Abstract
MicroRNAs (miRNAs/miRs) belong to a class of small non-coding RNAs that can negatively regulate messenger RNA (mRNA) expression of target genes. miRNAs are involved in multiple aspects of ovarian cancer cell dysfunction and the phenotype of ovarian cancer cells can be modified by targeting miRNA expression. miRNA profiling has detected a number of candidate miRNAs with the potential to regulate many important biological functions in ovarian cancer, but their role still needs to be clarified, given the remarkable heterogeneity among ovarian cancers and the context dependent role of miRNAs. This review summarizes the data collected from The Cancer Genome Atlas (TCGA) and several other genome-wide projects to identify dysregulated miRNAs in ovarian cancers. Copy number variations (CNVs), epigenetic alterations, and oncogenic mutations are also discussed that impact miRNA levels in ovarian disease. Emphasis is given to the role of particular miRNAs in altering expression of genes in human ovarian cancers with the potential to provide diagnostic, prognostic and therapeutic targets. Particular attention has been given to TP53, BRCA1/2, CA125 (MUC16), HE4 (WFDC2), and imprinted genes such as ARHI (DIRAS3). Better understanding of the abnormalities in miRNA expression and downstream transcriptional and biological consequences will provide leads for more effective biomarkers and translational approaches in the management of ovarian cancer.
Keywords: microRNA, ovarian cancer, biomarker
INTRODUCTION
In the United States, ovarian cancer is the most lethal gynecologic malignancy in women with 22,240 estimated new cases and 14,030 estimated deaths in 2013 (1). With advances in diagnosis and treatment, 5-year survival has improved significantly over the last three decades, but the overall cure rate remains at 30% (2). Poor outcomes relate to late diagnosis and persistence of dormant, drug resistant cancer cells (2). If we are to improve clinical outcomes, we must take advantage of contemporary technologies to identify the molecular alterations that occur in ovarian cancer and to define the heterogeneity that is observed within and between cancers from different patients. Having identified these changes, we can better develop strategies for earlier diagnosis or more effective therapy.
MicroRNAs (miRNAs) are small non-coding RNAs of 19–25 nucleotides that can modulate gene expression by hybridizing to complementary target mRNAs, resulting in either mRNA degradation or direct inhibition of translation. miRNA can also activate gene expression by interacting with complementary regions found in the promoter and coding region, as well as the 3’UTR of mRNA targets (3). Expressed miRNAs provide a novel layer of regulation for human gene expression and play important roles in diverse biological processes (4) including carcinogenesis (5). Alterations in miRNAs have been detected in human ovarian cancers (6). The Cancer Genome Atlas (TCGA) project has recently published an integrated analysis of nearly 500 high-grade serous ovarian cancers that clearly documents multiple changes in miRNA levels (7). In this review, we focus on changes in miRNA expression in ovarian cancers and their potential application for earlier detection, more accurate prognostication and more effective treatment of the disease. Several miRNAs are predicted to regulate a number of clinically relevant genes in ovarian cancer such as MUC16 (CA125), WFDC2 (HE4), and several imprinted tumor suppressor genes such as DIRAS3 (ARHI) that are downregulated in ovarian cancer.
Dysregulation of miRNAs has been detected by miRNA profiling of ovarian cancers
Several studies have compared expression of miRNAs in ovarian cancers to whole normal ovaries, primary ovarian surface epithelial cells (OSE) and immortalized OSE (8–11). Among these reports, 310 dysregulated miRNAs in ovarian cancers have been reported. Of these 310 miRNAs, 34 miRNAs were found to be consistently dysregulated in ovarian cancers from at least three independent studies (Table 1.1 and Table 1.2) (8, 9, 12–15). Several miRNAs that regulate growth in other cancer types are downregulated in ovarian cancers (Table 1.1 and Table 1.2), including let-7a/b/d/f, miR-31, miR-34abc, miR-125b, and miR-127. Other oncogenic miRNAs such as miR-20a, miR-23a/b, and miR-200b/c are up-regulated in ovarian cancers (Table 1.1 and Table 1.2).
Table 1.1.
Consistently deregulated miRNAs in ovarian cancers.
| miRNAs | Alteration | Counterpart | Effect | Mechanism of Deregulation |
Targets | Associated References |
|---|---|---|---|---|---|---|
| Let- 7a/b/d/f |
Down- regulated |
HOSE, IOSE, ovary, fallopian tube from fimbriated end |
tumor suppressor |
promoter methylation, copy number variations |
KLK10, HMAG2 |
10,11,16,17,20,42,62,63 |
| miR-22 | HOSE | tumor suppressor |
ARRB1, CLIP2, EVI1,FRAT2, EDC3 |
8–10, 17, 20 | ||
| miR-31 | HOSE | tumor suppressor |
copy number variations |
E2F2, STK40, CEBPA |
9, 10 | |
| miR- 34a/b/c |
IOSE, ovary, fallopian tube from fimbriated end |
tumor suppressor |
promoter methylation, copy number variations and p53 mutation |
MET, CDK4 | 16, 17, 20, 23, 63 | |
| miR-125b | HOSE, IOSE, ovary, fallopian tube from fimbriated end |
putative tumor suppressor |
BCL3, VEGF, HIF-1α, HER3 |
10, 14, 16, 17, 41, 42, 62, 65 | ||
| miR-127- 3p |
HOSE, NOSE, ovary, serum |
related to drug-resistant |
imprinting, copy number variations, promoter methylation |
8, 10, 17, 20 | ||
| miR-152 | HOSE, IOSE, fallopian tube from fimbriated end |
putative tumor suppressor |
promoter methylation |
9, 10, 16, 20 | ||
| miR-155 | IOSE, blood, serum |
putative tumor suppressor |
20, 63 | |||
| miR- 181a-3p |
HOSE, ovary, blood |
copy number variations, promoter methylation |
10, 17 | |||
| miR-382 | HOSE | copy number variations, promoter methylation |
8–10 |
Table 1.2.
Consistently deregulated miRNAs in ovarian cancers.
| miRNAs | Alteration | Counterpart | Effect | Mechanism of Deregulation |
Targets | Associated References |
|---|---|---|---|---|---|---|
| miR- 15a/16 |
Up- regulated |
HOSE, fallopian tube from fimbriated end |
promoter methylation |
Bmi-1 | 7–10, 13, 16, 41, 42 | |
| miR-20a | HOSE, fallopian tube from fimbriated end |
oncogenic miRNA |
APP | 8, 9, 16, 42, 66 | ||
| miR-23a/b | ovary | copy number variationspromoter methylation |
7, 41, 42 | |||
| miR- 30a/b/c |
HOSE, IOSE, fallopian tube from fimbriated end |
related to drug-resistant |
copy number variations |
AVEN, GALNT1 |
8–10, 16–17, 41 | |
| miR-92 | HOSE, fallopian tube from fimbriated end |
putative oncogenic miRNA |
8–10, 16, 41 | |||
| miR-93 | ovary | putative oncogenic miRNA |
copy number variations, promoter methylation |
41, 42 | ||
| miR-106a | HOSE, fallopian tube from fimbriated end |
putative oncogenic miRNA |
7, 10, 16, 41 | |||
| miR-146b | HOSE, IOSE, fallopian tube from fimbriated end |
putative oncogenic miRNA |
copy number variations |
16, 63 | ||
| miR-182 | HOSE, IOSE, ovary fallopian tube from fimbriated end |
putative oncogenic miRNA(67) |
copy number variations, promoter methylation |
PDCD4 | 7–8, 16–17, 20, 68 | |
| miR-200 | HOSE, ovary fallopian tube from fimbriated end |
oncogenic miRNA |
copy number variations |
ZEB, c-Myc, TUBBIII, FN1, NTRK2, QKI |
7, 10, 12, 16, 17, 42, 62, 69 | |
| miR-203 | HOSE, ovary fallopian tube from fimbriated end |
promoter methylation |
9–10, 16–17 | |||
| miR205 | HOSE, ovary fallopian tube from fimbriated end |
putative oncogenic miRNA |
promoter methylation |
10, 16–17 | ||
| miR-223 | HOSE | putative oncogenic miRNA |
SEPTIN6, MMP9, USF2 |
9–10, 41 |
High grade serous ovarian cancers exhibit distinctive changes in miRNA expression
Ovarian cancers are remarkably heterogeneous at the cellular and molecular level and can be divided into type I low-grade and type II high-grade cancers based on histologic appearance and molecular profile. More than 70% of ovarian cancer related deaths occur in patients with advanced stage, high grade serous ovarian cancer (7). High grade cancers are characterized by multiple copy number abnormalities, TP53 mutation and epigenetic changes. When alterations in BRCA1 and BRCA2 occur, they are most frequently associated with high grade serous ovarian cancers.
Mining the TCGA data, Miles et al identified seventeen miRNAs that were dysregulated in high grade serous cancers when compared to normal ovarian samples, including eight up-regulated miRNAs (miR-183-3P, miR-15b-3p, miR-15b, miR-590-5p, miR-18a, miR-16, miR-96, and miR-18b) and nine down-regulated miRNAs (miR-140-3p, miR-145-3p, miR-143-5p, miR-34b-5p, miR-145, miR-139-5p, miR-34c-3p, miR-133a and miR-34c-5p) (16). In other reports that compared miRNA expression in ovarian cancers and normal ovarian tissues (17–19), five miRNAs were down-regulated (miR-140-3p, miR-143-5p, miR-34b-5p, miR-34c-5p and miR-145) and three were up-regulated (miR-96, miR-15b and miR-16) and these were among the top ten miRNAs from TCGA data listed in Table 1.1 and Table 1.2. These miRs could well contribute the pathogenesis of high-grade serous ovarian cancers, but their dysregulation needs to be confirmed in larger data sets and their functional roles need to be elucidated. Use of whole normal ovaries as a control in profiling is problematic. As epithelial cells comprise the majority of cells within a cancer but only a small subpopulation among cells within the normal ovary, apparent differences in miRNA expression could reflect differences in miRNA profiles between normal epithelial cells, granulosa-theca cells and germ cells. Epithelial cells that cover the ovary or that line the fallopian tube would provide more relevant as a control.
Copy number alterations regulate miRNAs
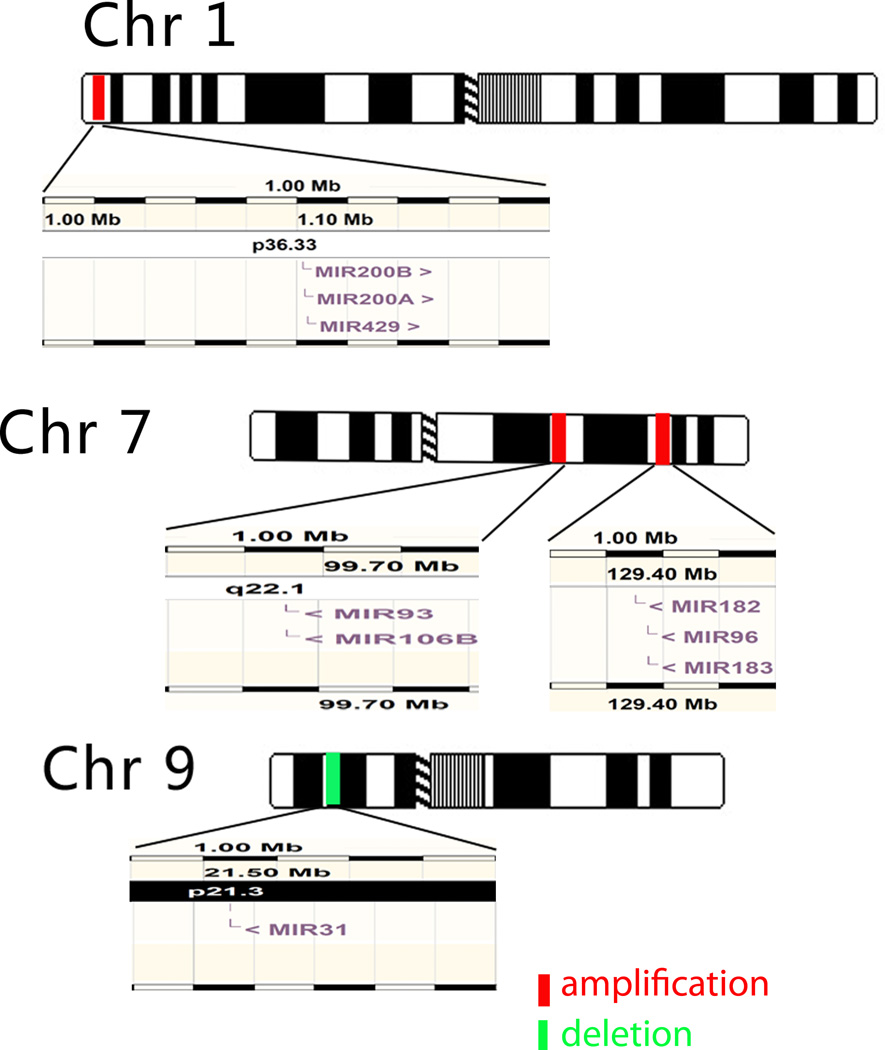
One of the characteristics of ovarian cancer is genomic instability (7). Chromosomal abnormalities are common in high grade serous ovarian cancers, as are alterations in DNA copy number (8). Overall, about 50% of miRNAs are found at fragile sites of chromosomes, as well as at the minimal regions of deletion, amplification or common chromosome breakpoints associated with different cancers (20). Chromosome abnormalities that involve miRNAs are not random events (4). Alterations of DNA copy number could account for much of the miRNA dysregulation in ovarian cancers (21). Through a high-resolution array-based genomic hybridization study of 227 human cancer samples, Zhang et al found that certain genomic loci containing miRNA genes were frequently altered in human ovarian cancers, breast cancers and melanomas (22). There were 26 miRNAs consistently associated with copy number gains and 15 miRNAs consistently associated with copy number losses in all three-cancer types (22). Down-regulation of eight potential tumor suppressor miRNAs (miR337, mir376a, miR376b, miR432, miR368, miR495, miR377 and miR410) mapped to a deletion in chromosome 14 (Dlk1-Gtl2 domain) and correlated with poor survival in epithelial ovarian cancer (23). Furthermore, the positive correlation between copy number and dysregulation of five miRNAs has been repeatedly confirmed by a number of studies involving miR-31 in 9p21; miR-93 in 7q22.1; miR-182 in 7q32.2; and miR-200b/429 in 1p36 (Fig. 1) (10, 21, 22, 24).
Fig 1.
Positive correlation between DNA copy number and dysregulation of miRNAs. The up-regulated miRNAs mir-200b/429, mir-93 and mir-182 are located on 1p36, 7q22.1, in 7q32.2, respectively. These regions are amplified in some ovarian cancers. mir-31 is located in a deleted region 9p21.3. A positive correlation between the alteration of copy number and the miRNAs listed above has been confirmed. The diagrams for miRNA genomic location come from http://www.ensembl.org.
In the TCGA data, several miRNAs are located in amplified or deleted genomic regions (25). Downregulation of let-7b is related to recurrent hemizygous genomic loss (86% of samples) and homozygous deletion (7% of samples). miR-31 is another frequently deleted miRNA. By contrast, miR-30 family members, located at two different focally amplified loci (8q24 and 1p34), are the most frequently amplified miRNAs, and copy number correlates with the expression of mature miRNA (25). Creighton et al computed the correlation between miRNA and its host gene expression and indicated that miRNA-host gene pairs tended to be highly correlated with each other, with 52% of the miRNA-host gene pairs showing significant positive correlation (25). Cyclin E1 (CCNE1), Notch3, HBXAP/Rsf-1, AKT2 and PIK3CA are among the most frequently amplified genes in high grade serous ovarian cancer (26). No known miRNA is found within 2Mb downstream of CCNE1, HBXAP/Rsf-1 or PIK3CA. Downstream of Notch3, however, miR-23a (19p13, negative strand, −13947483:−13947389) has been shown to be consistently up-regulated in ovarian cancers in different studies (11). MiRNA-641 (19q13.2, negative strand, −40788533: − 40788510) is located near the amplicon that contains AKT2, but miRNA-641 is not overexpressed in high grade serous ovarian cancers.
Epigenetic alterations regulate miRNAs
In addition to copy number changes, Iorio et al found that miR-21, miR-203, and miR-205 were overexpressed in ovarian cancers and that levels could be further increased in the ovarian cancer cell line OVCAR3 by incubation with the demethylating agent 5-aza-2-deoxycytidine (5-AZA), suggesting that these miRNAs might be regulated by methylation (18). Zhang and colleagues treated five ovarian cancer cell lines with 5-AZA and a histone deacetylase inhibitor 4-phenylbutyric acid and found that 16 of 44 (36.4%) miRNAs down-regulated in advanced stage ovarian cancer could be restored using these drugs (21). Recently, the hypermethylation of tumor suppressor mir-34a and mir-34bc has also been confirmed in ovarian cancer patients with decreased mir-34 (19, 24). Thus, epigenetic alteration is also an important mechanism for miRNA dysregulation.
TP53 regulates miRNAs
TP53 mutation is found in at least 96% of high grade serous ovarian cancers and can regulate miRNAs. As the miRNA-34 family is upregulated by wild type TP53, expression of miRNA 34a was decreased in 100% and 34b and 34c in 72% of cancers with TP53 mutation (17). In addition to genomic deletion, TP53 mutation may also be responsible for the underexpression of miR-31 (10).
miRNAs can downregulate BRCA1 and BRCA2 expression
Approximately 15% of ovarian cancer patients have a strong family history associated with germ line mutations of BRCA1, BRCA2, mismatch repair genes or, on rare occasions, TP53 (27). While mutations of BRCA1 and BRCA2 can affect gene expression profiles, at least one study found that a fraction of high grade serous ovarian cancers exhibited BRCA1/2 associated abnormalities in the absence of mutation (17). miRNAs can downregulate wild-type BRCA1 expression. A G to C polymorphism (rs2910164) in the miR-146a precursor leads to mismatch in its stem region. This variant allele can increase miR-146a expression as well as the binding capacity between miR-146a and the 3'-untranslated region (UTR) of BRCA1. Thus, miR-146a can bind to the 3'-UTRs of BRCA1 and BRCA2 mRNAs and potentially modulate their expression. The rs2910164 polymorphism of miR-146a may affect the age of cancer onset. Patients who had at least one miR-146a variant allele were diagnosed at a younger age than women without a variant allele (28).
Low grade serous ovarian cancers exhibit distinctive changes in miRNA expression
Only 10% of ovarian cancers are low grade. Clinically, these cancers grow slowly and present in early stage, but are not as responsive as high grade cancers to platinum and taxane based therapy (29). More than half of low grade serous ovarian cancers are associated with mutations of KRAS, and smaller fractions exhibit mutations of BRAF, PTEN and PIK3CA (2). Less than 20 miRNAs differ in expression between low grade and high grade serous ovarian cancers (17). By analyzing the miRNA profiles of the NCI-60 panel of 60 human cancer cell lines, Patnaik et al found that mutation of BRAF and PTEN affects miRNA expression, but mutation of KRAS did not (30). Among the miRNAs related with mutant BRAF, 4 miRNAs (miR-509-3p, miR-30d, miR-30b-3p, miR-30b) have been reported to be upregulated in low grade serous ovarian cancers, when compared to normal fallopian tube (17). The BRAFV600E mutation can increase MAPK signaling, lead to higher levels of mature miRNAs and enhance miRNA processing in undifferentiated pleomorphic sarcoma (31), but its role in low grade serous cancer still needs to be explored. PTEN mutation is responsible for downregulation of miR-29b and miR-769-3p in cancer cell lines, PIK3CA mutation has not yet been linked to any change in miRNA expression.
Dysregulation of miRNAs and predicted targets tend to be anti-correlated in ovarian cancer gene expression
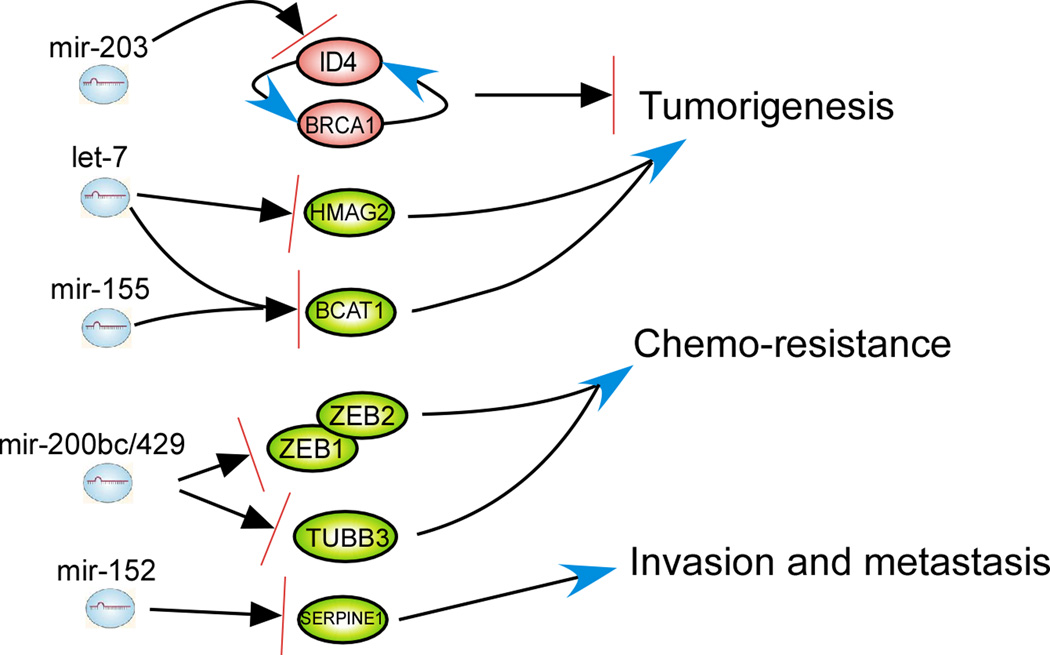
The function of miRNAs is determined by the genes and signaling pathways regulated by each miRNA. Having assembled the published transcriptome profiling data in ovarian cancer, we have integrated the results from both miRNA and transcriptome profiling to identify genes that are regulated by miRNAs. A number of differentially expressed genes in ovarian cancers have been reported (32). Approximately 200 miRNAs were anti-correlated with the 186 most differentially expressed genes using the miRNA algorithm Targetscan (http://www.targetscan.org/ Release 5.1 Apr 2009) (Table S1 and Table S2). Among these 186 genes, ten differentially expressed genes have been reported in at least two independent studies (33, 34). These ten gene changes are thought to associate with ovarian cancers and are summarized in Table 2 along with their potential regulating miRNAs. Three pairs of miRNA/mRNA association are of particular interest: 1) downregulation of tumor suppressor gene ID4 and miR-203 upregulation in ovarian cancers; 2) BCAT1 upregulation and downregulation of let-7, miR-125b & miR-155; and 3) SERPINE1 upregulation and miR-152 downregulation (Fig. 2). ID4 has a regulatory loop with another tumor suppressor BRCA1, which enables appropriate normal cycling during cell division. There is a modestly correlated downregulation between BRCA1 and ID4 (35). The BRCA1–ID4 regulatory loop might be disrupted in many breast and ovarian cancers (35). Upregulation of miR-203 may be responsible for this disruption. Since downregulation of ID4 has been indicated as a potential biomarker of recurrence in breast cancer (36), both ID4 and miR-203 might serve as disease biomarkers in ovarian cancers as well.
Table 2.
Ten consistently deregulated genes and their regulating miRNAs in ovarian cancers
| Genes | Gene ID | Alteration | Description | Effect | Regulating miRNAs |
Associated References |
|---|---|---|---|---|---|---|
| CLU | 1191 | clusterin | chemoresistance, prognosis |
70, 71 | ||
| ID4 | 3400 | down | inhibitor of DNA binding 4, dominant negative helix- loop-helix protein |
oncogenesis | miR-203 | 72 |
| BCAT1 | 586 | branched chain amino-acid transaminase 1, cytosolic |
chemo-resistance, oncogenesis |
let-7, miR-125b, miR-155 |
36, 73 | |
| FN1 | 2335 | fibronectin 1 | chemoresistance, prognosis |
74 | ||
| MAL | 4118 | mal, T-cell differentiation protein |
chemoresistance, prognosis |
31–32 | ||
| SERPINE1 | 5054 | up | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
invasion and metastasis |
miR-152 | 33 |
| SERPINA5 | 5104 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
putative tumor suppressor |
75 | ||
| SOX17 | 64321 | SRY (sex determining region Y)-box 17 |
||||
| TOP2A | 7153 | topoisomerase (DNA) II alpha 170kDa |
chemoresistance | 73 | ||
| THBS2 | 7058 | thrombospondin 2 |
metastasis | 33 |
Fig 2.
Potential function of dysregulated miRNA in ovarian cancers. Upregulation of mir-203 can inhibit the expression of ID4, disrupting the BRCA1–ID4 regulatory loop, which assures appropriate cycling of expression for both genes during normal cell division. Down-regulated let-7 family and mir-155 expression may be responsible for the overexpression of the oncogene BCAT1, a direct target of c-Myc. Low levels of let-7 also induce upregulation of embryonic genes such as HMAG2. These are also important in oncogenesis. Downregulation of ZEB1/2 and TUBB3 by mir-200b/c and 429 can increase sensitivity to paclitaxel of ovarian cancer cells. mir-152 may play a potential tumor suppressor role by targeting SERPINE1, an inhibitor of fibrinolysis, involved in tumor cell invasion and metastasis. An arrow indicates activation and an arrow with bar indicates inhibition.
BCAT1 is a direct target of c-myc, which is important in oncogenesis and amplified in a fraction of ovarian cancers (37). The let-7 family also performs a tumor suppressor role in many cancer types including ovarian cancers. MiR-125b is a putative tumor suppressor in ovarian cancers and can suppress cancer proliferation by targeting BCL3 (15). Although miR-155 levels are increased in B cell lymphoma (38), miR-155 has been consistently down-regulated in ovarian cancers with a potential tumor suppressor role in targeting BCAT1. SERPINE1 (also known as PIA-1) is an inhibitor of fibrinolysis and involved in tumor cell invasion and metastasis. MiR-152 is down-regulated in hepatocellular and gastric carcinomas, and acts as a tumor suppressor by targeting DNMT1 (39). In ovarian cancers, miR-152 might serve as a tumor suppressor by targeting SERPINE1 based on predictions of the Targetscan algorithm.
miRNAs regulate ovarian cancer-associated imprinted genes
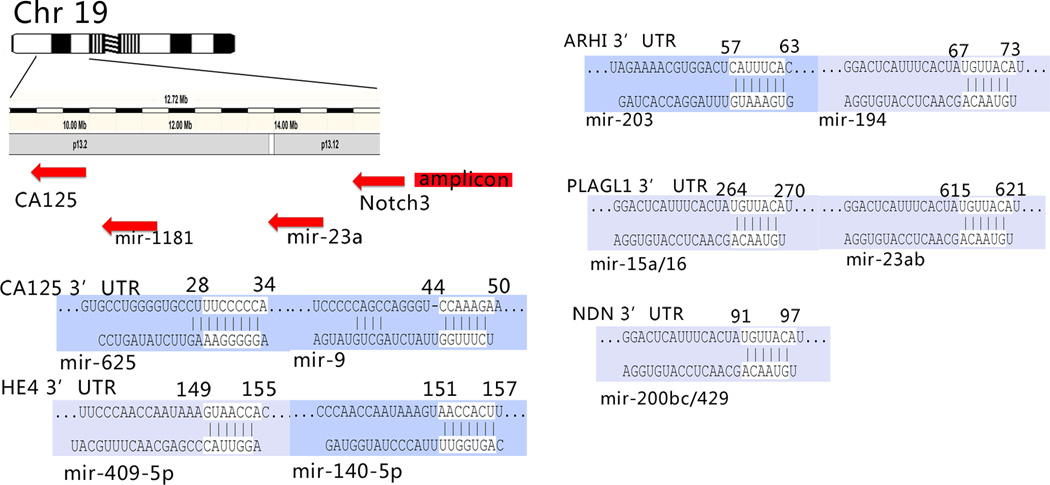
Genomic imprinting represents another level of regulation in gene expression where one allele of each autosomal gene pair is preferentially silenced depending upon its parent-of-origin, leaving only a single functional allele. Disruption of imprinted gene expression is linked to the initiation of malignancy (40). Of the candidate imprinted genes identified to date, four tumor suppressor genes including ARHI (DIRAS3), LOT1 (also known as PLAGL1/ZAC1), PEG3, and NDN are consistently down-regulated in ovarian cancers (2, 41). A Targetscan algorithm predicted that there are 53 miRNAs with poorly conserved binding sites in the 3’ UTR of ARHI, 2 conserved sites in PEG3, 15 conserved sites in PLAGL1, and 4 conserved sites in NDN. As shown in Fig. 3, ARHI-targeting miR-203 and miR-194, PLAGL1-targeting miR-15a/16 and miR-23a/b, and NDN-targeting miR200b/c/429 have been reported to be overexpressed in ovarian cancers (9–11, 17, 18, 42, 43). Among these miRNAs, miR-194 and miR-23a/b are overexpressed in a number of human cancers (44, 45). NDN-targeting miR200b/c/429 is associated with decreased progression-free survival and overall survival in ovarian cancer patients (43). ARHI-targeting miR-203 and PLAGL1-targeting miR-15a/16 have been reported as putative tumor suppressors in other cancer types (46). MiRNA-221 and 222 are also predicted to target ARHI and their negative regulation effects on ARHI gene has been confirmed in prostate cancer cells (47). However, dysregulation of miR-221 and 222 in ovarian cancers has not been observed in all studies (8, 10, 11, 18, 21, 42, 43). In addition, ARHI-targeting miR-371 and miR-181b/c are reported to be overexpressed in chemo-resistant biopsies and cell lines (48). MiRNA can be imprinted in normal physiological development and in oncogenesis (49).
Fig 3.
Predicted interaction of dysregulated miRNA and protein coding genes. CA-125 and HE4 are well characterized ovarian cancer biomarkers. An amplicon upstream of CA125 in Chr19 is responsible for the overexpression of the oncogene Notch3. mir-23a, mir-27a, mir-24-2, mir-199a-1 and mir-1181 are located between Notch 3 and CA-125, mir-23a has been reported to be up-regulated in different studies. Also shown are the predicted binding sites of dysregulated miRNAs in the 3' UTR of imprinted tumor suppressor genes. The annotation for the binding sites is taken from www.tagetscan.org.
miRNAs may serve as biomarkers and also regulate levels of protein biomarkers
Detection of altered levels of miRNA dysregulation in blood, serum and tumor-derived exosomes of cancer patients might provide biomarkers for early detection (50). Among the consistently dysregulated miRNAs in ovarian cancers listed in Table 1.1 and Table 1.2, two down-regulated miRNAs, let-7 family and miR-155, and five up-regulated miRNAs, miR-15/16 cluster, miR-20a, miR-92, miR-203 and miR-205 are found in the peripheral circulation of patients with ovarian cancers and represent promising biomarkers for early diagnosis (50). The miR-15/16 cluster, miR-20a, and miR-205 were also identified as the top ten up-regulated miRNAs in the TCGA data set (Table 1.1 and Table 1.2) (7).
CA125, also known as MUC16, is a well-characterized biomarker that is used to monitor the progression and regression of epithelial ovarian cancer (51). The complete sequence of the cDNA-encoding MUC16 has been determined (52). Based on our in silico analysis of the TCGA data base and literature reports, a number of miRNAs could potentially regulate the MUC16 gene (Table 3). Several miRNAs in this list have already been shown to be low in cancers. For example, miR-9 and miR-584 are downregulated in ovarian cancers when compared to normal ovary (18, 42, 53). miR-124 and miR-637 were also downregulated in ovarian cancer cell lines compared to IOSE (18, 21). If miRNAs in Table 3 are true regulators of CA125, down-regulation of these miRNAs could be one of mechanisms that lead to abnormal CA125 levels in ovarian cancers and could be potential biomarkers along with CA125 for patients with ovarian cancers.
Table 3.
Putative CA125-targeting miRNAs in ovarian cancers
| miRNAs | Chromosome Location |
Targets | Deregulation in cancers |
Potential link with chromosome abnormality in ovarian cancer |
|---|---|---|---|---|
| miR-9-2 | 5q14.3 | CA125, NFkB | Down- regulated |
Yes, deletion of 5q14 tumor suppressor loci |
| miR-9-3 | 15q26.1 | CA125, NFkB | Down- regulated |
Yes, deletion of 15q26 region |
| miR-124-1 | 8p23.1 | CA125, EZH2 | Down- regulated |
Yes, deletion of 8p23 region |
| miR-324-3p | 17p13.1 | CA125 | NA | Yes, LOH of 17p13 region |
| miR-544 | 14 | CA125, cMYC | NA | Yes, loss of 14q region |
| miR-584 | 5q32 | CA125, Rock1 | Down- regulated |
Yes, loss of 5q32 region |
| miR-625 | 14q23.3 | CA125 | NA | Yes, loss of 14q region |
| miR-637 | 19p13.3 | CA125, LIF | Down- regulated |
Yes, LOH of 19p13 region |
Additionally, the Notch3 amplicon is located upstream of the CA125 (MUC16) gene on Chromosome 19, in high grade serous ovarian cancer.(26) In addition to miR-23a mentioned above, several miRNAs, including miR-27a (−13947342:−13947241), miR-24-2(−13947183:−13947089), miR-199a-1 (−10928182:−10928090) and miR-1181 (−10514225:−10514121) map between Notch 3 and the CA125 (MUC16) gene (Fig. 3). miR-1181 increased serially in blood samples from patients diagnosed with relapsed ovarian cancers (22 serous and 2 endometrioid) when compared to age- and sex-matched volunteers without a history of cancer. So far, there is no report that links these miRNAs with CA125 levels.
Human epididymis secretory protein 4 (HE4) is a member of a family of whey acidic four-disulfide core proteins (WFCD2) that are secreted at high levels by normal endometrium and by endometrial and epithelial ovarian cancers. HE4 overexpression in ovarian cancers has been confirmed through microarray studies (54). Gene therapy targeting the promoter of HE4 can reduce the xenograft growth, block primary and metastatic tumors and prolong life span of mice with ovarian cancer (55). The HE4 ELISA assay has been shown to have a potential advantage over the CA125 assay in that it is less frequently positive in pre-menopausal women with non-malignant gynecologic conditions.(56) HE4 protein can also provide a useful biomarker in a small fraction of ovarian cancers that have little or no CA125 expression (57). A predictive model - ROMA (Risk of Ovarian Malignancy Algorithm) - utilizes the combination of HE4 and CA125 to triage patients with pelvic masses to gynecologic oncologists (58). The HE4 gene is located at 20q12-q13.2 without any known miRNAs that map within or near this region. After analysis with the Targetscan algorithm, seven miRNA binding sites were found in the 3’-UTR of the HE4 gene. Among these miRNAs, miR-140-5p and miR-409-5p are downregulated in ovarian cancers (Fig. 3) (9, 11). These two miRNAs that regulate the HE4 gene might serve as candidate miRNA biomarkers for detecting or monitoring ovarian cancers.
miRNAs might serve as potential prognostic biomarkers
Based on currently available data, some miRNAs have the potential to be prognostic biomarkers for ovarian cancer. Overexpression of miR-200 family members and miR-519a, under-expression of let-7 family members and miR-153 have been linked to a poor prognosis of patients with ovarian cancers (42, 43, 59). Under-expression of a tumor suppressor miR-9 in recurrent ovarian cancers is reported as a signature for recurrence (42). Low expression of miR-31 in ovarian cancers could also be an indicator for earlier disease recurrence and metastasis. Upregulation of miR-15a, miR-21 and miR-92 has been reported to signal recurrent disease. Let-7 family members have also served as prognostic biomarkers in ovarian cancer (43). Downregulation of miR-34a/b/c, miR449b, miR-503 and miR-507 has been observed in late stage and high grade ovarian cancers (17, 21, 24). However, at the present time the mechanism that underlies the change in the expression of miRNA in ovarian cancer recurrence and metastasis remains elusive.
miRNAs can provide predictive biomarkers for response or lack of response to treatment
miRNAs have been implicated in the initiation, progression, metastasis and chemoresistance of cancers at different sites. In theory, targeting miRNAs could provide a therapeutic strategy. Transfection of paclitaxel-resistant A2780 ovarian cancer cells with miR-27a inhibitors has been reported to reduce the expression of MDR1 mRNA and P-gp protein, to increase HIPK2 protein expression, and to enhance paclitaxel sensitivity (60). Exogenous expression of miR-31 has been shown to inhibit proliferation and induce significant p53-independent apoptosis in ovarian cancers (10). Considering that virtually all high-grade serous ovarian cancers exhibit p53 deficiency, miR-31-based therapy might be particularly effective in this subset of ovarian cancers (10). miR-152 and miR-185 can increase cisplatin sensitivity ovarian cancer in vitro and in vivo by targeting DNMT1 (61).
miRNAs could provide targets for therapy
miRNA mimics, adenovirus-associated vectors that express miRNAs, miRNA masks and miRNA sponges, are being developed to modulate the gain or loss of miRNA function (62). Many miRNA targets have been identified. Among the most consistently deregulated miRNAs in ovarian cancers are listed in Table 1.1, Table 1.2 and in Fig. 2 & 3. Targeting miR-203 may restore both the BRCA1–ID4 regulatory loop and expression of ARHI in ovarian cancers. Delivery of let-7, miR-125b, miR-155 and miR-152 might suppress tumorgenesis and metastasis by targeting BCAT1 and SERPINE1 (Table 1.1, Table 1.2 and Fig. 2). Additionally, about 50 dysregulated miRNAs have been linked to chemo-resistance or chemo-sensitivity to taxanes or platinum compounds (48). Overexpression of miR-27a and miR-514 and downregulation of let-7e have been related to development of resistance to taxanes and/or platinum. MiR-214 expression can induce platinum resistance by affecting PTEN function (63). Conversely, the upregulation of miR-378 and miR-625 has correlated with sensitivity to platinum based therapy.
Conclusions
Epithelial ovarian cancers are remarkably heterogeneous and this heterogeneity is reflected in dysregulation of multiple miRNAs. Different miRNA profiles are observed in high grade and in low grade cancers. miRNAs can be regulated by abnormalities in DNA copy number, methylation, histone acetylation and mutation in TP53. In turn, miRNAs can regulate BRCA1, BRCA2 and imprinted tumor suppressor genes such as ARHI (DIRAS3), Loss of certain miRNAs may upregulate ovarian cancer biomarkers such as CA125 (MUC16) and HE4 (WFCD2). Altered levels of miRNAs may also serve as potential biomarkers for detecting, monitoring, estimating resectability, determining prognosis and predicting response to conventional therapy. As methods are developed to manipulate miRNAs in the clinic certain miRs may also serve as targets for therapy.
Supplementary Material
Acknowledgments
This work was supported by the Anne and Henry Zarrow Foundation, the Kwang-Hua Education Foundation of Xi'an Jiaotong University, the Program for Changjiang Scholars and Innovative Research Team in University, the MD Anderson SPORE in Ovarian Cancer NCI P50 CA83639, MD Anderson CCSG NCI P30 CA16672, the National Foundation for Cancer Research, and kind gifts from Stuart and Gaye Lynn Zarrow and from Mrs. Dolores Wilkenfeld.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors declare no conflict of interest.
References
- 1.Latha TS, Panati K, Gowd DS, Reddy MC, Lomada D. Ovarian Cancer Biology and Immunotherapy. International reviews of immunology. 2014 doi: 10.3109/08830185.2014.921161. [DOI] [PubMed] [Google Scholar]
- 2.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions - beyond repression of gene expression. Nature reviews Genetics. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 4.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer. 2010;17:F77–F89. doi: 10.1677/ERC-09-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Network. TCGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumor progression in ovarian carcinoma. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;24:447–463. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahajan A, Liu Z, Gellert L, Zou X, Yang G, Lee P, et al. HMGA2: a biomarker significantly overexpressed in high-grade ovarian serous carcinoma. Mod Pathol. 2010;23:673–681. doi: 10.1038/modpathol.2010.49. [DOI] [PubMed] [Google Scholar]
- 13.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69:9090–9095. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan Y, Yao H, Zheng Z, Qiu G, Sun K. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer. 2011;128:2274–2283. doi: 10.1002/ijc.25575. [DOI] [PubMed] [Google Scholar]
- 16.Miles GD, Seiler M, Rodriguez L, Rajagopal G, Bhanot G. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes. 2012;5:164. doi: 10.1186/1756-0500-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CH, Subramanian S, Beck AH, Espinosa I, Senz J, Zhu SX, et al. MicroRNA profiling of BRCA1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PLoS One. 2009;4:e7314. doi: 10.1371/journal.pone.0007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 19.Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flavin R, Smyth P, Barrett C, Russell S, Wen H, Wei J, et al. miR-29b expression is associated with disease-free survival in patients with ovarian serous carcinoma. Int J Gynecol Cancer. 2009;19:641–647. doi: 10.1111/IGC.0b013e3181a48cf9. [DOI] [PubMed] [Google Scholar]
- 24.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creighton CJ, Hernandez-Herrera A, Jacobsen A, Levine DA, Mankoo P, Schultz N, et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One. 2012;7:e34546. doi: 10.1371/journal.pone.0034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama K, Nakayama N, Jinawath N, Salani R, Kurman RJ, Shih Ie M, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–2617. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 27.Aletti GD, Gallenberg MM, Cliby WA, Jatoi A, Hartmann LC. Current management strategies for ovarian cancer. Mayo Clin Proc. 2007;82:751–770. doi: 10.4065/82.6.751. [DOI] [PubMed] [Google Scholar]
- 28.Pastrello C, Polesel J, Della Puppa L, Viel A, Maestro R. Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. Carcinogenesis. 2010;31:2124–2126. doi: 10.1093/carcin/bgq184. [DOI] [PubMed] [Google Scholar]
- 29.Gershenson DM, Sun CC, Bodurka D, Coleman RL, Lu KH, Sood AK, et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecol Oncol. 2009;114:48–52. doi: 10.1016/j.ygyno.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Patnaik SK, Dahlgaard J, Mazin W, Kannisto E, Jensen T, Knudsen S, et al. Expression of microRNAs in the NCI-60 cancer cell-lines. PLoS One. 2012;7:e49918. doi: 10.1371/journal.pone.0049918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mito JK, Min HD, Ma Y, Carter JE, Brigman BE, Dodd L, et al. Oncogene-dependent control of miRNA biogenesis and metastatic progression in a model of undifferentiated pleomorphic sarcoma. J Pathol. 2013;229:132–140. doi: 10.1002/path.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berchuck A, Iversen ES, Luo J, Clarke JP, Horne H, Levine DA, et al. Microarray analysis of early stage serous ovarian cancers shows profiles predictive of favorable outcome. Clin Cancer Res. 2009;15:2448–2455. doi: 10.1158/1078-0432.CCR-08-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee PS, Teaberry VS, Bland AE, Huang Z, Whitaker RS, Baba T, et al. Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int J Cancer. 2010;126:1378–1389. doi: 10.1002/ijc.24797. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Sun MZ, Liu S, Yeh D, Yu L, Song Y, et al. Smad4 mediates malignant behaviors of human ovarian carcinoma cell through the effect on expressions of E-cadherin, plasminogen activator inhibitor-1 and VEGF. BMB Rep. 2010;43:554–560. doi: 10.5483/bmbrep.2010.43.8.554. [DOI] [PubMed] [Google Scholar]
- 35.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, et al. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci U S A. 2002;99:7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, et al. Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer. 2008;8:154. doi: 10.1186/1471-2407-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Yosef T, Yanuka O, Halle D, Benvenisty N. Involvement of Myc targets in c-myc and N-myc induced human tumors. Oncogene. 1998;17:165–171. doi: 10.1038/sj.onc.1201939. [DOI] [PubMed] [Google Scholar]
- 38.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, et al. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170–1179. doi: 10.1007/s11605-010-1202-2. [DOI] [PubMed] [Google Scholar]
- 40.Lim DH, Maher ER. Genomic imprinting syndromes and cancer. Adv Genet. 2010;70:145–175. doi: 10.1016/B978-0-12-380866-0.60006-X. [DOI] [PubMed] [Google Scholar]
- 41.Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, et al. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112:1489–1502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 42.Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 44.Mees ST, Mardin WA, Wendel C, Baeumer N, Willscher E, Senninger N, et al. EP300--a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. Int J Cancer. 2010;126:114–124. doi: 10.1002/ijc.24695. [DOI] [PubMed] [Google Scholar]
- 45.Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, et al. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010;277:3726–3734. doi: 10.1111/j.1742-4658.2010.07773.x. [DOI] [PubMed] [Google Scholar]
- 46.Viticchie G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Zaman MS, Deng G, Majid S, Saini S, Liu J, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila) 2011;4:76–86. doi: 10.1158/1940-6207.CAPR-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boren T, Xiong Y, Hakam A, Wenham R, Apte S, Chan G, et al. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol Oncol. 2009;113:249–255. doi: 10.1016/j.ygyno.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 49.Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 50.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 51.Scholler N, Urban N. CA125 in ovarian cancer. Biomark Med. 2007;1:513–523. doi: 10.2217/17520363.1.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin BW, Lloyd KO. Molecular cloning of the CA125 ovarian cancer antigen: identification as a new mucin, MUC16. J Biol Chem. 2001;276:27371–27375. doi: 10.1074/jbc.M103554200. [DOI] [PubMed] [Google Scholar]
- 53.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–5546. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 54.Schummer M, Ng WV, Bumgarner RE, Nelson PS, Schummer B, Bednarski DW, et al. Comparative hybridization of an array of 21,500 ovarian cDNAs for the discovery of genes overexpressed in ovarian carcinomas. Gene. 1999;238:375–385. doi: 10.1016/s0378-1119(99)00342-x. [DOI] [PubMed] [Google Scholar]
- 55.Huang YH, Zugates GT, Peng W, Holtz D, Dunton C, Green JJ, et al. Nanoparticle-delivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Res. 2009;69:6184–6191. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hellstrom I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, et al. The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res. 2003;63:3695–3700. [PubMed] [Google Scholar]
- 57.Rosen DG, Wang L, Atkinson JN, Yu Y, Lu KH, Diamandis EP, et al. Potential markers that complement expression of CA125 in epithelial ovarian cancer. Gynecol Oncol. 2005;99:267–277. doi: 10.1016/j.ygyno.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 58.Anastasi E, Marchei GG, Viggiani V, Gennarini G, Frati L, Reale MG. HE4: a new potential early biomarker for the recurrence of ovarian cancer. Tumour Biol. 2010;31:113–119. doi: 10.1007/s13277-009-0015-y. [DOI] [PubMed] [Google Scholar]
- 59.Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, et al. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology. 2010;57:734–743. doi: 10.1111/j.1365-2559.2010.03686.x. [DOI] [PubMed] [Google Scholar]
- 60.Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L, et al. MiR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol. 2010;119:125–130. doi: 10.1016/j.ygyno.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2013 doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 62.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.