 wells) and 4T1shScrbl (n=6 wells) cells. a: initial OCR; b and b’: percent OCR used for ATP synthesis in 4T1shScrbl and 4T1shPGC-1α cells respectively; c and c’: percent OCR associated with proton leakage in 4T1shScrbl and 4T1shPGC-1α cells respectively; d and d’: maximum mitochondria respiratory capacity 4T1shScrbl and 4T1shPGC-1α cells respectively. C. Mitochondrial OCR (delta OCR pre and post rotenone with or without atpenin A5 treatment) in 4T1shPGC-1α (n=4) and 4T1shScrbl (n=6 wells) cells, one-way ANOVA. D. OCR measurements in permeabilized 4T1 shScrbl and shPGC1-α cells (n=3 wells per cell line). Data is represented as mean +/− SEM. ns: not significant. * p<0.05, **** p< 0.0001.
PGC-1α expression determines invasive capacity and metastasis potential of cancer cells
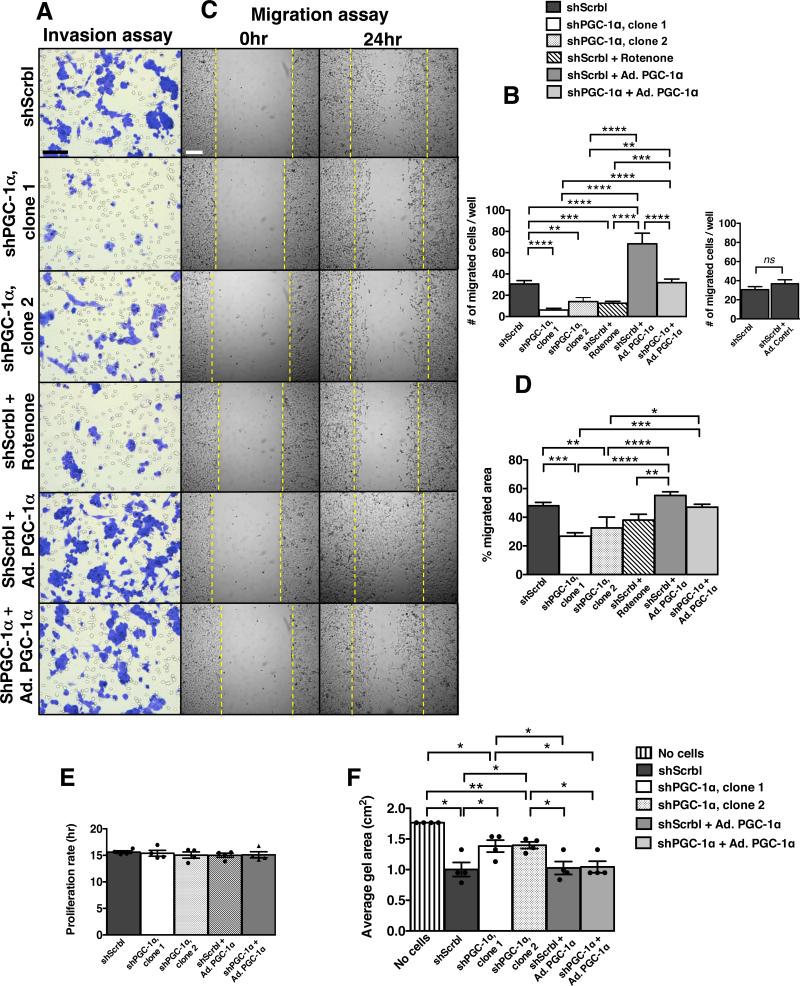
Next, we questioned whether PGC-1α-mediated induction of mitochondria biogenesis and respiration was associated with invasive capacity of CCC. We measured invasion, migration, and cell division of 4T1, B16F10 and MDA-MB-231 cells with stable suppression, rescued expression, or over-expression of PGC-1α or with rotenone treatment. While suppression of PGC-1α revealed specific down-regulation of mitochondrial respiration, over expression of PGC-1α in cancer cells resulted in mixed metabolic response, likely resulting from hyper induction of many metabolic processes due to supra-physiological levels of PGC-1αin cultured cells(12) (Fig. 3A). Nevertheless, over-expression of PGC-1αin shPGC-1α cancer cells partially reversed the gene expression pattern associated with suppression of mitochondrial biogenesis and respiration (Fig. 3A). PGC-1α knockdown and rotenone treatment significantly reduced invasion of cancer cells, while over-expression of PGC-1α enhanced invasion and restored invasive properties to shPGC-1α cells (Fig. 5A-B; Supplementary Fig. 3E, 4G-H & 5G-H). Migration was also dependent on PGC-1α expression levels (Fig. 5C-D; Supplementary Fig. 3E, 4I-J & 5I-J), whereas alterations in PGC-1α expression had no effect on cell division (Fig. 5E). Loss of PGC-1α expression reduced cancer cells’ ability to tighten type I collagen in contraction assays, suggestive of compromised actin cytoskeleton remodeling and associated ATP-fueled molecular motors (Fig. 5F, Supplementary Fig. 4K & 5K). Over-expression of PGC-1α alone did not enhance collagen I contraction, but rescued shPGC-1α reduced type I collagen contractility (Fig. 5F, Supplementary Fig. 4K & 5K). Taken together, these results indicate that loss of PGC-1α expression diminishes invasive and migratory properties of cancer cells and these features are restored with the rescue of PGC-1α gene expression in shPGC-1α cancer cells.
Figure 5. PGC-1α expression modulates cancer cell invasion and migration.
A. Hematoxylin stained 4T1 cells (scale bar: 50 μm) following invasion and B. quantitation of invasion assay. Ad. PGC-1α: adenoviral induction of PGC-1α expression (n=6 wells/group, one-way ANOVA). Ad. Contrl.: adeonovirus with empty pcDNA vector control (n=6 wells/group, unpaired two-tailed Student's t-test) C. Light microscopy imaging (scale bar: 50 μm) of migrated cells in scratch assay and D. quantitation of migration assay (n=5 wells/group, one-way ANOVA). E. Proliferation rate (n=4 cell numbers measured over time/group, one-way ANOVA) and. F. type I collagen gel area reflecting gel contraction by indicated cells (n=4 wells/group, unpaired two-tailed Student's t-test). Data is represented as mean +/− SEM. ns: not significant. * p<0.05, ** p<0.01, *** p< 0.001, ****p < 0.0001.
PGC-1α promotes cancer cell metastasis
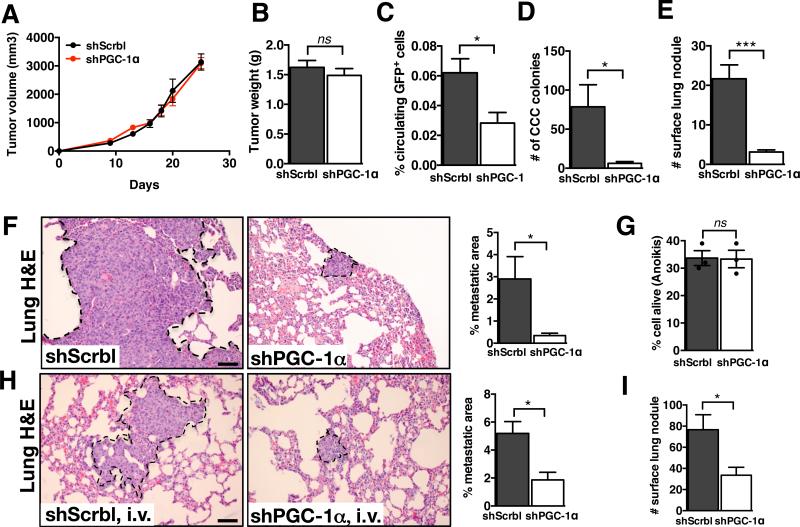
Expression levels of PGC-1α had no affect on the growth of primary 4T1 tumors (Fig. 6A-B). MDA-MB-231shPGC-1α tumors showed similar tumor growth kinetics and weight when compared to control MDA-MB-231shScrbl tumors (Supplementary Fig. 8A-B). Similarly, PGC-1α gene expression knockdown did not impact the growth of primary B16F10 melanomas (Supplementary Fig. 8I-J). The number of CCC was significantly reduced in mice with 4T1shPGC-1α tumors compared to mice with control 4T1shScrbl, as assessed by the reduced number of GFP+ cancer cells in the blood by FACS analysis and also by the decreased number of blood-derived cancer cell colonies (colony formation assay) (Fig. 6C-D). The decreased dissemination of cancer cells was associated with a significant reduction in the computed percent metastatic lung area and number of surface lung nodules of mice with 4T1shPGC-1α tumors compared to mice with control 4T1shScrbl tumors (Fig. 6E-F). These findings were reproduced using a second knockdown clone of PGC-1α in 4T1 cells (Supplementary Fig. 7B-E). CCC numbers (Supplementary Fig. 8CD) and metastasis (Supplementary Fig. 8E-F) were also significantly reduced in mice bearing MDA-MB-231shPGC-1α tumors in contrast with mice bearing MD-MB-231shScrbl tumors. Decreased cancer cell dissemination and reduced metastatic disease were also observed when PGC-1α expression was suppressed in B16F10 melanoma cells (Supplementary Fig. 8K-N). These results indicate that suppression of PGC-1α reduces cancer cells metastasis.
Figure 6. Loss of PGC-1α expression suppresses cancer cell dissemination and metastasis.
A. 4T1shScrbl and 4T1shPGC-1α cells were implanted in the mammary fat pad of mice and tumor volume was measured over time (4T1shScrbl, n=6 mice; 4T1shPGC-1α, n=7 mice). B. Tumor weight at experimental endpoint (4T1shScrbl, n=6 mice; 4T1shPGC-1α, n=7 mice, unpaired two-tailed Student's t-test). C. Percent of GFP+ cancer cells per 200μl blood collected at experimental endpoint 4T1shScrbl, n=6 mice; 4T1shPGC-1α, n=6 mice, unpaired two-tailed Student's t-test). D. Number of CCC colonies formed (4T1shScrbl, n=6 mice; 4T1shPGC-1α, n=7 mice, unpaired two-tailed Student's t-test). E. Number of lung surface nodules in of mice with orthotopic 4T1shScrbl and 4T1shPGC-1α tumors (4T1shScrbl, n=6 mice; 4T1shPGC-1α, n=7 mice, unpaired two-tailed Student's t-test). F. Representative images of H&E stained lung sections of mice harboring orthotopic 4T1shScrbl and 4T1shPGC-1α tumors and percent metastatic lung surface area relative to total lung surface area. Scale bar: 50μm. Metastatic lung nodules are encircled. (4T1shScrbl, n=6 mice; 4T1shPGC-1α, n=7 mice, unpaired two-tailed Student's t-test). G. Percent alive cells in anoikis assay (n=3 percent live cell measurements, unpaired two-tailed Student's t-test). H. Representative images of H&E stained lung sections of mice with intravenous injection of 4T1shScrbl and 4T1shPGC-1α cells and percent metastatic lung surface area relative to total lung surface area (4T1shScrbl, n=5 mice; 4T1shPGC-1α, n=6 mice, unpaired two-tailed Student's t-test). Scale bar: 50μm. Lung nodules are encircled. I. Number of lung surface nodules (4T1shScrbl, n=5 mice; 4T1shPGC-1α, n=6 mice, unpaired two-tailed Student's t-test). Data is represented as mean +/− SEM. ns: not significant. * p<0.05, *** p< 0.001.
Our studies pointed to the possibility that PGC-1α expression is essential for intravasation of the cancer cells into the circulation. Similar anchorage-independent survival (anoikis) rates in cells with suppressed PGC-1α was observed compared to control lines (Fig. 6G). We next questioned whether extravasation of cancer cells is similarly impaired when PGC-1α is suppressed. We monitored lung colonization and lung metastatic nodule formation in mice following intravenous injection of shPGC-1α and control shScrbl cells. Metastatic lung colonization and nodule formation was significantly impaired when PGC-1α expression was suppressed (Fig. 6H-I, Supplementary Fig. 8G-H, O-P). Taken together, our results support an important role for PGC-1α-mediated mitochondrial biogenesis and oxidative phosphorylation in facilitating migration, invasion, and intravasation/extravasation of cancer cells.
Mitochondrial respiration fuels cancer cell motility
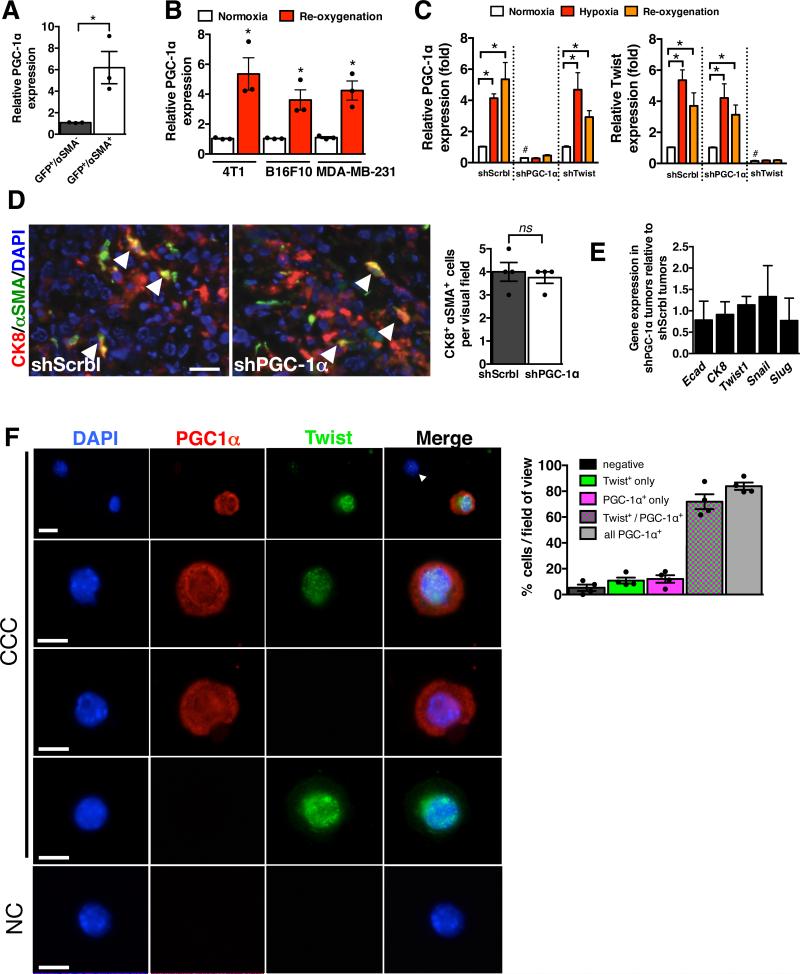
To evaluate PGC-1α expression in invading cancer cells in vivo, we FACS purified PCC based on GFP expression and αSMA labeling. Cancer cells exhibiting an EMT program (GFP+/αSMA+) express significantly higher levels of PGC-1α when compared to cancer cells without EMT program (GFP+/αSMA−) (Fig. 7A). Because hypoxia is a potent inducer of cancer cells’ invasive properties, we evaluated PGC-1α expression in 4T1, B16F10 and MDA-MB-231 cells following re-oxygenation from hypoxia. PGC-1α expression was significantly increased following re-oxygenation (Fig. 7B). Cells in hypoxia as well as after re-oxygenation showed elevated PGC-1α and Twist expression (Fig. 7C). However, when PGC-1α was suppressed, hypoxia had no effect on PGC-1α expression, but did up-regulate Twist expression, suggesting that Twist expression associated with hypoxia-induced EMT was independent of PGC-1α up-regulation (Fig. 7C). Similarly, in cells with knockdown of Twist, PGC-1α expression was elevated under hypoxic condition, while Twist was not up-regulated (Fig. 7C). Furthermore, double immunolabeling for CK8 (epithelial marker) and αSMA (mesenchymal marker) revealed a similar number of double positive cancer cells in shPGC-1α and control shScrbl primary tumors, suggesting an equal frequency of cancer cells acquiring EMT program (Fig. 7D). Quantitative PCR analyses for mesenchymal and epithelial genes showed comparable levels of EMT program related genes in both shPGC-1α and control shScrbl primary tumors (Fig. 7E). Immunolabeling for Twist and PGC-1α in CCC indicated that a majority of the CCC were PGC-1α+, and nearly all PGC-1α+ CCC were Twist+ (Fig. 7F). Taken together these results suggest that acquisition of EMT and invasive phenotype is associated with Twist expression, and that PGC-1α up-regulation in invasive cells is independent of Twist. Invasive feature of migrating cancer cells, which may be precipitated by hypoxia, was associated with elevated PGC-1α expression, however suppression of PGC-1α did not impact the expression of EMT related genes (Fig. 3A). Collectively, these findings suggest that induction of PGC-1α expression is not mutually exclusive to acquisition of mesenchymal features (EMT), and modulating mitochondrial respiration in cancer cells via hypoxia-driven induction of PGC-1α expression does not limit induction of EMT-associated genes, but functionally impairs cell motility and invasive properties.
Figure 7. PGC-1α expression is co-induced with an EMT program.
A. PGC-1α expression in FACS purified GFP+/αSMA− and GFP+/αSMA+ cells from 4T1 primary tumor (n=3 RNA samples from FACS purified cells of 3 mice, unpaired two-tailed Student's t-test). * p<0.05. B PGC-1α expression in indicated cells cultured in normoxia and in re-oxygenation (24 hrs) following 48 hrs of hypoxia (n=3 RNA samples/cell line, unpaired two-tailed Student's t-test). * p<0.05. C. PGC-1α and Twist expression in 4T1shScrbl, 4T1shPGC-1α and 4T1shTwist cells cultured in normoxia, hypoxia (48 hrs), and re-oxygened (24 hrs) following 48 hrs of hypoxia (n=3 RNA samples/cell line, unpaired two-tailed Student's t-test). * p<0.05. #: p <0.05 for PGC-1α expression in 4T1shPGC-1α compared to 4T1shScrbl in normoxic condition, and for Twist expression in 4T1shTwist compared to 4T1shScrbl in normoxic condition. D. Representative CK8 (red) and αSMA (green) immunolabeling of the primary tumor. Nuclear staining (DAPI, blue). Arrowheads point to double positive (CK8+/αSMA+) cells. Scale bar: 100 μm. Bar graph: quantitation of number of CK8+/αSMA+ cells per field of view (n=4 stained slides of tumors from 4 mice, unpaired two-tailed Student's t-test). E. Relative expression of indicated genes in 4T1shPGC-1α tumors normalized to 4T1shScrbl tumors (n=5 RNA samples from tumors of 5 mice, unpaired two-tailed Student's t-test). F. Immunolabeling of cytopsin of 4T1 CCC for Twist and PGC-1α (n=4 mice). Scale bar: 10μm. NC: negative control, secondary antibody used only. Bar graph: Relative percentage of CCC negative for both Twist and PGC-1α, positive for Twist only (PGC-1α−), positive for PGC-1α only (Twist−), positive for both Twist and PGC-1α, and all PGC1a positive CCC (regardless of Twist status). Scale bars: 25μm (upper panel), 10μm (lower panel). Data is represented as mean +/− SEM. ns: not significant.
Enhanced PGC-1α expression is associated with distant metastasis and poor outcome in patients with invasive breast cancer
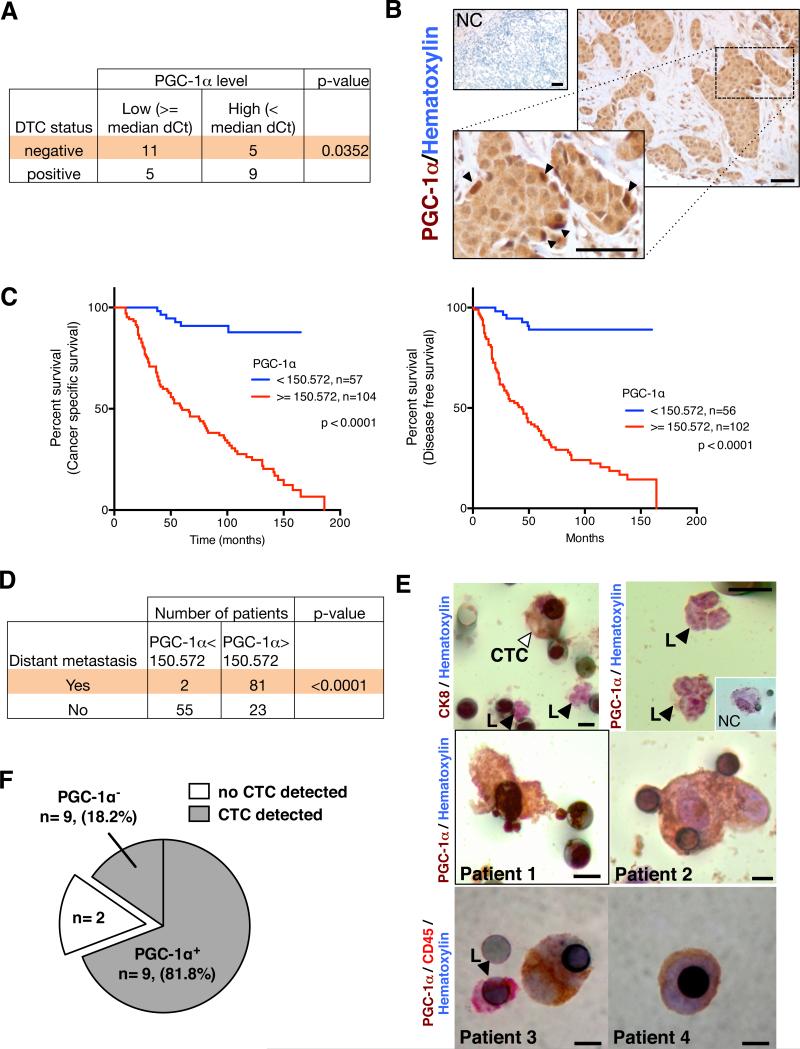
We measured PGC-1α gene expression in laser microdissected neoplastic cells harvested from the periphery of breast tumors in patients diagnosed with invasive ductal carcinomas (IDC) [further categorized based on the presence of disseminated tumor cells (DTC) in the bone marrow (BM+: positive DTC detection, n=14; BM−: negative DTC detection, n=16) (20)]. High PGC-1α transcript levels in cancer cells were significantly correlated with positive DTC status (Fig. 8A, Supplementary Table 1). These data suggest a possible role for PGC-1α in mediating cancer cell dissemination to the bone in patients with IDC.
Figure 8. PGC-1α expression in circulating cancer cells correlates with invasion and distant metastasis in patients with IDC.
A. Correlation of relative PGC-1α expression (median dCt value used as cut-off) in laser dissected neoplastic cells from resected tumors of patients with IDC and status of disseminated tumor cells (DTC) in the bone marrow (BM+: bone marrow is positive for DTC; BM−: bone marrow is negative for DTC). BM+ status, n=16 RNA samples from 16 patients; BM−status, n=14 RNA samples from 14 patients. (Supplementary Table 1, one-sided χ2 analysis). B. Immunohistochemistry staining for PGC-1α (haematoxylin counter stain) in breast tumors from patients with IDC. NC: negative control, secondary antibody and haematoxylin counter stain only. Arrowheads in zoomed-in insert point to the strongly positive PGC-1α cells found on the invasive edge. Scale bar: 50μm. C. Cancer specific survival and disease free survival of IDC patients grouped based on PGC-1α expression (Supplementary Table 2, log-rank test). D. Correlative analysis of the number of IDC patients with and without distant metastases categorized based on the indicated PGC-1α expression threshold (Supplementary Table 2, Fisher's exact test). Kaplan-Meier curves were drawn and differences between the curves were calculated by the log-rank test. E. Circulating cancer cell (“CTC”, right arrowhead) stains for the epithelial marker CK8, while leukocytes (“L”, left arrowhead) are negative for CK8 (upper left panel). Leukocytes are negative for PGC-1α (upper right panel). Insert shows (NC) negative control staining of a CTC (secondary antibody only). PGC-1α (patient 1 and 2) or CD45 and PGC-1α (patient 3 and 4) immunostaining of CTC (lower panels, Supplementary Table 3). Scale bar: 8μm. F. Representation of the PGC-1α+ CTC status evaluated by immunohistochemistry staining in IDC patients with lung metastasis (Supplementary Table 4).
We next probed tissue microarrays (TMAs) for the expression of PGC-1α in breast tumors from 161 patients with IDC (Fig. 8B, Supplementary Table 2). There was a significant correlation between high PGC-1α expression at the invasive tumor front and distant metastasis and, moreover, poor overall- and cancer-specific survival (Fig. 8C-D). We also detected PGC-1α expression in CCC (‘CTC’) harvested from patients with metastatic IDC, and PGC-1α+ CTCs were negative for leukocyte marker CD45 (Fig. 8E, Supplementary Table 3). In addition, CTCs were detected in 11 out of 13 patients with IDC who had confirmed lung metastases and over 80% of the CTCs were PGC-1α+ (Fig. 8F, Supplementary Table 4). These results illustrate a positive correlation between PGC-1α expression and formation of distant metastases in patients with invasive breast cancer.
Discussion
New therapeutic strategies are beginning to focus on the unique patterns of metabolism observed in cancer cells. Therefor, determining the metabolic requirements of invasive cancer cells could be of therapeutic value. We show that the PGC-1α-mediated mitochondrial biogenesis and respiration in cancer cells is functionally relevant for metastatic dissemination. Invading cancer cells rely on PGC-1α to stimulate mitochondrial biogenesis and oxidative phosphorylation during their transit to target organs of metastasis. Indeed, PGC-1α suppression significantly impaired mitochondrial biogenesis and oxidative phosphorylation and, moreover, decreased the frequency of metastasis. The enhanced mitochondrial respiration/oxidative phosphorylation did not impact glycolytic and anabolic rates in the CCC, and did not affect cancer cell proliferation or primary tumor growth kinetics. Although it must be pointed out that more studies are required to specifically connect PGC-1α induced oxidative phosphorylation and metastasis, these results demonstrate that invasive and migratory properties of cancer cells are dependent on mitochondrial respiration and identify PGC-1α as a potential target for therapeutic intervention.
Mitochondrial proficiency and ROS detoxification are critical for cancer cell viability(21, 22). While diminished oxygen levels feed back to reduce mitochondrial biogenesis to avoid metabolic crisis in normal cells(3, 4), zonal hypoxia and focal nutrient bioavailability in different areas of a growing tumor can differentially impact the energetic needs of cancer cells. ROS accumulation in hypoxic cancer cells induces expression of PGC-1α/β to promote detoxification via direct induction of superoxide dismutase 2 (SOD2), catalase, and glutathione peroxidase(23-25). Focal hypoxia may induce an EMT program(16, 26), and our results indicate that while cancer cells acquire a mesenchymal phenotype (EMT) independently of PGC-1α-induced pathways, both pathways coexist and are associated with cancer cell migration, invasion and metastasis(27). We propose that functional EMT program in cancer cells, with acquisition of enhanced migratory and invasive properties, is synergistically coupled with mitochondrial biogenesis and respiration. Altering mitochondrial function could also impact other fundamental cellular processes, in part via retrograde mitochondria-nucleus signaling, and contribute to changes in nuclear transcriptome associated with survival and acquisition of cancer stem cell properties(28). Additionally, dynamic changes in the energy processing of cancer cells in disease progression likely impact the surrounding tumor stroma, which in turn may impact the migratory phenotype of cells.
We speculate that cells in the hypoxic areas of tumor trigger acquisition of migratory features via EMT and PGC-1α mediated mitochondrial respiration. It is conceivable that invading cancer cells enhance their mitochondrial content and up-regulate oxidative phosphorylation as they approach more oxygenated areas of the tumor, which ensures the increased production of ATP required for trafficking to distal tissues. Mitochondrial proficiency and ROS detoxification are critical for cancer cell viability(21, 22), and ATP generation (a feature of non-dividing migratory cells(3, 29)) and antioxidant production ensure cancer cell survival when detaching from their basement membrane(30). The high ATP production found in CCC may be used to promote their retention in target organs. A recent study showed that ATP secreted from platelets stimulates endothelial cell retraction that promotes transendothelial migration of cancer cells (31).
Our clinical analysis of human invasive breast cancers revealed a significant correlation between PGC-1α expression in invasive cancer cells and formation of distant metastases. Of note, expression of ERRα (PGC-1α-coupled transcription factor) in IDC patients also correlates with poor prognosis(32). Furthermore, PGC-1α expression was detected in a high percentage of CTCs of metastatic breast cancer patients. Our studies shed uncovered dynamic shifts in the metabolic program of cancer cells to facilitate diverse steps associated with cancer progression and metastasis.
Material and Methods
Animal studies
Orthotopic (mammary fat pad for 4T1 and MDA-MB-231, under the renal capsule for 786-0, and subcutaneous for B16F10, MDA-MB-235(33)) and intravenous (i.v.) injections of cancer cells were performed as previously described(34, 35). MMTV-PyMT mice were previously described(36) and disease progression in these mice and experimental endpoint at which PCC, MCC and CCC was determined as previously described(34). Adult (at least 8 weeks of age) BALB/c female mice were used for 4T1 injections, adult C57BL/6 male or female mice were used for B16F0 injections, and adult female and male Nu/Nu mice were used for MDA-MB-231, 786-0, and MDA-MB-235 injections. n=5-7 mice were used per experimental groups. Metastatic surface area was computed as previously described(34). No statistical method was used to predetermine sample size and the experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment. Blood volume collection to harvest CCC was 200 μl. Blood was incubated with ACK lysis buffer (2 ml/200 μl blood for 15 minutes at 4°C) before FACS purification based on GFP expression. For CCC colony formation, ACK lysis buffer treated 200 μl blood was plated in 10 cm2 dishes in DMEM tissue culture media supplemented with 10% FBS and penicillin/streptomycin. All animal experiments were reviewed and approved by the Institutional Care and Use Committee at the Beth Israel Deaconess Medical Center and University of Texas MD Anderson Cancer Center.
Cell lines, stable transfection of shPGC-1α, and over-expression of PGC-1α
4T1 (mouse mammary adenocarcinoma), B16F10 (mouse melanoma), MDA-MB-231 (human mammary adenocarcinoma) and MDA-MB-435 (human melanoma) cell lines were obtained from ATCC and cultured in recommended tissue culture media. Partial gene mutations reported for these lines are listed below (WT: wild-type: no mutation; * known mutations): 4T1 (P53*)(37), B16F10 (P53WT/KrasWT/cMycWT)(38) and MDA-MB-231 (P53*/Kras*/cMyc*)(39). For stable transfection of PGC-1α, pre-designed shRNAs from Origene were used and puromycin resistant clones subsequently propagated. For over-expression of PGC-1α, recombinant adenovirus expressing PGC-1α or empty pcDNA control vector was kindly provided by Dr. Bruce Spiegelman, Dana-Farber Cancer Institute, Boston, MA). For proliferation rate, cells growing exponentially were counted twice at 12 hrs intervals and respective proliferation rate calculated. Hypoxia experiments were performed as previously described, and reoxygenation included a 24 hrs incubation in normoxia (21% oxygen) following a 48 hrs exposure to hypoxia (1-2% oxygen).
Gene expression array and real-time PCR validation
RNA was extracted from PCC, MCC and CCC using RNeasy Plus Mini Kit (Qiagen) and submitted to the Molecular Genetics Core Facility at Children's Hospital (Boston, MA). Microarray analysis was performed using Mouse Ref8 Gene Expression BeadChip (Illumina platform) and Metacore (GeneGo) and Knowledge Based Pathway (IPA) (rank invariant normalization with subtracted background). Gene expression validation by real-time PCR was performed as previously described(34) using the primers listed in Supplementary Table 5. Heat maps were drawn using R software. For PGC-1α expression in the laser micro-dissected cancer cells from human IDC tumor specimen, the SuperScript™ III One-Step RT PCR System from Invitrogen.
Relative mtDNA content measurements
Mitochondria DNA (mtDNA) was measured by PCR analysis of total DNA extracted from mouse and human CCC by assessing the relative levels of cytochrome oxydase 1 (mouse mtCO1: 5’-TTGGTCCCCTCCTCCAGC-3’ and 5’-CCAGTGCTAGCCGCAGGCA-3’) vs. β-actin (Supplementary Table 5) and Arp/36b4 (5’-GGAGCCAGCGAGGCCACACTGCTG-3’ and 5’-CTGGCCACGTTGCGGACACCCTCC-3’) and human mtCO1 (5’-TGGAGCCTCCGTAGACCTAA-3’ and 5’-TGCGAAGCCTGGTAGGATAA-3’) vs. β-globin (Supplementary Table 5). For mitochondria protein content, mitochondria were isolated as previously described(40), with the following modifications: cells plated in 10cm2 tissue culture dishes were counted and pelleted at 1,500 rpm for 5 minutes at room temperature. The cell pellet was re-suspended in 2 ml of mitochondria isolation medium (MIM) with BSA (MIM: 300mM sucrose, 10mM NA-HEPES, 0.2mM EDTA, pH 7.2; 1 mg ml−1 BSA) and dounced homogenized. Lysate were spun at 2,000 rpm for 10 minutes at 4°C. The supernatant was then transferred to new tubes and spun at 13,000 rpm for 15 mintues at 4°C. The mitochondria pellet was then washed once with 1 ml MIM+BSA and 1 ml MIM. The final pellet was re-suspended in 20 μl MIM. For visualization, 3 μl were spread under a cover slip and visualized under high magnification light microscopy. For protein content, 1:100 dilution of 20 μl MIM resuspension was used with microBCA assay kit (PierceNet) following the manufacturer's direction. Protein content was normalized to cell count.
Measurement of oxygen consumption rate
Oxygen consumption rate (OCR) were measured using the Seahorse XF24 and XF96 instruments (Seahorse Bioscience) under standard conditions and after addition of 0.1 μM oligomycin, 0.05 μM FCCP, 1 μM of rotenone and 10 μM of atpenin A5. Real time measurements (triplicates) of OCR in pMoles per minutes in tissue culture medium above cells plated in a microplate were plotted over time prior addition of rotenone to the culture media (basal oxygen consumption rate), and after addition of rotenone to specifically measure mitochondrial respiration. The difference in OCR prior and after rotenone addition to the culture media reflects the oxygen consumption by mitochondria (mitochondrial OCR). The OCR measurements were adjusted to cell numbers plated. To this end, the cells were stained with crystal violet (0.1% weight/volume of water) following paraformaldehyde (PFA) permeabilization (4% PFA) and spectrophotometric measurements of 10% acetic acid-solubilized cells were performed with a standard plate reader (Molecular Device). For OCR measurements of permeabilized cells, the Seahorse plasma membrane pemeabilizer kit was used, allowing cells to permeabilize for 30 min, with 10 mM malate and 10 mM glutamate supplemented media, and with sequential injection of 4 mM ADP, 2 μM rotenone, 10 mM succinate and 2.5 μM antimycin A.
ATP measurements
ATP/ADP measurements were obtained using the ATP Determination kit (Life Technologies). For the later, cells were homogenized in lysis buffer (1% Triton X-100, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCL pH 7.5, 1% NaDOC) supplemented with protease cocktail inhibitor, Complete (Roche) and phenylmethylsulfonyl fluoride (Sigma-Aldrich). Protein was quantified by using the BCA Protein Assay (Thermo Scientific) which is used for normalization. Measurements were obtained using the fluorescence plate reader, FLUOstar Omega (BMG Labtech). The samples were normalized to the protein concentration of the corresponding shScrbl cell line.
Transmission electron microscopy
Transmission electron microscopy imaging was performed at the High Resolution Electron Microscopy Facility at UTMDACC. Samples fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.3, were washed in 0.1 M cacodylate buffer and treated with 0.1% Millipore-filtered buffered tannic acid, postfixed with 1% buffered osmium tetroxide for 30 min, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were washed several times in water, then dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for about 2 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica, Deerfield, IL), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques Corp, Danvers, MA). For quantification, we examined 3 to 9 cells magnified at 5000x per group and counted the number of mitochondria per cell.
Targeted Mass Spectrometry Analysis
For cultured cells as well as FACS purified cells, 4 ml or 400 ml of 80% LC-MS grade methanol was added to each 10 cm2 dish or FACS samples respectively and incubated at −80°C for 15 minutes. Cells were scrapped off the dish using a sterile cell scrapper and collected from plate to be centrifuged at 14,000 rpm for 5 minutes in cold room to pellet cell debris and proteins. Supernatants were saved. Pellets were resuspended in 500 μl 80% methanol by vortexing and subsequently centrifuged as before. Supernatants were centrifuged one final time at 14,000 rpm for 10 minutes at 4°C. Metabolite extractions were dried to a pellet by SpeedVac with no heat. Samples were re-suspended using 20 μL LC-MS grade water and 10 μL were injected and analyzed using a 5500 QTRAP hybrid triple quadrupole mass spectrometer (AB/Sciex) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (SRM). 254 endogenous water-soluble metabolites were targeted for steady-state analyses of samples. Some metabolites were targeted in both positive and negative ion mode via positive/negative polarity switching for a total of 289 SRM transitions. ESI voltage was +4900V in positive ion mode and –4500V in negative ion mode. The dwell time was 3ms per SRM transition and the total cycle time was ~1.56 seconds. Approximately 10-12 data points were acquired per detected metabolite. Samples were delivered to the MS via normal phase chromatography using a 4.6 mm i.d × 10 cm Amide XBridge HILIC column (Waters) at 350 μL min−1. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 35% B from 0-3.5 minutes; 35% B to 2% B from 3.5-11.5 minutes; 2% B was held from 11.5-16.5 minutes; 2% B to 85% B from 16.5-17.5 minutes; 85% B was held for 7 minutes to re-equilibrate the column. Buffer A was comprised of 20 mM ammonium hydroxide/20 mM ammonium acetate (pH 9.0) in 95:5 water:acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/Sciex). Metabolomics data analysis was done in part using Metaboanalyst software (www.metaboanalsyst.ca <http://www.metaboanalsyst.ca>). For glucose isotopic tracer experiments, cells were placed in glucose-free media supplemented with 10% dialyzed serum and with uniformly labeled [U-13C6] glucose (Cambridge Isotope Laboratories) for 12 hrs before extraction for LC-MS/MS analyses. A set of SRM transitions was used to target the heavy form of each metabolite.
Invasion and migration assays
For PGC-1α gene expression analysis of collected cells directly following migration, uncoated polycarbonate membranes (8 μm pore) were used. The cells were seeded in the upper chamber and the migrated cells in the lower chamber were collected 12 hrs following seeding. For invasion assays, the polycarbonate membranes were coated on both sides with Matrigel and cells on the basal side of the membrane (post migration) were fixed in 100% ethanol and stained with hematoxylin before microscopic evaluation. For the scratch/migration assay, the cell free area was measured 24 hrs after scratching the dish, and 0.1 μM rotenone was used.
Anoikis assay
5.106 cells were starved in 0.5% FBS for 24 hrs. The cells were then counted and resuspended in 13 ml serum free DMEM in 15ml-Falcon tube and allowed to rock at 37°C for 24 hrs. The cells were then pelleted and counted using a hemocytometer. Triplicate measurements of the two cell counts were used to determine the percent viability.
Type I collagen contractibility assay
5.104 cells per well of 24-well plates were seeded on 3 mg ml−1 type I collagen gel. Stressed matrix is allowed to contract for 48 hrs and released. Collagen gel size change (average gel area) was measured with a ruler 24 hrs following release of stressed matrix.
FACS
Tumors were resected, minced, and digested in 400U/ml type II collagenase at 37°C while shaking. Single cell suspension following filtering through 75 μm mesh were fixed in BD Cytofix/Cytoperm (BD Biosciences) and stained in 2% FBS containing PBS with DMEM with anti mouse αSMA primary antibody and TRITC conjugated secondary antibody. All FACS analyses were performed at the Joslin Diabetes Center Flow Cytometry Core, Boston, MA. FACS purified cells were spun down at 5,000 rpm for 10 minutes at 25°C and cell pellet processed for quantitative PCR analysis using Cells-to-cDNA kit (Ambion) according to the manufacturer's direction. CCC and PCC were also FACS purified and cytospin (onto glass slides at 800rpm for 5 min) stained following a 10 min acetone fixation step at 4°C.
Immonostaining
Thin frozen sections (5 μm) were immunolabeled and quantitation of immunolabeling was performed as previously described(34). Antibodies related information for stainings on mouse tissues and cells: PGC1-a (1:200; clone 4C1.3, Calbiochem™ ST1202), Twist (1:200; Santa Cruz Biotechnology, inc. H-81), CK8 (1:200; DSHB TROMA-I) and αSMA (1:200; clone 1A4, Sigma-Aldrich® F3777).
Western blot analyses
Western blot analyses were performed as previously described(34), using anti-PGC-1α antibody (1 μg ml−1; clone 4C1.3, Calbiochem™ ST1202) as recommended by the manufacturer. In Figure 3B, the results were obtained with samples from the same experiment and on two separate blots that were processed in parallel.
Patient information and data collection
For data presented in Fig. 7A, patients were diagnosed with breast cancer and tumors were surgically resected at the Department of Gynecology, University Medical Center Hamburg-Eppendorf (Hamburg, Germany). Written informed consent was obtained and the study was approved by the University Medical Center Hamburg-Eppendorf institutional review board. Material collection and processing was previously described(41) and RNA was characterized from patients diagnosed with invasive ductal carcinoma (IDC, all early stage estrogen receptor responsive primary tumors) with known bone marrow aspirate positivity status. Detection of disseminated tumor cells in bone marrow was performed with anti-cytokeratin monoclonal antibody A45-B/B3 as previously described(42) and according to international standards(43). Patient information details are provided in Supplementary Table 1. For data presented in Fig. 7B-D, 161 de-identified samples were collected from patients with IDC recruited for biopsy at the A. C. Carmargo Hospital in Sao Paulo, Brazil, after informed consent from patients and approval by the institutional review board. The tissue microarrays (TMAs) were constructed from 1.50 mm cores of formalin-fixed paraffin-embedded breast tissue. Immunohistochemistry was performed using the anti-PGC1α antibody (Calbiochem ST1202, clone 4C1.3, 1 μg ml−1) as recommended by the manufacturer. Immunohistochemistry evaluation was performed by selecting invasive cancer cells, as defined by isolated cells, the smallest isolated nests of cells, cells from the border of solid tumors, cells which were invading adipose tissue, stroma or micro emboli. The staining intensity for PGC1α in these cells was automatically measured using APERIO system (Vista, CA, USA). Martingale residual plot was used for assessing a threshold for PGC1α, and this threshold was used for correlative analyses with clinical variables (Supplementary Table 2 and Fig. 7D) and survival (Fig. 7C). In Fig. 7C, clinical information was missing for 3 patients to define disease free survival. The observers were blinded when performing these analyses. For data presented in Fig. 7E, four patients (Supplementary Table 3) were recruited for circulating cancer cells enrichment at the A. C. Carmargo Hospital in Sao Paulo, Brazil, after patient informed consent and approval by the institutional review board was obtained. Patient peripheral blood sample (8 ml) was collected into an EDTA tube and diluted 1:10 with ISET Buffer™ (RareCell Diagnostics, Paris, France) (44). An additional 13 IDC patients with lung metastasis were recruited for CTC evaluation as described above (Supplementary Table 4). After 10 minutes at room temperature the sample was loaded into a well of ISET Block™ (RareCell Diagnostics, Paris, France), with 8 μm-diameter cylindrical pores, above a polycarbonate membrane. The blood sample was filtered by aspiration under vacuum of -10kPa and then the membrane was washed by aspiration with PBS buffer. The membrane was disassembled from the ISET Block™, allowed to air-dry and stored at -20°C. The ISET membranes were then hydrated with 70μl of Tris-Saline Buffer (TBS) for 10 min followed by vertical draining on absorbent paper towel. In a moist chamber, the cells were permeabilized with 70 μl of TBS 0.2% Triton X-100, 5 min at room temperature. The membranes were rinsed with TBS, incubated for 15 min in the dark at room temperature with 3% hydrogen peroxide solution, and rinsed again with TBS. The membranes were then stained using antibodies against CK8 (1:50; clone NCL-L-CK8-TS1, Novocastra™), PGC-1α (1:200; clone 4C1.3, Calbiochem™) or CD45 (clone 2B11 + PD7/26, Dako™). For negative controls, primary antibodies were omitted. The primary antibodies were allowed to incubate for 1 hour at room temperature. The membranes were then rinsed with TBS and developed using the Dual long system HRP (Dako™) (30 min, room temperature) and DAB chromogen (Dako™) and Permanent Red (Dako™) (5 min, room temperature). The membranes were then rinsed with deionized water twice and counterstained with Harrys hematoxylin for 1 minute. On average 1.73 CTC in 1 ml of blood were analyzed.
Statistical Analyses
Significance was determined by One-way ANOVA or unpaired two-tailed Student's t-test and p<0.05 was considered statistically significant. Correlative analysis for PGC-1α transcript levels in cancer cells laser-microdissected from breast tumors of IDC patients (the median dCt was cut off to define “high” and “low” PGC-1α transcript level) with bone marrow DTC status (positive or negative) employed a one-sided χ2 test. Association between clinical characteristics and PGC-1α expression levels was verified by a two-tailed Fisher's exact test. For survival analysis, Kaplan–Meier curves were drawn and differences between the curves were calculated by the log-rank test(45) using R Foundation for Statistical Computing (Vienna, Austria, 2010) software. *p<0.05 was considered statistically significant. Analysis of microarray data was performed using Metacore (GeneGo) and Knowledge Based Pathway (IPA) (p<0.05).
Supplementary Material
Acknowledgements
This study was primarily supported by funds from the Cancer Prevention and Research Institute of Texas and funds from MD Anderson Cancer Center (MDACC). JTO was funded by the DoD Breast Cancer Research Predoctoral Traineeship Award (W81XWH-09-1-0008). R.K. is supported by NIH Grants CA125550, CA155370, CA151925, DK081576 and DK055001. Mass spectrometry work was partially supported by CA12096405 (JMA), CA00651646 (JMA). We wish to thank Dr. Bruce Spiegelman and Dr. Jennifer Estall (Dana Farber Cancer Institute, Boston, MA) for providing us with reagents related to PGC1α. We thank Dr. Marina Protopopova (MDACC, Houston, TX) and Dr. Florian Muller (MDACC, Houston, TX) for their help with the Seahorse experiments. We thank Dr. Lew Cantley (BIDMC, Boston, MA) for his critical reading of the manuscript. We also thank Min Yuan and Susanne Breitkopf (BIDMC, Boston, MA) for their help with mass spectrometry experiments, and Girijesh Buruzula and Joyce LaVecchio at the Joslin Flow Cytometry Core Facility (Joslin Diabetes Center, Boston, MA) for helping with all flow cytometry experiments. For electron microscopy imaging, the High Resolution Electron Microscopy Facility at UTMDACC is supported by the Institutional Core Grant CA16672.
Footnotes
Conflict of Interest: None
Authors Contribution
JTO performed data analyses and helped with the preparation of figures; KNGH performed experiments; HWK, KP, and MCH helped with data analyses; FMD, LTDC, RMR, JMA performed experiments and analyzed data, AD performed statistical analyses, VSL performed experiments, analyzed the data and contributed to the design of the experiment, writing of the manuscript and preparation of figures, RK contributed to the conceptual design of the study and provided advise regarding experiments and writing of the manuscript.
Competing Financial Interest statement:
The authors have no competing financial interests.
Gene expression array accession number:
The gene expression array data was deposited in Gene Expression Omnibus database (accession number GSE37344).
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956 Feb 24;123:309. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009 May 22;324:1029. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012 Mar 20;21:297. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008 Jan;7:11. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011 Oct 5;14:443. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004 Nov;4:891. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 7.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992 Mar 15;52:1399. [PubMed] [Google Scholar]
- 8.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009 Jun;119:1420. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002 Jun;2:442. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999 Jul 9;98:115. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 11.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998 Mar 20;92:829. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 12.Girnun GD. The diverse role of the PPARgamma coactivator 1 family of transcriptional coactivators in cancer. Semin Cell Dev Biol. Jan;21:2012. doi: 10.1016/j.semcdb.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhalla K, et al. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011 Nov 1;71:6888. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Errico I, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proceedings of the National Academy of Sciences of the United States of America. 2011 Apr 19;108:6603. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992 Mar;12:954. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooke VG, et al. Pericyte depletion results in hypoxia-associated epithelial to-mesenchymal transition and metastasis mediated by met signaling pathway. Cancer cell. 2012 Jan 17;21:66. doi: 10.1016/j.ccr.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007 Jul;104:13. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 18.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha). J Biol Chem. 2003 Mar 14;278:9013. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 20.Woelfle U, et al. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003 Sep 15;63:5679. [PubMed] [Google Scholar]
- 21.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim Biophys Acta. 2011 Jun;1807:552. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011 May;11:325. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 23.Hu J, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012 Feb 17;148:651. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006 Oct 20;127:397. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 25.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006 Oct 19;443:787. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 26.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008 Mar;10:295. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 27.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013 Feb 1;339:580. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones AW, Yao Z, Vicencio JM, Karkucinska-Wieckowska A, Szabadkai G. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion. 2012 Jan;12:86. doi: 10.1016/j.mito.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Gibbins JR. Epithelial cell motility: the effect of 2-deoxyglucose on cell migration, ATP production, and the structure of the cytoplasmic ground substance in lamellipodia of epithelial cells in culture. Cell Motil. 1982;2:25. doi: 10.1002/cm.970020104. [DOI] [PubMed] [Google Scholar]
- 30.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009 Sep 3;461:109. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumacher D, Strilic B, Sivaraj KK, Wettschureck N, Offermanns S. Platelet-derived nucleotides promote tumor-cell transendothelial migration and metastasis via P2Y2 receptor. Cancer cell. 2013 Jul 8;24:130. doi: 10.1016/j.ccr.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004 Jul 1;64:4670. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- 33.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007 Jul;104:13. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 34.Cooke VG, et al. Pericyte depletion results in hypoxia-associated epithelial-to-mesenchymal transition and metastasis mediated by met signaling pathway. Cancer cell. 2012 Jan 17;21:66. doi: 10.1016/j.ccr.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell JT, et al. VEGF-A and Tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proceedings of the National Academy of Sciences of the United States of America. Sep 20;108:16002. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992 Mar;12:954. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992 Mar 15;52:1399. [PubMed] [Google Scholar]
- 38.Castle JC, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012 Mar 1;72:1081. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 39.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012 Mar 29;483:603. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm EA, Jones BE, Radda GK, Veech RL, Clarke K. Increased uncoupling proteins and decreased efficiency in palmitate-perfused hyperthyroid rat heart. Am J Physiol Heart Circ Physiol. 2001 Mar;280:H977. doi: 10.1152/ajpheart.2001.280.3.H977. [DOI] [PubMed] [Google Scholar]
- 41.Woelfle U, et al. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003 Sep 15;63:5679. [PubMed] [Google Scholar]
- 42.Braun S, et al. Cytokeratin-positive cells in the bone marrow and survival of patients with stage I, II, or III breast cancer. N Engl J Med. 2000 Feb 24;342:525. doi: 10.1056/NEJM200002243420801. [DOI] [PubMed] [Google Scholar]
- 43.Fehm T, et al. A concept for the standardized detection of disseminated tumor cells in bone marrow from patients with primary breast cancer and its clinical implementation. Cancer. 2006 Sep 1;107:885. doi: 10.1002/cncr.22076. [DOI] [PubMed] [Google Scholar]
- 44.Hofman V, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology. 2012 Feb;23:30. doi: 10.1111/j.1365-2303.2010.00835.x. [DOI] [PubMed] [Google Scholar]
- 45.Balakrishanan N, Rao CR. Handbook of Statistics 23. Advances in Survival Analyses. Handbook of Statistics 23. Advances in Survival Analyses. 2004;27 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.