ABSTRACT
MicroRNAs are 19–22 nucleotide RNAs involved in such important processes as development, proliferation, differentiation and apoptosis. Different miRNAs are uniquely expressed in lymphoid T cells, and play a role indevelopment and differentiation of various subtypes by targeting their target genes. Recent studies have shown that aberrant miRNA expression may be involved in T cell leukemogenesis and lymphogenesis, and may function as tumor suppressor (such as miR-451, miR-31, miR-150, and miR-29a) or oncogene (e.g. miR-222, miR-223, miR-17-92, miR-155). MiRNAs can be used as new biomarkers for prognosis and diagnosis or as an index of disease severity in T-cell leukemia and lymphoma. This article presents a review of studies in recent years on the role of miRNAs in T-cell development and their aberrant expression in pathogenesis of T-cell leukemia and lymphoma. Characterizing miRNAs can help recognize their role as new important molecules with prognostic and therapeutic applications.
Keywords: MicroRNA, Leukemia, Lymphoma, Tumor Suppressor, Oncogene
INTRODUCTION
MicroRNAs (miRNA) are small 19–22 nucleotide non-coding, post-transcriptional regulatory RNA molecules involved in regulation of gene expression and a variety of biological processes like cell development, proliferation, differentiation, apoptosis and hematopoiesis1,2. For miRNA biogenesis, Pri-miRNA with more than 1kb in length is first transcribed by RNA polymerase II, and is then converted to 70 nucleotide Pre-miRNA using a protein complex including nuclease Drosha and DiGeorge syndrome critical region gene 8 (DGCR8)3. Pre-miRNA is transported from nucleus to cytoplasm using exportin 5, and is converted to a double stranded 21–22 nucleotide miRNA by RNase III enzyme Dicer4. Only one strand of this mature miRNA is loaded on and incorporated with RNA-induced silencing complex (RISC). Single-strand mature miRNA ultimately drives RISC towards the 3'-UTR of the target mRNA to inhibit translation of mRNA or decrease stability5,6.
T cells are differentiated in thymus, and can be categorized by expression of CD4 and CD8 phenotypes. Development of thymocytes begins from double negative [DN (CD4- CD8-)] stage, continues with double positive [DP (CD4+ CD8+)] stage and ends in single positive [SP (CD4+ or CD8+)] stage circulating in blood and peripheral lymph nodes7. When confronted with an infectious agent, naive CD4+ T-cell can be differentiated to at least four effector lineages including T helper type 1 cells (Th1), Th2 cells, Th17 cells and regulatory T-cells (Treg cells), while naive CD8+ T-cell differentiates to cytotoxic effectors(8). Each of these populations has specific miRNA expression profiles, which participate in regulation of development from DN stage and in differentiation to different subtypes9. Dysregulated expression of miRNAs has been found to be involved in many cancers, including cancers of the immune cells10. In this paper, we first evaluate the role of different miRNAs in T-cell development and then altered expression of miRNAs in T-cell leukemia and lymphoma will be considered. Finally, prognostic and diagnostic importance and therapeutic use of miRNA will be discussed.
MicroRNAs and T-lymphocyte differentiation
Recent studies have shown that distinctive miRNAs are expressed in innate and acquired immune cells, and are involved in regulation of their development and function. Differentiation of various T-cell subgroups is regulated by targeting different proteins/molecules of signaling pathways by a variety of miRNAs, resulting in initiation or inhibition/termination of differentiation (Figure 1).
Figure 1.
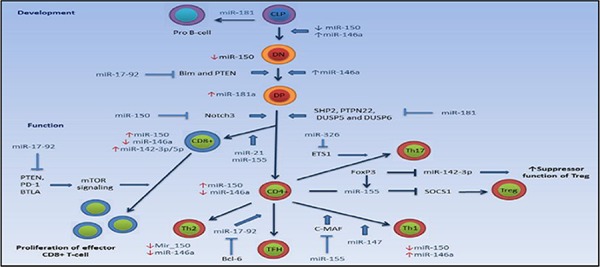
Involvement of microRNAs in regulation of T-cell development and function
T-cell development in thymus is influenced by miR-17-92 and 181a. MiR-17-92 is involved in DN thymocyte to DP thymocyte transition by inhibiting the expression of PTEN and Bim, and miR-181 playes a role in transition of DP to SP thymocyte by inhibiting the expression of PTPN22, SHP2 and DUSP5/6. MiR-181a shows the highest expression in DP thymocyte stage. MiR-146a expression rises during the DN to DP transition, while its expression is decreased in SP stage. MiR-146a expression is increased and decreased in Th1 and Th2 cells by differentiation of naïve CD4+ T-cells, respectively. Expression of miR-150 is decreased during the evolution of T-cells, while it is increased in the final stage of development by formation of CD4+ and CD8 + T-cells. Expression of miR-150 is decreased after differentiation of naïve CD4+ T-cells to Th1 and Th2 subtypes. Bcl-6 plays a role in CD4+ T-cell differentiation to TFH by inhibiting expression of miR-17-92. MiR-155 inhibits SOCS1 and c-MAF, causing the differentiation of naïve CD4 + T-cells to Treg and Th1, respectively. MiR-17-92 expression is increased upon contact with antigen by naïve CD8 + T-cells, leading to proliferation of effector CD8+ cells by inhibiting the expression of PTEN, PD-1 and BTLA.
Abbreviation: CLP: common lymphoid progenitor, DN: double negative, DP: double positive, Bim: Bcl-2-interacting mediator of cell death, PTEN: phosphatase and tensinhomologe, SHP2: SH2-domain-containing protein tyrosine phosphatase 2, DUSP5/6: dual-specificity protein phosphatase 5/6, PTPN22, protein tyrosine phosphatase, non-receptor type 22; Notch 3: Neurogenic locus notch homolog protein 3, ETS1: v-ets avian erythroblastosis virus E26 oncogene homolog 1, PD-1, programmed cell death, BTLA, B and T- lymphocyte associated, mTOR: signaling, mammalian target of rapamycin; FoxP3: forkhead box P3, SOCS1: Suppressor of cytokine signaling 1, Bcl-6, B-cell lymphoma 6 protein, c-MAF, macrophage-activating factor, Th1: T helper type 1 cells, Th2: T helper type 2 cells, TH17: T helper type 17 cells, Treg: regulatory T-cells, TFH: T follicular helper cell.
MiRNA-17-92
MiRNA-17-92 is highly expressed in T precursor cells, and is decreased after maturation. Bcl-2-interacting mediator of cell death (Bim) is the target gene of miR-17-92. MiR-17-92 also regulates the expression of phosphatase and tensin homolog (PTEN) tumor suppressor gene, which has been associated with lymphoproliferation in miR-17-92-transgenic models. All lymphocyte types in mice transfected with this miR-17-92 undergo expansion and show enhanced proliferation and survival, especially for CD4+ T-cells11, 12.
B-cell lymphoma 6 protein (Bcl-6)is involved in the development and function of T follicular helper cells (Tfh), inhibits the expression of miR-17-92 which silences CXCR5 and differentiation to Tfh results13. Increased expression of miR-17-92 at DN stage 1 results in autoimmunity, leading to increased proliferation and survival of T-cells particularly effector CD4+ T-cells14. Expression of this miRNA is highly induced after T cell activation, decreased after clonal expansion and silenced during memory development15. Increased expression of miR-17-92 in effector CD8+ T cells enhances mTOR signaling. The results of Western Blot and FACS on miR-17-92-MIT-transduced P14 cells have confirmed decreased expression of negative regulators of PI3-Akt-mTOR signaling pathways such as phosphatase and tensin homolog (PTEN), programmed cell death 1 (PD1), B and T-lymphocyte associated (BTLA), resulting in increased mTOR signaling. This increased signaling directs differentiation towards short-lived terminal effector cells (Figure 1)15. Decreased expression of miR-17-92 has been observed during differentiation towards CD8+ T-cells(16), while its increased expression leads to increased expansion and proliferation of effector CD8+ T-cells(15), indicating its role in CD8 + T-cell effector activity in the face of virus.
MiRNA-125
MiRNA-125 regulates the genes encoding molecules involved in differentiation of naive CD4 + T-cells in humans but not in mice. These genes include IFN-γ, IL-2 receptor β, IL-10 receptor α and the transcriptional repressor Blimp-1 which undergo reduced expression by long-term expression of this miRNA. Transfecting naive CD4+ T-cells by synthetic miR-125 caused these cells to express lower levels of Th1 and Th2 memory surface molecules and show overexpression of naive CD45RA+, CD45RO-, indicating the role of mi-125 in maintenance of naive T-cell status. In fact, it prevents generation of the effector function in T-cells, which is indicated by reduced intracellular production of IFN-γ and IL-13. Reduced expression of miR-125 is associated with effector memory CD4+ T-cell phenotype17. MiR-125a is also involved in differentiation to Treg cells, and increased expression of it has been observed in Treg cells18. MiR-125 is highly expressed in hematopoietic stem cell (HSC), and is associated with increased self-renewal and Survival19. Increased expression of miR-125 in HSC deviates differentiation toward the lymphoid lineage, which has a higher level of expression in common lymphoid progenitor (CLP) than common myeloid progenitor (CMP). Increased expression of miR-125b is related to lymphoproliferative disease associated with an increase in CD8+ expansion20. MiR-125b has an important role in T-ALL and B-ALL leukemogenesis, and facilitates leukemogenesis in lymphoma through down regulation in interferon regulatory factor-4(IRF4) expression, which plays a role in inhibiting proto-oncogene Bcl-621.
MiRNA-146a
MiRNA-146a is an immune system regulator and is highly expressed in Treg and Th1 cells. The target genes of miR-146a include IL-1 receptor associated kinase 1 (IRAK1) and tumor necrosis factor receptor-associated factor 6 (TRAF6), which are downregulated by this miRNA(11). It is expressed in immature megakaryocytes, primitive hematopoietic stem/progenitor cells (HSPC) and thymocytes, such that miRNA-146a has the highest expression in DP and DN thymocytes and the lowest expression level in SP thymocytes, and has also a low level of expression in myeloid and erythroid series22. Expression and phosphorylation of Signal transducer and activator 1 (STAT1) is increased in mice with miR-146a deficient Tregs, which functions as target gene for miR-146a. STAT1 is the essential transcription factor for Th1 differentiation. MiR-146a by decreasing the level of STAT1 enables Treg cells to suppress Th1 responses, i.e., it is involved in Treg cell-mediated Th1 regulation. Increased IFN-γ in Tregs is essential for controlling IFN-γ dependent Th1 immune responses at least by targeting STAT1, which is the key molecule in IFN-γ signaling pathways23. The absence of miR-146a will disrupt the suppressor function of Treg cells and non-regulated IFN-γ responses by increasing the expression and activation of STAT124. MiR-146a functions as a negative regulator in toll-like receptor (TLR)/IL-1 signaling pathway by targeting TRAF6 and IRAK123. IL-1 stimulates the production of IL-17 in CD4+ T-cells including FoxP3 Treg25. IRAK1 causes Th17 differentiation in presence of TGF-β, and the absence of miR-146a could exacerbate Th17 responses and result in autoimmunity26. TRAF6 is essential in NF-κB activation and downstream of antigen receptor in T-cells(11). NF-κB (p50) is required for expression the induction of miR-146a during T-cell receptor (TCR) stimulation, and plays a role in memory T cell formation27. High expression of miR-146a downregulates IRAK1 expression with decreased NF-κB signaling28.
Studying rheumatoid arthritis patients has indicated that miR-146a plays an important role in differentiation of IL-17 producing T-cells (Th17), and its high expression is directly associated with the expression of IL-17 in peripheral blood mononuclear cells (PBMC) and synovium in these patients29. MiR-146a has low and high expression in naive and memory T cells, respectively, such that by stimulation of primary T lymphocytes by phorbol myristate acetate (PMA) and ionomycin, the expression level of miR-146a is sharply increased after 8 days, which coincides with increased CD45R0 as a marker of memory cells30. In addition, the memory T cells producing miR-146a are involved in increased expansion by stimulation with anti-CD3 and anti-CD2827. Fas-associated death domain (FADD) is also a target for miR-146a, and increased level of this miRNA in T cells protects them against activation-induced cell death (AICD)(30). MiR-146a expression level remains high in Th1 following T naive activation, while it is decreased in Th231. Ectopic expression of miR-146a leads to significant up-regulation in the expression of transcription factors PU.1, C-fos, GATA3 and CCAAT/enhancer-binding protein alpha (C/EBPα), and slight up-regulation in expression of FoxP3 and Runx1, while leads to down-regulation in the expression of the transcription factors Notch-1, Lmo2, SOS1, Ikaros and STAT3. These results suggest that ectopic overexpression of miR-146a does not induce differentiation of T-lymphoblasts, and only upregulates the molecules involved in T-cell activation32. In the study of Fallah P et al, increased expression of miR-146a in CD133+ cells, significant upregulation of Ikaros genes and specific T-cell markers CD2, CD4 and CD25 indicate the role of this miRNA in differentiation of CD133+ cells to T-lymphoid lineage33.
MiRNA-150
This miRNA is required for differentiation of lymphocytes from their progenitors, and has a negative effect in proliferation and survival of T-cell acute lymphoblastic leukemia (T-ALL) cell line by targeting Notch3, the transcription factor involved in T cell differentiation and leukemogenesis9. Reduced level of mRNA and Notch3 protein and increased miR-150 expression has been observed during T cell maturation from DP to SP (CD4+, CD8+) phenotype9. Increased expression of this miRNA in both B and T lymphocytes and lack of its expression in monocytes has been observed34. High levels of this miRNA are expressed in mature T and B cells, and such that the miR-150 expression in thymic DN and SP cells is low and high respectively. Increased expression of miR-150 in hematopoietic stem and progenitor cells will cause no change in T cell development.
Expression of miR-150 is silenced in naive T-cells following TCR engagement and differentiation into Th1 and Th2 cells11, 31. CXCR4 is another target for miR-150. Expression of CXCR4 protein in mononuclear cells (MNCs) is increased following simultaneous knockdown of miR-150, and their migration is also enhanced, indicating the relationship between this miRNA and stem cell migration36. Increased expression of miR-150 in CD133+ cells upregulates the expression of Ikaros and CD4 marker genes, indicating its role in differentiation of CD4 T-lymphoblasts33.
MiRNA-155
MiRNA-155 is involved in regulation of T cell lineage fate by targeting C-maf, and causes differentiation of T cells to Th111. Gene expression studies in miR-155 knockout mice indicate increased expression of Th2 cytokines such as IL-4, IL-5 and IL-10, the transcription factor C-maf and Th2 differentiation14, 37. Expression of miR-155 is increased in response to inflammatory stimuli such as TRL-ligand and proinflammatory cytokines. It is important in hemostasis and overall survival in Treg by targeting suppressor of cytokine signaling (SOCS1)38. MiR-155 is constitutively expressed in a large number of Tregs in a FoxP3 dependent manner, so that there is an increased expression of miR-155 when Foxp3 is bound to promoter region of BIC gene which encodes miR-155, while the cells lacking FoxP3, B cells and myeloid cells temporarily display an increased expression of miRNA when activated11,23. FoxP3 is involved in differentiation, homeostasis and function of Treg, and is essential in differentiation and proliferation but not immunosuppression of Treg cells by inducing the miR-155 expression. Expression of miR-155 causes increased proliferation through IL-2, STAT5 and IFN-γ signaling pathways by downregulation of SOCS1 expression14, 23. BIC/miR-155 plays a key role in homeostasis and function of the immune system, and its expression is increased in B and T lymphocytes and activated dendritic cells39. Studies in mice deficient in BIC-miR-155 have indicated that BIC-miR-155 is essential for regulating the development of Foxp3 + Tregs, and will not affect the Treg-suppressive activity40. In the study of Banerjee A et al, the use of miR-155 antagonism has increased the expression of IFN-γ receptor α chain in CD4 + T-cells, which has been introduced as the second target gene of miR-155. In fact, miR-155 causes differentiation towards Th1 by inhibiting IFN-γ signaling41. MiR-155 is also involved in differentiation and antiviral activity of CD8+ T-cells, so that increased expression of miR-155 has been observed in primary effector and effector memory CD8+ T-cells, and its decreased expression has been observed in naive and central memory cells42. In comparison to naive cells, expression of miR-155 along with miR-21 during differentiation of primary CD8+ T cells has been increased, indicating their role in CD8 + T-cell differentiation(16). Antiviral response of CD8+ T-cells is reduced in miR-155 deficient mice, while increased expression of miR-155 results in increase in such response in vivo42. The inhibition of miR-155 expression by IL-10 in response to LPS leads to increased expression of Src homology-2 domain-containing in-ositol 5-phosphatase 1(SHIP1) as a miR-155 target. SHIP1 coverts phosphatidylinositol triphosphate (PIP3) to phosphatidylinositol biphosphate (PIP2), reducing PIP3 levels and thus the pro-inflammatory response43. Increased level of miR-155 has been observed in Hodgkin's and Burkitt's lymphoma44.
MiRNA-181
MiRNA-181 is preferentially expressed in thymus, lungs and brain, and is expressed in detectable levels in the bone marrow and spleen4. High expression level of miR-181 in the spleen indicates its requirement and role in T-cell development45. MiR-181a and miR-181c have distinct functions in early T cell development. IL-2 plays a role in T-cell activation and proliferation, and has a binding site for miR-181c in its 3' UTR. Therefore, ectopic expression of this miRNA will negatively regulate CD4+ T-cell activation by inhibiting IL-2, whereas miRNA expression is reduced in effector state46. Mir-181a, which plays a role in differentiation and function of T-cells, is present during maturation of T-cell from DN stage1 to DN stage 3 before CD4+ and CD8+ stages, and is reduced in later development stages47. It reaches maximum expression in DP T-cell stage, resulting in DP thymocyte differentiation to SP thymocyte, but miR-181c does not play this role48, 49. MiR-181a functions as a regulator of TCR signaling after transcription, such that increased expression of miR-181a results in increased TCR signaling in T-cells and vice versa. This regulation is done by decreased expression of such tyrosine phosphatases as SH-2 domain containing protein tyrosine (SHP-2), non-receptor type 22 protein tyrosine phosphatase (PTPN22) and dual-specificity protein phosphatase 5 (DUSP5/6), leading to increased activity of lymphocyte cell specific protein (LCK) and extracellular signal-regulated kinase (ErK) as two TCR signaling molecules(38, 49). PTPN22 is a negative regulator of LCK in downstream TCR signaling. DUSP5 and DUSP6 inactivate ERK1/2 by dephosphorylating in different sites in nucleus and cytoplasm50. SHP-2 also mediates negative co-stimulatory signals elicited by CTLA-451. Activity of the mentioned phosphatases inhibits TCR signaling, and is involved in regulation of TCR sensitivity to antigen during positive selection49. DP cells have a high level of miR-181a. Expression of miR-181a is inversely related with the expression of Bcl-2, CD69 and TCRα as its targets, indicating its role in regulating positive selection in the thymus12. There is an increased expression of this miRNA in thymocytes, indicating its specific role in the early development of thymocytes in the thymus52. This miRNA directly drives HSC differentiation towards B cells5. Blocking miR-181a in DP cells inhibits both positive and negative selections44.
MiRNA-182
Foxo family of transcription factors is important in homeostasis and immunological tolerance by controlling the development and function of B and T lymphocytes53. Studies have shown that FoxO1 factor is the target of miR-182 in activated T helper cells54. Induction of the cell cycle inhibitor p27 and Bim protein by FoxO1 inhibits lymphocyte proliferation and apoptosis. STAT5 is activated by TCR stimulation, IL-2 secretion and IL-2R signaling, and increases miR-182 expression by binding regulatory region of miR-182. In fact, miR-182 expression is directly dependent upon STAT5 activation. Continued expression of miR-182 in activated T helper cells inhibits higher levels of FoxO1 mRNA, resulting in increased clonal expansion of these cells54, 55.
Other miRNAs
MiR-142 inhibits production of CD4+ and CD8+ T-cells, but has no effect upon B cells45. MiR-142-3p and 5p are among miRNAs with the highest expression in CD8+ T-cells16. Expression of miR-142-3p is inhibited by FoxP3, leading to increased production of cAMP and suppressor function of Treg cells56. MiR-142-3p targets the mRNA of glycoprotein A Repetitions Predominant (GARP), which is highly expressed when CD25 + CD4 T-cell activation occurs, thereby controlling this cell proliferation57. MiR-142-3p expression is reduced in effector T-cells but not in naive and memory T cells (58). MiR-147 is induced to differentiate naive CD4+ cells to Th1 cells59. Increased miR-106a expression in activated Jurkat T cells is responsible for downregulating the anti-inflammatory cytokine IL-1014. The use of antagomirs for miR-126 reduces Th2 cell responses24. MiR-21 and miR-31 affect FoxP3 expression in Treg cells, so that miR-31 binds the 3' UTR of FoxP3 gene, resulting in direct and negative regulation of its expression, while miR-21 indirectly causes increased expression of it60. MiR-21 expression plays an important role in antigen induced CD8 T-cell differentiation, such that its expression is higher in effector and memory CD8+ T-cells compared to naive T-cells58. MiR-326 promotes Th17 differentiation by targeting ETS-1 as a negative regulator of Th17, and its increased expression is strongly correlated with the severity of multiple sclerosis (MS) disease61. Finally, it can be concluded that the miRNAs 17-92,-125 and -181a are involved in T-cell development up to SP T-cell, and the miRNAs -150, -155,-146a, -182 and -326 play a role in function and differentiation of SP T-cells to various T-cell subtypes (Table 1).
Table 1.
MicroRNAs relevant to T-lymphocyte differentiation, their target genes and functions
| MicroRNAs | Expression | Target genes | Functions | References |
|---|---|---|---|---|
| miR-17-92 | Highly expressed in T-cell precursor and decreased after maturation | Bim, PTEN | Modulates DN to DP transition. Skews differentiation toward short-lived terminal effector cells by increasing mTOR signaling, promotes effector CD8+ T-cell expansion |
(11, 12, 15) |
| PTEN, PD1, BTLA | ||||
| miR-125 | Highly expressed in Treg cells | IFN-γ, IL-2R β, IL-10Rα, Blimp1 | Maintains the Naive state of T-cell. Involved in Treg differentiation |
(17, 62) |
| miR-126 | ….. | …. | Modulates Th2 responses | (24) |
| miR-142-3p | Highly expressed in CD8+ t-cells, downregulated in effector T-cells | ….. | Repressed by FoxP3 leading to increased suppressor function of Treg cells, highly expressed in CD8+ T-cells | (16, 56, 58) |
| miR-146a | Highly expressed in DN, DP thymocytes, low expressed in SP thymocyte and Th2 | IRAK1, TRAF6 | Highly expressed in Treg and Th1 cells. Plays a role in Treg cell-mediated Th1 responses |
(11, 22) (23) |
| STAT1 | ||||
| miR-147 | …… | ….. | Modulates Th1 differentiation | (59) |
| miR-150 | Low expression in DN but high expression in SP, Silenced after differentiation | Notch3 | Modulates maturation from DP to SP stage of thymocyte | (9, 11, 31, 35) |
| miR-155 | ….. | c-MAF, IFN-γRα SOCS1 | Knockout mice have increased Th2 cell generation and reduced Th17 and Th1 cells. Regulates the development of Treg |
(11, 41) (14) |
| miR-181a | Highly expressed in DP cells | SHP2, PTPN22, DUSP5, DUSP6 | Increases TCR signaling in thymocytes and peripheral T-cells, Modulates T cell sensitivity in response to | (11, 38, 49) |
| Bcl-2, CD69, TCR alpha | Antigens, Regulates T cell and B cell development Positive selection in thymocyte | (12) | ||
| miR-181c | ….. | IL-2 | Negatively regulates CD4+ T-cell activation | (46) |
| miR-182 | ….. | FoxO1 | Increases clonal expansion of activated T helper | (54) |
| miR-326 | Increased expression is associated with severity of MS | Ets-1 | Promotes Th17 differentiation | (61) |
Bim: Bcl-2-interacting mediator of cell death, PTEN: phosphatase and tensinhomologe, PD1: programmed cell death 1, BTLA: B and T- lymphocyte associated, DN: double negative; DP, double positive, SP: single positive, mTOR: mammalian target of rapamycin; Blimp1, B lymphocyte-induced maturation protein-1, FoxP3: forkhead box P3, IRAK1: IL-1 receptor-associated kinase-1, TRAF6: tumor necrosis factor receptor-associated factor 6, Notch3: Neurogenic locus notch homolog protein 3; c-MAF, macrophage-activating factor, SOCS1, Suppressor of cytokine signaling 1, SHP2: SH2-domain-containing protein tyrosine phosphatase 2, DUSP5: dual-specificity protein phosphatase 5, PTPN22: protein tyrosine phosphatase, non-receptor type 22, FoxO1: Forkhead box protein O1, MS: multiple sclerosis: Ets-1: v-ets avian erythroblastosis virus E26 oncogene homolog 1.
MicroRNA and T-cell leukemia
T-ALL is a malignant disease of T-cell lymphoid progenitor cells, and is responsible for 15% of childhood ALL and 25% of adult ALL cases63. Juxtaposition of promoter and enhancer elements of TCR genes with transcription factor genes during VDJ recombination is among the cytogenetic changes causing T-ALL64. Cytogenetic changes play an important role in leukemogenesis in cancers of immune cells including T-ALL by altering the expression and function of miRNA, which can function as tumor suppressors or oncogenes (Table 2)65. For example, downregulation of miR-151-5p and miR-708 and upregulation of miR-196b in MLL-rearranged ALL and T-ALL subtype has been observed(66). In addition, t(8;14), t(5;10) and t(7;7) chromosomal translocations occurring in ALL affect different gene regions of various miRNAs including miR-196b (chr7p15), miR-148a (chr7p15) and miR-151 (chr8q24), leading to their upregulation or downregulation63. Comparison of miRNA expression in ALL subtypes has also indicated the effect of these chromosomal translocations. Each ALL subtype has its own specific miRNA expression profile. For example, reduced expression of miR-151 and increased expression of miR-424 and miR-148 is related to T-lineage ALL, while increased expression of miRNA-425-5p, -191, -128, -629,-146b and -126 is associated with B-lineage ALL67. Increased expression of miR-196b is associated with increased activity of HOX gene family members in T-ALL patients with inversion in chromosome 7, and the relationship between this increased expression and leukemogenesis has not been elucidated and requires further studies68. T-cell acute lymphocytic leukemia protein 1(TAL1) complex is ectopically expressed after t(1;14) translocation in T-ALL64. This complex causes ectopic expression of miR-223 in transformed DP thymocytes by regulation of miR-223 expression as a direct transcriptional target in normal and malignant T cells. As a tumor suppressor, F-box/WD repeat-containing protein 7 (FBXW7) ubiquitin ligase is repressed through the mentioned pathway. Eventually, important oncoproteins such as c-MYC, MYB and Notch, which are involved in pathogenesis of T-ALL will be stabilized, and the malignant phenotype will appear in T-ALL69. Myeloid cell leukemia sequence1 (MCL1) is a target for FBXW7 E3 ligase, the level of which is increased in mouse leukemia expressing miR-22370. MiR-223 is involved in myeloid differentiation, and is considered a myeloid miRNA. Overexpression of this miRNA is observed in T-ALL cases with similar gene profiles and phenotypic characteristics of acute myeloid leukemia (AML) with unfavorable outcomes71. Insulin-like growth factor 1 receptor (IGF1R) which plays a role in T-ALL cell growth and leukemia-initiating activity is miR-223 target in its 3'UTR, and its expression is inhibited by miR-223. On the other hand, Notch signaling negatively regulates the expression of mir-223. As a result, Notch signaling supports IGF1R expression directly during increased expression of IGF1R and indirectly through antagonizing miR-22372. MiR-221 and miR-222 are involved in erythropoiesis and erythroid differentiation73. MiR-221 and miR-222 show the highest level of expression in early T-cell acute lymphoblastic leukemia (ETP-ALL), and miR-222 affects leukemogenesis by changing the expression of ETS1 proto-oncogene74. MiR-222 expression correlates with immature immunophenotype of T-ALL and ectopic expression of CD13 and CD33 myeloid markers, and association with these myeloid characters is due to decreased expression of ETS174.cAMP/protein kinase A (PKA) causes deactivation of T cells and reduced cell proliferation through cyclin D3 downregulation and p27 upregulation75. MiR-142 has the highest level of expression in myeloid series but increased expression of it (miR-142-3p) increases leukemogenesis and proliferation by lowering cAMP levels and reducing PKA activity in T-leukemic cells73, 76. Hence, activating the cAMP/PkA pathway can be a therapeutic target. This same miRNA causes resistance of T-leukemic cells to glucocorticoid treatment by targeting glucocorticoid receptor (GRα) Mrna76. Glucocorticoids are among apoptosis inducing drugs in a wide variety of malignancies77. As a result, miR-142-3p plays an important role in T-ALL leukemogenesis, and may be a useful therapeutic target. Increased or decreased expression of miRNAs can be useful in determining prognosis and overall survival of leukemia. For example, increased expression of miR-221, miR-16 and miR-142-3p is related to shorter overall survival in T-ALL, and miR-221 has a poor prognosis66, 76,78. In addition, increased expression of miR-33 in T-ALL causes unfavorable prognosis compared with normal thymocytes79. MiR-146a family has been recognized as immune system regulator11. In T-ALL, increased expression of miR-146a and miR-146b is associated with an immature immunophenotype and expression of CD34 and CD33. These two miRNAs play the role of ERG regulators in T-ALL and AML80. ETS transcription factor ERG plays an important role in hematopoiesis, and its increased expression is an adverse prognosis factor in T-ALL and AML(81). MiR-19 is highly expressed both in T-ALL patients and in cell lines. Its ectopic expression causes silencing of CYLD (Cylindromatosis (turban tumor syndrome) expression which is a negative regulator of NF-κB and JNK signaling, and thus NF-κB expression is induced82.
Table 2.
MicroRNAs relevant to T-cell malignancie, their targets and functions
| Micro RNA | Aberrant Expression | Targets | T-cell malignancy relevance functions | Prognosis | Biological function | Ref. |
|---|---|---|---|---|---|---|
| miR-222 | ↑ in ETP-ALL | Ets-1 | Associate with immature immunophenotype of T-ALL and aberrant expression of myeloid markers CD13 and CD33 | NM | Oncogene | (74) |
| miR-223 | ↑ in T-ALL | FBXW7 ubiquitin ligase | Up-regulated by TAL1 complex and promotes the malignant phenotype. | poor | Oncogene | (69, 70) |
| miR-142-3p | ↑ in T-ALL | cAMP/PKAGRα | Increases T-leukemic cell proliferation and leukemogenesis. leading to glucocorticoid resistance | Poor | Oncogene | (76) |
| miR-221 | ↑ in T-ALL | …. | Associated with shorter OS | Poor | Oncogene | (78) |
| miR-19 | ↑ in T-ALL | CYLD | Induces NF-kB expression, promotes leukemogenesis | NM | Oncogene | (82) |
| miR-451 | ↓ in T-ALL | MYC | Its reppression is essential for Notch-induced T-ALL | NM | Tumor suppressor | (83) |
| miR-17-92 | ↑ in T-ALL | E2F1 | Activated by NK like homodomain proteins, suppresses apoptosis and increases cell survival | NM | Oncogene | (84) |
| miR-146a | ↑ in T-ALL | Associated with immature immunophenotypeabd expression of CD33 and CD34, acts as an ERG regulator in T-ALL | Increased ERG is related to adverse prognosis | Oncogene | (80) | |
| ↑ in ATL | Induced by viral Tax protein through activation of NF-kB, Increases HTLV-1 positive T-cell growth | Increased NF-kB is related to prolonged survival | Oncogene | (88) | ||
|
Plasma miR-155 + miR-126 |
↑ in ATL ↓ in ATL |
….. | Novel biomarkers of progression and prognosis in ATL | Poor | Oncogene Tumor suppressor |
(86) |
| miR-31 | ↓ in ATL | NIK | Suppresses ATL Cell Growth and Promotes Apoptosis by Inhibiting NF-kB | NM | Tumor suppressor | (90) |
| miR-155 | ↑ in ATL | Induced by viral Tax protein through activation of NF-kB and plays a role in biology and pathogenesis of ATL. | Increased NF-kB is related to prolonged survival | Oncogene | (87) | |
| ↑ in CTCL and MF | STAT5/BIC/miR-155 pathway promotes the malignant T-cell proliferation in CTCL, most significant differentially expressed miRNA | NM | Oncogene | (99–100) | ||
| miR-122 | ↑ in CTCL | Akt (PKB) | Induces phosphorilation of Akt and maintains the CTCL cells from apopotosis induced by chemptherapy | Poor (in advanced stage of MF) | Oncogene | (94) |
| miR-21/miR-486/miR-214 | ↑ in SS | ….. | Promote cell survival and apoptotic resistance of CTCL cell line, can be novel diagnostic/prognostic biomarkers | NM | Oncogenes | (98) |
| miR-155+miR-21 | ↑ in NK cell leukemia/lymphoma | PTEN, PDCD4, SHIP1 | Play a NK-cell lymphomagenesis by downregulation target genes and upregulation pAkt | NM | Oncogene | (103) |
| miR-150 | ↓ in NKTL | DKC1, AKT2 | Reduces levels of pAKT and increases tumor suppressors such as Bim and p53 | NM | Tumor suppressor | (104) |
| miR-146a | ↓ in NKTL | TRAF6 | miR-146a downregulates NFkB Activity via Targeting TRAF6, can be strong prognostic factor | Poor | Tumor suppressor | (108) |
| Plasma miR-221 | ↑ ENKTCL | ….. | Can be a prognostic factor for NK/Tcell lymphoma | Related to shorter OS after treatment | Oncogene | (105) |
| miR-29a | ↓ in ENKTCL | MCL1 | Participates in tumorigenesis and development of ENKTCL | NM | Tumor suppressor | (106) |
| miR-16 | ↑ in T LBL/ALL | …. | Can be a potential prognostic marker | Associated with longer OS | Oncogene | (109) |
| miR-193b | ↓ in T-LBL | SMO | Activates the GLI/Hh pathway and promotes survival and cell proliferation | NM | Tumor suppressor | (110) |
ETP/ALL: Early T-cell precursor acute lymphoblastic leukemia, Ets-1: v-ets avian erythroblastosis virus E26 oncogene homolog 1, T-ALL: T-cell acute lymphoblasti leukemia, NM: not mentioned, FBXW7: F-box/WD repeat-containing protein 7, TAL1: T-cell acute lymphocytic leukemia protein 1, IGF1R: insulin growth factor 1 receptor, c-MYC: v-myc avian myelocytomatosis viral oncogene homolog, cAMP/PKA: cyclic adenosin monophosphate/protein kinase A, GRα: glucocorticoid receptor-a, OS: overall survival, CYLD: Cylindromatosis (turban tumor syndrome); NF-kB: nuclear factor kappaB, E2F1: E2F transcription factor 1, ERG: v-etserythroblastosis virus E26 oncogene homolog ATL, Adult T-cell leukemia; HTLV-1; human T-cell leukemia virus type-I, NIK: NF-kB inducing kinase, CTCL, cutaneous T-cell lymphoma, MF, mycosis fungoides, BIC: B-cell integration cluster; STAT5, Signal Transducer and Activator of Transcription 5, Akt, also known as Protein Kinase B (PKB), NK cell: natural killer cell, PTEN: phosphatase and tensin homologue, PDC4: programmed cell death 4, SHIP1: Src homology-2 domain-containing inositol 5-phosphatase 1, SS: Sezarysyndrom; NKTL: natural killer/T- cell lymphoma, DKC1: dyskeratosiscongenita 1; Bim: Bcl-2-interacting mediator of cell death, p53: protein 53 or tumor protein 53, TRAF6, tumor necrosis factor receptor-associated factor 6, ENKTCL: extranodal NK/T-cell lymphoma; MCL1: myeloid cell leukemia sequence 1 (BCL2-related), T LBL/ALL, T-lymphoblastic lymphoma/leukemia, T-LBL: T-cell lymphoblastic lymphoma, SMO: smoothened, frizzled family receptor, Gli/Hh: Gli/hed
NF-κB binds their upstream promoter regions and regulates their expression, and is one of the mechanisms regulating gene expression in T-ALL82. Expression of miR-451 as a tumor suppressor and direct inhibitor of MYC expression is inhibited in T-ALL, and is important in T-ALL oncogenesis by Notch83. T-ALL development by Notch oncogene is delayed through deletion in miR-181ab1, indicating the crucial role of miR-181ab in activating Notch oncogenic signals52. Lack of miR-181ab causes 32 % increase in median survival of T-ALL mice from 41 days to 54 days52. Activation of miR-17-92 by NK like homodomain proteins suppresses apoptosis in T-ALL by reducing the level of E2F1, and the cell survival is therefore increased84. Adult T-cell leukemia is a T-cell neoplasm in which the lymphocytes are transformed by type 1 Human T-cell leukemia virus (HTLV-1) with leukemia as the result85. Increased miR-155 in plasma and decreased miR-126 levels is correlated with poor prognosis in adult T-cell leukemia (ATL), and can be used as a new biomarker for assessing disease stage and progression86. MiR-155 undergoes increased expression in HTLV-1-positive T-cell line, and Tax protein is involved in this increased expression. Indeed, Tax leads to binding of NF-κB and activator protein-1 (AP-1) transcription factors to their respective sites in miR-155 gene promoter and transcription of miRNA gene, increasing growth of these cells, and is therefore involved in biology and pathogenesis of lymphoma type of ATL, adult T-cell leukemia/lymphoma (ATLL)87. Tax also induces the activity of miR-146a promoter via the NF-κB pathway, eventually enhancing the growth of HTLV-1 positive T-cells (88). Activation of NF-κB pathway is a hallmark of HTLV-1 infected cells and leukemic cells, and its abnormal activation is associated with prolonged survival, proliferation and invasiveness of ATL cells89. Tax is a 40-kDa transactivator protein and the main protein encoded by HTLV-1, stimulating and activating the expression of host cell genes through different signaling pathways such as NF-κB and AP-1, and is also involved in leukemogenesis87, 88. MiR-31 has been identified as a tumor suppressor because of inhibiting ATL cell growth and apoptosis through inhibition of NF-κB. NF-κB pathway is inhibited by inhibiting the expression of NF-κB inducing kinase (NIK) as target gene, and NIK and miR-31 expression is increased and decreased in ATL, respectively90. In the study of Bellon M et al, evaluation of miRNA expression levels in HTLV-1 infected cells (ATL) showed that the expression of -150, -155, -223, 142-3p,-142-5p miRNAs was increased, while the expression of -181a, -132,-125a and-146b miRNAs was decreased18. But, contrary to previous reports, Yamagishi M and Watanabe T observed decreased expression of miR-223 in HTLV-1 infected cells89.
MicroRNA and T-cell lymphoma
T-cell lymphomas form a heterogeneous group of T-cell derived lymphoid neoplasms comprising 10-15% of non-Hodgkin's lymphomas. Natural killer cell lymphoma is also classified under T-cell category, and is known as T-cell and NK-cell lymphoma91, 92. By comparing miRNA expression in cutaneous T-cell lymphoma (CTCL) and benign skin Dx, increased expression of miR -326,-663b, -711 and decreased expression of miR - 203 and -205 can distinguish these two diseases with an accuracy of 90%93. Increased expression of miR-122 in CTCL induces AKT phosphorylation, and its activation regulates apoptosis by inhibiting PTEN, and the cell is thus protected against GSI-induced cytotoxicity. Increased expression of this miRNA in CTCL prevents from chemotherapy-induced apoptosis94. Therefore, targeting miR-122 may improve the outcome of chemotherapy94. AKT is an oncogene with a vital role in regulating the pathways involved in cell survival, proliferation and death95. Increased expression of miR-122 is associated with poor prognosis in an advanced stage of Mycosis Fungoides (MF)94. By evaluating the expression level of miR-223, Sézary syndrome (SzS) can be distinguished from MF with over 90% reliability. Downregulation of miR-342 and upregulation of miR-17-5p will decrease and increase cell apoptosis in SzS, respectively, which is involved in pathogenesis of the disease96. MiR-199a2 and -214 are inserted as a tandem in single transcript so-called-dynamin 3 opposite strand (DNM3oS). DNM3os is activated in SzS cells with increased expression of transcription factor TWIST, leading to increased expression of both miRNAs in SzS97. Upregulation of -21, -486, and -214 miRNA is involved in promoting cell survival in SzS, and contributes to resistance to apoptosis in CTCL cell lines, and even may be a new diagnostic/prognostic biomarker for this type of incurable CTCL98. Expression of miR-155 and its host gene BIC in malignant T-cells is increased in CTCL, and STAT5 plays an important role in regulation of increased expression of (BIC/miR-155). STAT5/BIC/miR-155 pathway promotes the proliferation of malignant T cells99. By comparing miRNA expression in benign and malignant stages of MF, it has been found that miR-155 and miR-92a are the most significant differentially expressed miRNAs, while miR-93 is expressed 5.8 times higher in malignant stage100. MiR-155 is upregulated in MF101. There seems to be a relationship between MYC activation and increased target proteins of downregulated miRNAs such as MUM1, Blimp-1 and STMN1 involved in pathogenesis of natural killer T-cell lymphoma (NKTL). MiR-30b, -101,-26a/-b and -363 are among these miRNAs. PRDM1 (the gene encoding BLIMP1) and STMN1 are targets of the first two miRNA, respectively102.
Expression level of miR-21 and miR-155 is specifically higher in NK-cell lymphoma line than normal NK cells. The transduction of these two miRNAs causes downregulation of PTEN, PDCD4 and SHIP1 and upregulationof phosphorylated AKT (pAKT), indicating their important role in NK-cell lymphoma genesis via activation of AKT signaling103. Expression level of mir-150 in NKTL is much lower than that in normal NK cells. This miRNA directly downregulates the expression of dyskeratosis congenita 1(DKC1) and AKT2, decreases pAKT level and increases the level of BimandP53, which functions as a tumor suppressor as mentioned in NK/T-cell lymphoma104. Research by Guo HQ et al., has indicated the level of plasma miR-221 may have a diagnostic and prognostic significance in extranodal NK/T-cell lymphoma (ENKTCL). Indeed, there is an indirect relationship between plasma levels of miR-221 and overall survival after treatment in this lymphoma105. There is a negative correlation between the expression of miR-29a and MCL1, which is overexpressed in extranodal NK/T-cell lymphoma, indicating that MCL1 is the target gene for miR-29a. Therefore, they participate in tumorigenesis and development of ENKTCL106. Expression of miR-223 and miR-886-3p is increased in nasal type NK/T-cell lymphoma, and miR-34c-5p expression is decreased in ENKTCL107. Expression evaluation of miR-146a in NKTL tissue has indicated that patients with a low level of miR-146a expression are associated with chemo resistance and poor prognosis. This miRNA functions as a tumor suppressor, so it can be used to determine prognosis in NKTL108. Evaluation of the expression levels of miR-16 on paraffinized lymphnode samples of patients with T-lymphoblastic lymphoma/leukemia (T-LBL/ALL)has shown that patients with high levels of miR-16 have higher overall survival than patients with low levels of miR-16, and it can be used as a marker of prognosis in T LBL/ALL108. Decreased expression ofmiR-193bin humans and miR-141and miR-30a in mouse increases smoothened (SMO)gene expression in T-cell lymphoblastic lymphoma, resulting inactivation of GLI/hedgehog (Hh) pathway and promoting cell proliferation and survival109. A summary of the role of aberrant expression of miRNA in leukemogenesis and lymphogenesis has been presented in (Figure 2.)
Figure 2.
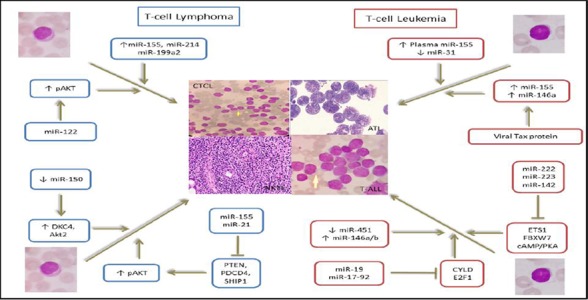
Involvement of microRNAs in T-cell Leukemia and Lymphoma development MicroRNAs known to be involved in pathogenesis of T-cell related leukemias and lymphomas through different mechanisms, including functioning as oncogenes or tumor suppressors. Increased expression of miR-155 and miR-146a plays an oncogene role by involvement of Tax protein and decreased expression of miR-31 as a tumor suppressor in ATL leukemogenesis. The expression of MiR -222, -223, -142, -19 and -17-92 as oncogenes is increased in T-ALL, leading to decreased expression of their target genes ETS1, FBXW7, cAMP/PKA, CYLD and E2F1, respectively. Decreased expression of miR-451 and increased expression of miR-141a/b are also involved in T-ALL leukemogenesis. Concurrent increase in the expression of miR-155 and miR-21, through reduced expression of PTEN, PDCD4 and SHIP1 along with eventual induction of Akt phosphorylation and decreased expression of miR-150 through increase in DKC4 and Akt2 have oncogene and tumor suppressor roles in NKTL, respectively, resulting in reduced apoptosis. Increased expression of micro-RNA -122, -155, -214, and-199a2 is also involved in CTCL lymphomagenesis.
ATL: adult T-cell leukemia, T-ALL: T-cell acute lymphoblastic leukemia, CTCL: cutaneous T-cell lymphoma, NKTL: natural killer/T-cell lymphoma, pAkt: phosphorylated Akt or protein kinase B, PTEN: phosphatase and tensin homologue, PDC4: programmed cell death 4, SHIP1: Src homology-2 domain-containing inositol 5-phosphatase 1, DKC1: dyskeratosiscongenita 1, FBXW7: F-box/WD repeat-containing protein 7,cAMP/PKA: cyclic adenosin monophosphate/protein kinase A, CYLD: Cylindromatosis (turban tumor syndrome), E2F1: E2F transcription factor 1.
DISCUSSION
According to presented contents, miRNAs are essential factors in regulation of normal lymphopoiesis; therefore, deregulation of their expression is associated with development of T-cell malignancies and pathogenesis of leukemia and lymphoma (Table 1, Table 2). Consequently, considering the studies, miRNAs can function as oncogenes or tumor suppressor genes in cancer cells65. The role of miRNAs in B-cell malignancies has also been reported, including miR-15a and miR-16-1, which induce apoptosis by inhibiting Bcl-2, and play the role of tumor suppressor. However, in chronic lymphocytic leukemia (CLL), reduced expression or deletion of these two miRNAs and increased Bcl-2 protein expression has been observed110. In addition, miR-29b and miR-181b play the role of tumor suppressor in CLL by targetingTCL1, playing the role of oncogene in CLL pathogenesis (95). Increased expression of miR-155 in diffuse large B-cell lymphomas and miR-181in AML, (M1 and M2) indicate their oncogenic role in leukemogenesis111. Dicer enzyme, which is involved in biogenesis of miRNA, is essential for T-cell biology as well as survival, proliferation and cytokine production during helper T-cell differentiation and its deletion can cause defects in T-cell development. As a result, disruption of miRNA biogenesis process could also be involved in development of T-cell malignancies59,112. For example, the role of Dicer in promoting effector CD8+ T-cell responses to antigen has been determined in vivo, and it is also involved In activation and proliferation of CD8 + T-cells113. Dicer deletion in T-cell lineage causes reduction in the number of thymocytes and peripheral T-cells which is due to increased apoptosis24. Mice having Tregs deficient in Dicer are defective in differentiation towards functional Tregs, and have been eventually destroyed due to autoimmune disease within a few weeks6. Despite numerous studies on the expression level of miRNAs and their role in cell differentiation, identification of important miRNAs and the target genes involved in differentiation of T cell in leukemia and lymphoma requires further studies. These studies can be conducted to determine the relationship between miRNA with T-cell leukemia and lymphoma, cytogenetics, chromosomal translocations, CD marker expression and relationship with prognosis and treatment. As mentioned in this article, the use of miRNA can be helpful for therapeutic purposes, but few studies have been conducted in this regard.
ACKNOWLEDGMENT
We wish to thank all our colleagues in Tarbiat Modares University, Tehran, Iran.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFRENCES
- 1. Zardo G, Ciolfi A, Vian L, et al. Transcriptional targeting by microRNA-Polycomb complexes: A novel route in cell fate determination. Cell Cycle. 2012; 11( 19): 3543– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. Journal of Clinical Oncology. 2009; 27( 34): 5848– 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianchi N, Zuccato C, Finotti A, Lampronti I, Borgatti M, Gambari R. Involvement of miRNA in erythroid differentiation. Epigenomics. 2012; 4( 1): 51– 65. [DOI] [PubMed] [Google Scholar]
- 4. Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Annals of the New York Academy of Sciences. 2010. January; 1183: 183– 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schickel R, Boyerinas B, Park SM, et al. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. [Review]. 2008. October 6; 27( 45): 5959– 74. [DOI] [PubMed] [Google Scholar]
- 6. Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmunity reviews. 2009. May; 8( 6): 520– 4. [DOI] [PubMed] [Google Scholar]
- 7. Staal FJ, Weerkamp F, Langerak AW, et al. Transcriptional control of T lymphocyte differentiation. Stem Cells. 2001; 19( 3): 165– 79. [DOI] [PubMed] [Google Scholar]
- 8. Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nature immunology. 2010; 11( 2): 114– 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011; 117( 26): 7053– 62. [DOI] [PubMed] [Google Scholar]
- 10. Lawrie CH. MicroRNAs and lymphomagenesis: a functional review. British journal of haematology. 2012. [DOI] [PubMed] [Google Scholar]
- 11. Baltimore D, Boldin MP, O'Connell RM, et al. MicroRNAs: new regulators of immune cell development and function. Nature immunology. 2008; 9( 8): 839– 45. [DOI] [PubMed] [Google Scholar]
- 12. Navarro F, Lieberman J. Small RNAs guide hematopoietic cell differentiation and function. The Journal of Immunology. 2010; 184( 11): 5939– 47. [DOI] [PubMed] [Google Scholar]
- 13. Yu D, Rao S, Tsai LM, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009; 31( 3): 457– 68. [DOI] [PubMed] [Google Scholar]
- 14. Tsitsiou E, Lindsay MA. MicroRNAs and the immune response. Current opinion in pharmacology. 2009; 9( 4): 514– 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu T, Wieland A, Araki K, et al. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proceedings of the National Academy of Sciences. 2012; 109( 25): 9965– 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salaun B, Yamamoto T, Badran B, et al. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. Journal of translational medicine. 2011; 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rossi RL, Rossetti G, Wenandy L, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nature immunology. 2011; 12( 8): 796– 803. [DOI] [PubMed] [Google Scholar]
- 18. Bellon M, Lepelletier Y, Hermine O, et al. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009. May 14; 113( 20): 4914– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012; 26( 9): 2011– 8. [DOI] [PubMed] [Google Scholar]
- 20. Ooi AL, Sahoo D, Adorno M, et al. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proceedings of the National Academy of Sciences. 2010; 107( 50): 21505– 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schotte D, Pieters R, Den Boer M. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2011; 26( 1): 1– 12. [DOI] [PubMed] [Google Scholar]
- 22. Starczynowski DT, Kuchenbauer F, Wegrzyn J, et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Experimental hematology. 2011; 39( 2): 167– 78. e4. [DOI] [PubMed] [Google Scholar]
- 23. Lu L-F, Boldin MP, Chaudhry A, Lin L-L, Taganov KD, Hanada T. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010; 142( 6): 914– 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Belver L, Papavasiliou FN, Ramiro AR. MicroRNA control of lymphocyte differentiation and function. Current opinion in immunology. 2011. June; 23( 3): 368– 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chung Y, Chang SH, Martinez GJ, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009; 30( 4): 576– 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maitra U, Davis S, Reilly CM, et al. Differential regulation of Foxp3 and IL-17 expression in CD4 T helper cells by IRAK-1. The Journal of Immunology. 2009; 182( 9): 5763– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rusca N, Deho L, Montagner S, et al. MiR-146a and NF-kappaB1 regulate mast cell survival and T lymphocyte differentiation. Mol Cell Biol. 2012. November; 32( 21): 4432– 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quinn EM, Wang JH, O'Callaghan G, et al. MicroRNA-146a is upregulated by and negatively regulates TLR2 signaling. PLoS One. 2013; 8( 4): e62232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niimoto T, Nakasa T, Ishikawa M, et al. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC musculoskeletal disorders. 2010; 11( 1): 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Curtale G, Citarella F, Carissimi C, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010; 115( 2): 265– 73. [DOI] [PubMed] [Google Scholar]
- 31. Monticelli S, Ansel KM, Xiao C, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005; 6( 8): R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saki N, Abroun S, Soleimani M, et al. The roles of miR-146a in the differentiation of Jurkat T-lymphoblasts. Hematology. 2013. June 22. [DOI] [PubMed] [Google Scholar]
- 33. Fallah P, Arefian E, Naderi M, et al. miR-146a and miR-150 promote the differentiation of CD133 (+) cells into T-lymphoid lineage. Mol Biol Rep. 2013. August; 40( 8): 4713– 9. [DOI] [PubMed] [Google Scholar]
- 34. Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008. October; 81( 4): 304– 10. [DOI] [PubMed] [Google Scholar]
- 35. Zhou B, Wang S, Mayr C, et al. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007. April 24; 104( 17): 7080– 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tano N, Kim HW, Ashraf M. microRNA-150 regulates mobilization and migration of bone marrow-derived mononuclear cells by targeting Cxcr4. PLoS One. 2011; 6( 10): e23114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dunand-Sauthier I, Santiago-Raber ML, Capponi L, et al. Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood. 2011. April 28; 117( 17): 4490– 500. [DOI] [PubMed] [Google Scholar]
- 38. O'Connell RM, Rao DS, Chaudhuri AA, et al. Physiological and pathological roles for microRNAs in the immune system. Nature Reviews Immunology. 2010; 10( 2): 111– 22. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007. April 27; 316( 5824): 608– 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kohlhaas S, Garden OA, Scudamore C, et al. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009. March 1; 182( 5): 2578– 82. [DOI] [PubMed] [Google Scholar]
- 41. Banerjee A, Schambach F, DeJong CS, et al. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010. January; 40( 1): 225– 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gracias DT, Stelekati E, Hope JL, et al. The microRNA miR-155 controls CD8 (+) T cell responses by regulating interferon signaling. Nature immunology. 2013. June; 14( 6): 593– 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCoy CE, Sheedy FJ, Qualls JE, et al. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem. 2010. July 2; 285( 27): 20492– 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008. March; 9( 3): 219– 30. [DOI] [PubMed] [Google Scholar]
- 45. Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004. January 2; 303( 5654): 83– 6. [DOI] [PubMed] [Google Scholar]
- 46. Xue Q, Guo ZY, Li W, et al. Human activated CD4(+) T lymphocytes increase IL-2 expression by downregulating microRNA-181c. Molecular Immunology. 2011. January; 48( 4): 592– 9. [DOI] [PubMed] [Google Scholar]
- 47. Labrecque N, Baldwin T, Lesage S. Molecular and genetic parameters defining T-cell clonal selection. Immunol Cell Biol. 2011. January; 89( 1): 16– 26. [DOI] [PubMed] [Google Scholar]
- 48. Liu G, Min H, Yue S, et al. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS One. 2008; 3( 10): e3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Laufer TM. T-cell sensitivity: a microRNA regulates the sensitivity of the T-cell receptor. Immunol Cell Biol. 2007. July; 85( 5): 346– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007. April 6; 129( 1): 147– 61. [DOI] [PubMed] [Google Scholar]
- 51. Marengère LE, Waterhouse P, Duncan GS, et al. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science. 1996; 272( 5265): 1170– 3. [DOI] [PubMed] [Google Scholar]
- 52. Fragoso R, Mao T, Wang S, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012; 8( 8): e1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haftmann C, Stittrich AB, Sgouroudis E, et al. Lymphocyte signaling: regulation of FoxO transcription factors by microRNAs. Annals of the New York Academy of Sciences. 2012. January; 1247: 46– 55. [DOI] [PubMed] [Google Scholar]
- 54. Stittrich AB, Haftmann C, Sgouroudis E, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nature immunology. 2010. November; 11( 11): 1057– 62. [DOI] [PubMed] [Google Scholar]
- 55. Alexander T, Haftmann C, Templin L, et al. A3. 22 Upregulated microRNA-182 Expression is Associated with Enhanced Conventional CD4+ T Cell Proliferation in SLE. Annals of the Rheumatic Diseases. 2013; 72( Suppl 1): A21– A. [Google Scholar]
- 56. Huang B, Zhao J, Lei Z, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009. February; 10( 2): 180– 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou Q, Haupt S, Prots I, Thummler K, Kremmer E, Lipsky PE, et al. miR-142-3p is involved in CD25+ CD4 T cell proliferation by targeting the expression of glycoprotein A repetitions predominant. J Immunol. 2013. June 15; 190( 12): 6579– 88. [DOI] [PubMed] [Google Scholar]
- 58. Wu H, Neilson JR, Kumar P, et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007; 2( 10): e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuchen S, Resch W, Yamane A, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010; 32( 6): 828– 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rouas R, Fayyad-Kazan H, El Zein N, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009. June; 39( 6): 1608– 18. [DOI] [PubMed] [Google Scholar]
- 61. Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nature immunology. 2009. December; 10( 12): 1252– 9. [DOI] [PubMed] [Google Scholar]
- 62. Bellon M, Lepelletier Y, Hermine O, et al. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009; 113( 20): 4914– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bhatia S, Kaul D, Varma N. Functional genomics of tumor suppressor miR-196b in T-cell acute lymphoblastic leukemia. Mol Cell Biochem. 2011. January; 346( 1–2): 103– 16. [DOI] [PubMed] [Google Scholar]
- 64. Iacobucci I, Papayannidis C, Lonetti A, et al. Cytogenetic and molecular predictors of outcome in acute lymphocytic leukemia: recent developments. Curr Hematol Malig Rep. 2012; 7( 2): 133– 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006. November; 6( 11): 857– 66. [DOI] [PubMed] [Google Scholar]
- 66. de Oliveira JC, Brassesco MS, Scrideli CA, et al. MicroRNA expression and activity in pediatric acute lymphoblastic leukemia (ALL). Pediatr Blood Cancer. 2012. October; 59( 4): 599– 604. [DOI] [PubMed] [Google Scholar]
- 67. Fulci V, Colombo T, Chiaretti S, et al. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009. December; 48( 12): 1069– 82. [DOI] [PubMed] [Google Scholar]
- 68. Schotte D, Lange-Turenhout EA, Stumpel DJ, et al. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010. October; 95( 10): 1675– 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mansour MR, Sanda T, Lawton LN, et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med. 2013. July 29; 210( 8): 1545– 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mavrakis KJ, Van Der Meulen J, Wolfe AL, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat Genet. 2011. July; 43( 7): 673– 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chiaretti S, Messina M, Tavolaro S, et al. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 2010. July; 95( 7): 1114– 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gusscott S, Kuchenbauer F, Humphries RK, et al. Notch-mediated repression of miR-223 contributes to IGF1R regulation in T-ALL. Leuk Res. 2012. July; 36( 7): 905– 11. [DOI] [PubMed] [Google Scholar]
- 73. Lawrie CH. MicroRNAs and haematology: small molecules, big function. British journal of haematology. 2007. June; 137( 6): 503– 12. [DOI] [PubMed] [Google Scholar]
- 74. Coskun E, Neumann M, Schlee C, et al. MicroRNA profiling reveals aberrant microRNA expression in adult ETP-ALL and functional studies implicate a role for miR-222 in acute leukemia. Leuk Res. 2013. June; 37( 6): 647– 56. [DOI] [PubMed] [Google Scholar]
- 75. van Oirschot BA, Stahl M, Lens SM, et al. Protein kinase A regulates expression of p27 (kip1) and cyclin D3 to suppress proliferation of leukemic T cell lines. J Biol Chem. 2001. September 7; 276( 36): 33854– 60. [DOI] [PubMed] [Google Scholar]
- 76. Lv M, Zhang X, Jia H, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-alpha and cAMP/PKA pathways. Leukemia. 2012. April; 26( 4): 769– 77. [DOI] [PubMed] [Google Scholar]
- 77. Greenstein S, Ghias K, Krett NL, et al. Mechanisms of glucocorticoid-mediated apoptosis in hematological malignancies. Clin Cancer Res. 2002. June; 8( 6): 1681– 94. [PubMed] [Google Scholar]
- 78. Gimenes-Teixeira HL, Lucena-Araujo AR, Dos Santos GA, et al. Increased expression of miR-221 is associated with shorter overall survival in T-cell acute lymphoid leukemia. Exp Hematol Oncol. 2013; 2( 1): 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schotte D, De Menezes RX, Akbari Moqadam F, et al. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011. May; 96( 5): 703– 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Coskun E, von der Heide EK, Schlee C, et al. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res. 2011. February; 35( 2): 208– 13. [DOI] [PubMed] [Google Scholar]
- 81. Baldus CD, Burmeister T, Martus P, et al. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol. 2006. October 10; 24( 29): 4714– 20. [DOI] [PubMed] [Google Scholar]
- 82. Ye H, Liu X, Lv M, Wu Y, et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012. July; 40( 12): 5201– 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li X, Sanda T, Look AT, et al. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J Exp Med. 2011. April 11; 208( 4): 663– 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nagel S, Venturini L, Przybylski GK, et al. Activation of miR-17-92 by NK-like homeodomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2009. January; 50( 1): 101– 8. [DOI] [PubMed] [Google Scholar]
- 85. Matsuoka M, Jeang K-T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nature Reviews Cancer. 2007; 7( 4): 270– 80. [DOI] [PubMed] [Google Scholar]
- 86. Ishihara K, Sasaki D, Tsuruda K, et al. Impact of miR-155 and miR-126 as novel biomarkers on the assessment of disease progression and prognosis in adult T-cell leukemia. Cancer Epidemiol. 2012. December; 36( 6): 560– 5. [DOI] [PubMed] [Google Scholar]
- 87. Tomita M. Important Roles of Cellular MicroRNA miR-155 in Leukemogenesis by Human T-Cell Leukemia Virus Type 1 Infection. ISRN Microbiol. 2012; 2012: 978607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tomita M, Tanaka Y, Mori N. MicroRNA miR-146a is induced by HTLV-1 tax and increases the growth of HTLV-1-infected T-cells. Int J Cancer. 2012. May 15; 130( 10): 2300– 9. [DOI] [PubMed] [Google Scholar]
- 89. Yamagishi M, Watanabe T. Molecular hallmarks of adult T cell leukemia. Front Microbiol. 2012; 3: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yamagishi M, Nakano K, Miyake A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012. January 17; 21( 1): 121– 35. [DOI] [PubMed] [Google Scholar]
- 91. Chan TS, Kwong YL, Tse E. Novel therapeutic agents for T-cell lymphomas. Discov Med. 2013. August; 16( 86): 27– 35. [PubMed] [Google Scholar]
- 92. Jaffe ES, Nicolae A, Pittaluga S. Peripheral T-cell and NK-cell lymphomas in the WHO classification: pearls and pitfalls. Mod Pathol. 2013. January; 26 Suppl 1: S71– 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ralfkiaer U, Hagedorn PH, Bangsgaard N, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood. 2011. November 24; 118( 22): 5891– 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Manfe V, Biskup E, Rosbjerg A, et al. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PLoS One. 2012; 7( 1): e29541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fabbri M, Garzon R, Andreeff M, et al. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008. June; 22( 6): 1095– 105. [DOI] [PubMed] [Google Scholar]
- 96. Ballabio E, Mitchell T, van Kester MS, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood. 2010. August 19; 116( 7): 1105– 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Qin Y, Buermans HP, van Kester MS, et al. Deep-sequencing analysis reveals that the miR-199a2/214 cluster within DNM3os represents the vast majority of aberrantly expressed microRNAs in Sezary syndrome. J Invest Dermatol. 2012. May; 132( 5): 1520– 2. [DOI] [PubMed] [Google Scholar]
- 98. Narducci MG, Arcelli D, Picchio MC, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell Death Dis. 2011; 2: e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kopp KL, Ralfkiaer U, Gjerdrum LM, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013. June 15; 12( 12): 1939– 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Van Kester MS, Ballabio E, Benner MF, et al. miRNA expression profiling of mycosis fungoides. Mol Oncol. 2011. June; 5( 3): 273– 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Maj J, Jankowska-Konsur A, Sadakierska-Chudy A, et al. Altered microRNA expression in mycosis fungoides. Br J Dermatol. 2012. February; 166( 2): 331– 6. [DOI] [PubMed] [Google Scholar]
- 102. Ng SB, Yan J, Huang G, et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood. 2011. November 3; 118( 18): 4919– 29. [DOI] [PubMed] [Google Scholar]
- 103. Yamanaka Y, Tagawa H, Takahashi N, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009. October 8; 114( 15): 3265– 75. [DOI] [PubMed] [Google Scholar]
- 104. Watanabe A, Tagawa H, Yamashita J, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011. August; 25( 8): 1324– 34. [DOI] [PubMed] [Google Scholar]
- 105. Guo HQ, Huang GL, Guo CC, et al. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers. 2010; 29( 5): 251– 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Huang HB, Zhan R, Wu SQ, et al. [ Expression of MCL-1 and microRNA-29a in extranodal NK/T-cell lymphoma tissue]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013. February; 21( 1): 95– 8. [DOI] [PubMed] [Google Scholar]
- 107. Ti HJ, Nong L, Wang W, et al. [ Expression of microRNA in extranodal NK/T cell lymphoma, nasal type]. Zhonghua Bing Li Xue Za Zhi. 2011. September; 40( 9): 610– 5. [PubMed] [Google Scholar]
- 108. Xi Y, Li J, Zan L, et al. Micro-RNA-16 expression in paraffin-embedded specimen correlates with overall survival of T-lymphoblastic lymphoma/leukemia. Hum Pathol. 2013. June; 44( 6): 1011– 6. [DOI] [PubMed] [Google Scholar]
- 109. Gonzalez-Gugel E, Villa-Morales M, Santos J, et al. Down-regulation of specific miRNAs enhances the expression of the gene Smoothened and contributes to T-cell lymphoblastic lymphoma development. Carcinogenesis. 2013. April; 34( 4): 902– 8. [DOI] [PubMed] [Google Scholar]
- 110. Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005. September 27; 102( 39): 13944– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Vasilatou D, Papageorgiou S, Pappa V, et al. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010. January 1; 84( 1): 1– 16. [DOI] [PubMed] [Google Scholar]
- 112. Muljo SA, Ansel KM, Kanellopoulou C, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005. July 18; 202( 2): 261– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci U S A. 2010. December 14; 107( 50): 21629– 34. [DOI] [PMC free article] [PubMed] [Google Scholar]


