Abstract
De novo generation of human hematopoietic stem cells (HSCs) from renewable cell types has been a long sought-after but elusive goal in regenerative medicine. Paralleling efforts to guide pluripotent stem cell differentiation by manipulating developmental cues, substantial progress has been made recently toward HSC generation via combinatorial transcription factor (TF)-mediated fate conversion, a paradigm established by Yamanaka's induction of pluripotency in somatic cells by mere four TFs. This review will integrate the recently reported strategies to directly convert a variety of starting cell types toward HSCs in the context of hematopoietic transcriptional regulation and discuss how these findings could be further developed toward the ultimate generation of therapeutic human HSCs.
Keywords: cell fate conversion, hematopoietic stem cells, induced reprogramming, transcription factors
Introduction
The process by which differentiated cell types arise from more primitive stem and progenitor cells generally proceeds down a strict lineal hierarchy defined by progressive functional specialization concomitant with restriction of lineage potential. From embryogenesis initiated by a single totipotent zygote to the lifelong homeostasis of organ parenchyma by tissue-specific stem cells, physiological differentiation of progenitor cells largely proceeds both unidirectionally and irreversibly, with differentiated cell types and even intermediate progenitors being remarkably fixed with respect to their cellular identity and functional potential. This paradigm, however, was challenged by the seminal works of Gurdon and others that demonstrated the sufficiency of factors present in oocyte cytoplasm to reverse differentiation of somatic nuclei and allow cloning of whole animals (Briggs & King, 1952; Gurdon, 1962; Wilmut et al, 1997). Subsequent efforts to identify the trans-acting factors capable of altering cell fate revealed the central role of transcription factors (TFs) in determining cellular identity. This was first demonstrated by the ability of a single TF MyoD to imbue myogenic identity on fibroblasts (Davis et al, 1987), which then established the foundation for the landmark discovery by Yamanaka that mere four TFs are sufficient to induce pluripotency in somatic cells (Takahashi & Yamanaka, 2006; Takahashi et al, 2007). These and other studies galvanized numerous investigators to harness the power of TFs in directly respecifying multiple cell fates: hepatocytes (Huang et al, 2011; Sekiya & Suzuki, 2011) cardiomyocytes (Ieda et al, 2010), cardiac pacemaker cells (Kapoor et al, 2013), oligodendrocytes (Najm et al, 2013; Yang et al, 2013), various types of neurons (Vierbuchen et al, 2010; Son et al, 2011; Liu et al, 2013), neural stem cells (Han et al, 2012), pancreatic beta cells (Zhou et al, 2008), sertoli cells (Buganim et al, 2012), thymic epithelium (Bredenkamp et al, 2014), endothelial cells (Han et al, 2014), and intestinal progenitors (Morris et al, 2014).
Studies of the hematopoietic system have borne profound insights into the transcriptional regulation of cellular identity. The ability to prospectively isolate hematopoietic stem, progenitor, and effector cells with defined lineage potentials has allowed dissection of molecular mechanisms underlying blood differentiation as well as identification of characteristic TFs governing diverse hematopoietic lineages (Orkin, 1995). However, the wave of breakthroughs from cell fate conversion studies transformed the hematopoietic system from a subject of scrutiny to a destination to be reached from alternative cell types. Initial studies focused on manipulating lineage potential by means of oncogenic transformation (Beug et al, 1979; Graf et al, 1992), lineage switching of hematopoietic progenitors (Heyworth et al, 2002), and direct cell fate conversion toward non-progenitor blood cells such as macrophages (Xie et al, 2004; Laiosa et al, 2006; Feng et al, 2008); however, substantial progress has been made recently toward de novo generation of hematopoietic stem cells (HSCs) (Szabo et al, 2010; Doulatov et al, 2013; Pereira et al, 2013; Batta et al, 2014; Pulecio et al, 2014; Riddell et al, 2014; Sandler et al, 2014). HSCs reside at the apex of the hematopoietic hierarchy and serve as the lifelong reservoir for all downstream blood cells. Their remarkable regenerative capacity to durably restore the entire hematopoietic system in transplant recipients has been harnessed as the standard of care for treatment of a number of morbid conditions including hematological malignancies, bone marrow failure syndromes, and immunodeficiency syndromes with ∽50,000 allogeneic or autologous transplants performed each year (Gratwohl et al, 2010). Many factors contribute to transplantation outcomes including relapse of primary disease, graft failure, and opportunistic infection. Moreover, in addition to the challenge of identifying histocompatible donors, allogeneic transplantation is further complicated by graft-versus-host disease (GVHD), which remains a significant cause of morbidity and mortality for a large number of patients who undergo allogeneic transplantation despite the use of prophylactic immunosuppressants (Petersdorf, 2013). Another major factor contributing to transplant success is the number of stem cells transplanted, with an increased number of CD34+ HSPCs being the strongest predictor of transplantation success as measured by rapid and durable hematopoietic recovery (Siena et al, 2000). Transplants that contain too few HSCs either fail to engraft altogether or result in delayed blood reconstitution post-transplantation that is associated increased morbidity and mortality. This is particularly clinically challenging for the 10–30% of patients from whom sufficient numbers of autologous stem cells cannot be harvested due to poor responsiveness to mobilizing agents such as GCSF (Bakanay & Demirer, 2012). Therefore, an ability to produce an inexhaustible supply of autologous HSCs from relatively dispensable and potentially expandable cell types via cell fate conversion represents an attractive solution to these challenges. This review will discuss the critical roles played by TFs in hematopoietic cell fate regulation and how this knowledge has propelled efforts to convert alternative cell types toward fully functional HSCs.
Transcriptional regulation of cellular identity
TFs dictate the specific gene expression pattern necessary for a cell to perform its unique functions. Mechanistically, TFs directly impact chromatin state by recruiting epigenetic modifiers to specific DNA sequence motifs present in gene regulatory regions such as promoters and enhancers (Rosenfeld et al, 2006). Depending on whether a TF recruits transcriptional coactivators or corepressors, it may either promote or suppress gene expression, respectively. Consequently, a TF may contribute to enforcing a particular cell fate by simultaneously activating genes required for maintaining the function and identity of that cell while antagonizing lineage inappropriate genes (Cantor et al, 2008; Pongubala et al, 2008; Schaffer et al, 2010; Qi et al, 2013). In addition to recruiting cofactors, TFs often bind cooperatively to DNA as components of multiprotein complexes (Huang et al, 2009; Ravasi et al, 2010; Kazemian et al, 2013). Thus, the same TF may exhibit completely different genome-wide binding patterns and regulate non-overlapping sets of target genes in different cell types (Hoffman et al, 2010; Pimkin et al, 2014). Indeed, cell fate and function is invariably the result of the combinatorial action of TF complexes that form interdependent nodes that comprise larger regulatory networks. The wiring of cell-type-specific TFs as self-reinforcing circuits allows robust, stable sustenance of specific transcriptional landscapes (Rao et al, 2002; Chew et al, 2005; Bonzanni et al, 2013). On the other hand, antagonistic relationships between TF sets governing alternate cell fates serve as barriers to cellular plasticity and thus provide a basis for their mutual exclusivity (Graf & Enver, 2009).
TFs exert differential spheres of influence over cell fate depending on their connectivity within gene regulatory networks. Some TFs function as critical hubs, and their loss may lead to the collapse of network integrity (Albert et al, 2000). The potency of TFs may also derive from their ability to act as pioneering factors that can directly trigger nucleosomal remodeling to grant chromatin access to additional TFs (Zaret & Carroll, 2011). Some of these factors can be so potent that they can single handedly convert cell fate as is the case for MyoD in establishing the myogenic transcriptome, Cebpa in activating the myeloid program (Xie et al, 2004), and Runx1 in coordinating the endothelial-to-hematopoietic transition during development (Feng et al, 2008). More often, however, multiple TFs must act in concert to access and activate cell-type-specific gene regulatory networks (Wilson et al, 2010a).
Reprogramming and cell fate conversion take advantage of the connectedness and interdependencies of TFs in orchestrating cell fate programs (Buganim et al, 2013). Ectopic expression of a subset of TFs enriched in a destination cell type can be sufficient to actuate the destination gene regulatory network in an alternate starting cell type. The destination cell gene regulatory network may then predominate over that of the starting cell type, thus altering its identity. This process depends on extensive chromatin reconfiguration including reversal of chromatin inaccessibility, installation of destination cell specific enhancers, and shutdown of regulatory elements specific to the starting cell type, the carryover of which could lead to retention of what has been termed ‘epigenetic memory’ (Hu et al, 2010; Kim et al, 2010; Apostolou & Hochedlinger, 2013; Vaskova et al, 2013).
The self-reinforcing nature of gene regulatory networks implies that a small number of TFs may be sufficient to trigger their establishment and maintenance of destination cell regulatory networks. Moreover, recent evidence has demonstrated that regulatory networks governing cell identity could potentially be seeded by multiple distinct combinations of TFs. Indeed, extensive studies of TFs regulating embryonic stem cells have yielded multiple TF combinations capable of iPS cell generation that are different and even completely distinct from that originally reported by Yamanaka (Montserrat et al, 2013; Shu et al, 2013; Buganim et al, 2014; Takashima et al, 2014).
Transcriptional regulation of hematopoietic cell fates
The hematopoietic system has been studied extensively as a model tissue hierarchy for dissecting transcriptional regulation of cellular identity and cell fate transitions. As cells descend from HSCs, they are subjected to tiers of decisions that successively commit them to their final effector function. The molecular events underlying developmental cell fate decisions have been attributed in part to cross-antagonism between lineage-specific TFs, perhaps best illustrated by the activities of Gata1 and Pu.1 in promoting erythroid and myeloid differentiation programs, respectively (Arinobu et al, 2007). Physical interaction between Gata1 and Pu.1 leads to mutual extinction of transcriptional activity (Nerlov et al, 2000; Zhang et al, 2000). While they are both expressed in multipotent progenitors, offset in their relative levels, modulated by parameters such as cell cycle length (Kueh et al, 2013) and instructive cytokine signaling (Sarrazin et al, 2009), allows the higher expressed factor to dominate cell fate decisions and hence tip the balance toward the respective lineage (Huang et al, 2007). Multiple such mutually antagonistic interactions could be assembled to form a greater landscape of attractors and transitional states that models multilineage differentiation (Krumsiek et al, 2011).
Pax5 is another TF whose role in hematopoietic lineage commitment has been studied extensively (Urbanek et al, 1994). Pax5 is essential in B-cell commitment, the lack of which arrests differentiation at the early pro-B stage (Nutt et al, 1999). Mechanistically, Pax5 locks progenitors into B-cell fate by both activating B cell promoting signaling pathways while silencing those important for the development of alternative lineages. Intriguingly, Pax5 null pro-B cells retain not only self-renewal potential but also multilineage differentiation potential spanning myeloid lineages and T cells (Mikkola et al, 2002) though Pax5-deficient progenitors do not give rise to B cells (Urbanek et al, 1994) and have very limited potential to give rise to erythrocytes or platelets (Nutt et al, 1999; Schaniel et al, 2002). The importance of Pax5 for limiting self-renewal and locking in B-cell fate is further illustrated by experiments in which deletion in CD19-positive B cells results in an aggressive and highly penetrant lymphoma in vivo (Cobaleda et al, 2007).
In addition to regulating developmental lineage commitment, TFs may impact subtype specification within hematopoietic lineages. For example, peripheral CD4 T-helper cells can develop into induced regulatory T cells (iTregs) with appropriate immunosuppressive functions upon induction of Foxp3, a TF critical for all regulatory T-cell development (Rudensky, 2011). However, inflammatory signals can extinguish Foxp3 expression in iTregs and convert them back to effector CD4 T-helper cells (Zhou et al, 2009). Furthermore, diverse subsets of macrophages are specified by the integration of Pu.1, a macrophage lineage determining TF, with transcriptional regulators downstream of tissue-specific environmental signals (Gosselin et al, 2014). These examples suggest that cellular identity can be a composite of multiple transcriptional modules each presided over by unique TFs or their combinations.
Artificial hematopoietic lineage conversions toward therapeutic application
Artificial manipulation of TFs has yielded important insights into the molecular underpinnings of lineage choice (Iwasaki et al, 2006). Overexpression together with the loss of function experiments has been used to confirm the nature of interaction between TFs and led to surprising discoveries on cellular plasticity. In addition to obtaining valuable mechanistic information, excitement in cell fate conversion research has been fueled by its enormous clinical potential. Cell fate conversion has the potential to generate patient-specific cells that are rare, inaccessible, or clinically useful from relatively dispensable autologous cells. If realized, such procedures could be applied to supplying cells for human disease modeling, therapeutic screening, and cell replacement therapy (Robinton & Daley, 2012; Cherry & Daley, 2013; Kamao et al, 2014; Nakamura et al, 2014; Stewart, 2014; Wainger et al, 2014). Thus, this strategy represents an attractive means to address both patient specificity and overcoming the rarity, and lack of means for expansion that currently limits the therapeutic use of HSCs.
A parallel strategy toward generating HSCs has been via stepwise differentiation of pluripotent stem cells such as embryonic stem cells or induced pluripotent stem cells (Sturgeon et al, 2013). However, attempts to direct differentiation of pluripotent stem cells toward HSCs by recapitulating the embryonic developmental trajectory in vitro has seen limited success. Simulating the temporal (Tober et al, 2013), spatial (Peeters et al, 2009; Wilkinson et al, 2009), mechanical (North et al, 2009), and cellular (Clements et al, 2011; Espin-Palazon et al, 2014) complexity of the embryonic milieu has proved technically challenging. Moreover, since the precise developmental intermediates en route to HSCs are only recently becoming elucidated (Rybtsov et al, 2011, 2014), the directed differentiation approach has thus far suffered from paucity of reliable developmental guideposts. For example, induction of T lymphoid potential has been used to guide directed differentiation as it correlates with definitive hematopoiesis, a temporal wave of embryonic hematopoiesis during which HSCs are specified (Kennedy et al, 2012). However, it is still unknown whether the in vitro derivatives with T lymphoid potential indeed possess the eventual capacity to produce HSCs or whether they might represent a developmental intermediate similar to embryonic T-cell progenitors that arise independently of HSCs (Yoshimoto et al, 2012). Due to such state of the field, little is known about the functional and molecular correlation between the developmental intermediates obtained in vitro and in vivo, and the knowledge is particularly lacking in the context of human HSC ontogeny (Ivanovs et al, 2014). Furthermore, the necessity to transit through multiple distinct intermediate cell types to reach HSCs means that culture conditions may need to be optimized for every intermediate and that deviation in any one step may extinguish the eventual potential to develop HSCs. Therefore, in spite of ‘forceful’ nature of TF manipulations, TF-mediated cell fate conversion may be a relatively simpler, direct, and even more tractable of a strategy for deriving HSCs as it only requires knowledge of the properties of the destination cell type.
Generation of hematopoietic stem cells via direct cell fate conversion
HSCs are functionally defined as cells capable of engrafting conditioned recipients and giving rise to all blood lineages (i.e., myeloid, thrombo-erythroid, and lymphoid), for an extended period (at least 4 months in mice). These functional hallmarks of HSCs, namely multilineage differentiation potential and extensive self-renewal capacity, are embodied at the clonal level such that only few HSCs are required to durably sustain the entire hematopoietic system (Holstege et al, 2014). Although fully functional human HSCs meeting the aforementioned criteria have yet to be produced in vitro, several strategies have recently been described that bring the goal of deriving fully function HSCs from alternative cell types within reach (Table1).
Table 1.
Summary of studies using transcription factor-mediated reprogramming to derive primitive blood progenitors
| Szabo et al (2010) | Pulecio et al (2014) | Pereira et al (2013) | Batta et al (2014) | Doulatov et al (2013) | Riddell et al (2014) | Sandler et al (2014) | |
|---|---|---|---|---|---|---|---|
| Species | Human | Human | Mouse | Mouse | Human | Mouse | Human |
| Starting cell type | Fibroblast | Fibroblast | Fibroblast | Fibroblast | ES cell-derived myeloid restricted progenitor | B-cell progenitor, myeloid progenitor, bone marrow myeloid effector | Umbilical vein endothelial cells, microvascular endothelial cells |
| Transcription factors | OCT4 | SOX2, mir125b | Gata2, Gfi1b, cFos, Etv6 | Erg, Gata2, Runx1c, Scl, Lmo2 | ERG, HOXA9, RORA, SOX4, MYB | Runx1t1, Hlf, Lmo2, Pbx1, Zfp37, Prdm5, Mycn, Meis1 | FOSB, GFI1, RUNX1, SPI1 |
| Factor inducibility | Constitutive | Constitutive | Inducible | Constitutive | Inducible | Inducible | Constitutive |
| Medium | In vitro | In vitro | In vitro | In vitro | In vitro | In vivo | In vitro with endothelial stroma |
| In vitro colony formation | + | + | + | + | + | + | + |
| Erythroid | + | + | Not shown | + | + | + | + |
| Myeloid | + | + | + | + | + | + | + |
| B | − | − | − | −a | − | + | + |
| T | − | − | − | −a | +b | + | +c |
| Engraftment | +d | +d | − | +d | + | + | + |
| Serial Transplantation | − | − | − | − | − | + | + |
| HSC? | No | No | No | No | No | Yes | Nof |
Lymphoid differentiation potential acquired with p53 deletion.
Although modest T-cell differentiation potential was confirmed in vitro, no T cells were detected in vivo.
Minimal in vitro T-cell differentiation possible when TFs are expressed using inducible system.
Engrafted cells express low levels of CD45, a pan-lympho-myeloid hematopoietic marker.
Very short-term (2 week), primarily erythroid engraftment.
In vivo function not assayed with cells derived using inducible system.
Szabo et al (2010) and Pulecio et al (2014) converted human fibroblasts to hematopoietic cells possessing multilineage myeloid potential aided by pluripotency-associated TFs, namely OCT4 and SOX2, respectively. The latter study also showed improved hematopoietic conversion with the addition of mir125b, a microRNA enriched in human hematopoietic progenitors. Since transient expression of pluripotency factors or OCT4 is sufficient to confer tri-germ layer differentiation potential on fibroblasts, fate conversion specifically to the blood lineage with OCT4 or SOX2 was likely mediated by the inductive effects of hematopoietic cytokines (Mitchell et al, 2014), which has previously been shown to be able to reprogram blood cell identity (Kondo et al, 2000). Although the resulting cells were able to engraft in vivo, the majority of them expressed low levels of the pan-hematopoietic antigen, CD45, and did not peripheralize, possibly suggesting incomplete hematopoietic conversion.
Instead of transitioning through a developmentally plastic state, a more direct, autonomous fate transition could be achieved by overexpressing lineage-specifying TFs enriched in destination cells. Toward this, Pereira et al (2013) screened 18 candidate TFs enriched in quiescent mouse HSCs that could activate exogenous human CD34 promoter inserted into mouse fibroblasts. The screen identified transient expression of Gata2, Gfi1b, cFos, and Etv6 to be sufficient for generating hematopoietic cells from fibroblasts via an intermediate cell type that coexpressed both endothelial and hematopoietic markers. Although the converted hematopoietic cells were similar to mouse hematopoietic stem/progenitor cells with respect to gene expression, they were devoid of in vitro clonogenic potential unless cocultured with placental stroma, suggesting that maturation into progenitor-like blood cells required additional signals. Clonal multilineage potential or in vivo functionality was not assayed. A similar fate conversion strategy from fibroblasts was employed by Batta et al (2014) who screened a curated set of 19 hematopoietic TFs for morphological change of murine fibroblasts to round hematopoietic cells. Five TFs, Erg, Gata2, Lmo2, Runx1c, and Scl, were found to robustly induce hematopoietic colonies from both embryonic and adult fibroblasts, and the reprogrammed cells were shown to possess erythroid, megakaryocytic, granulocytic, and macrophage differentiation potentials. Similar to Pereira et al, Batta et al also observed that fibroblasts converted to hematopoietic cells via an endothelial intermediate. In vitro clonogenic assays confirmed the presence of cells possessing multilineage potential; upon transplantation, however, these cells only gave rise to very short-term (2 weeks) erythroid chimerism. Interestingly, p53 nullizygosity not only enhanced the efficiency of reprogramming but also increased erythroid differentiation potential in addition to permitting production of receptor rearranged B and T lineage cells.
Although iPS cells have the developmental potential to be differentiated toward potentially transplantable autologous tissues, their hematopoietic differentiation has yielded progenitors with greatly restricted self-renewal and differentiation potentials quite unlike those of true HSCs. Doulatov et al (2013) sought to respecify iPS cell-derived myeloid restricted progenitors toward HSCs using TFs enriched in both human and mouse HSCs that appeared underexpressed in the blood progenitors cells derived from pluripotent cells. Screening nine candidate TFs and using serial plating as a readout, ectopic expressions of ERG, HOXA9, and RORA were found to instill robust in vitro clonogenic potential but not multilineage potential or engraftment capacity. However, additional ectopic expression of SOX4 and MYB enabled the acquisition of myelo-erythroid differentiation potential as well as short-term myeloid engraftment capacity in immunocompromised mice. Although modest T lineage potential was confirmed in vitro, the grafts failed to produce lymphoid lineage in vivo. Long-term engraftment was not achieved.
As opposed to respecifying embryonic-like hematopoietic cells derived from pluripotent stem cells, Riddell et al (2014) undertook reprogramming of primary adult lineage committed murine hematopoietic progenitors and effectors using gene regulatory factors exhibiting restricted expression in mouse HSCs relative to the majority of their differentiated progeny. An unbiased screen of 36 factors, which included 33 TFs and three translational regulators, was performed in the transplantation setting to take advantage of the sensitivity of the assay in reading out HSC activity at the single-cell level, and potentially co-opt signals present in the in vivo environment that might facilitate cell conversion. The screen identified six genes Hlf, Runx1t1, Pbx1, Lmo2, Prdm5, and Zfp37 whose transient ectopic expression was sufficient for instilling multilineage reconstituting potential on otherwise lineage committed hematopoietic cells. Inclusion of Meis1 and Mycn was found to improve reprogramming efficiency. Long-term multilineage reconstitution, serial transplantability, reconstitution of bone marrow progenitor compartments and secondary hematopoietic organs, and single-cell gene expression profiling confirmed that the reprogrammed cells possessed the functional and molecular properties of endogenous HSCs and thus were termed ‘induced HSCs’ (iHSCs).
HSCs and endothelial cells share an intimate ontological relationship as HSCs are specified from hemogenic endothelial-like intermediates during embryogenesis via endothelial-to-hematopoietic transition (EHT). Sandler et al hypothesized that EHT may be recapitulated in mature, non-hemogenic endothelial cells by ectopic expression of key TFs and provision of an inductive environment (Zovein et al, 2008; Bertrand et al, 2010; Boisset et al, 2010; Gordon-Keylock & Medvinsky, 2011; Sandler et al, 2014). Screening 25 TFs expressed at a higher level in human cord blood HSPCs relative to human umbilical vein endothelial cells (HUVECs) identified the minimal set of FOSB, GFI1, RUNX1, and SPI1 to be sufficient and necessary for robustly generating hematopoietic colonies from HUVECs and human adult dermal microvascular endothelial cells. The reprogramming was found to strictly depend on an endothelial stroma previously developed by the authors' group for maintaining human cord blood HSPCs (Butler et al, 2012). The reprogrammed cells possessed both in vitro and in vivo multilineage differentiation potential, long-term reconstitution potential, in vivo homing/engraftment capacity, and serial transplantability, only with the caveat of defective T-cell differentiation potential, thus earning the label of multipotent progenitors (MPPs).
Despite sharing the same destination identity, the studies summarized above show striking diversity with respect to experimental design and the TF combinations identified (Fig1). However, in totality, the reported TFs are highly enriched for both developmental genes involved in embryonic specification of hematopoiesis and those implicated in leukemogenesis. Classical developmental hematopoiesis genes such as Scl, Gata2, Gfi1, Runx1, and Spi1(Pu.1) (Wilson et al, 2010b) appear to be important in fate conversions that involve lineage switching, such as from fibroblasts or endothelial cells, whereas they do not appear to be important in reprogramming or respecification within the blood lineage. This could reflect that even post-embryogenesis, the same small set of TFs that specified hematopoiesis earlier in life can pioneer the establishment of hematopoietic program in non-hematopoietic adult cell types, though such activity may be redundant in conversions within hematopoietic lineage. The requirement for TFs such as HOXA9, MYB, Lmo2, Pbx1, Mycn, and Meis1 in reprogramming of committed blood cells is consistent with their classical roles in tumor development (Thorsteinsdottir et al, 2001; Kawagoe et al, 2007; Jin et al, 2010; McCormack et al, 2010). In particular, components of Hox effectors, namely HOXA9, Pbx1, and Meis1, are found in TF sets reported by both Doulatov et al and Riddell et al. This particular set of TFs has been shown to function as a heterotrimeric complex (Shen et al, 1997) and is frequently represented in regenerative processes (Mercader et al, 2005; Capellini et al, 2006; Chen et al, 2013; Roensch et al, 2013) and across multiple types of cancers (Morgan et al, 2007; Shears et al, 2008; Plowright et al, 2009; Sun et al, 2013), suggesting that it may regulate generic properties of stem/progenitors such as self-renewal, anti-apoptosis, and differentiation arrest. In spite of the apparently central role played by the Hox complex in regulating stemness, it is intriguing that ectopic stimulation of this pathway was not required to generate progenitor cells as highly functional as the MPPs directly from endothelial cells (Sandler et al, 2014). However, this could be explained by the fact that endothelial cells exhibit intrinsic HoxA cluster activity, which is integral to vascular development and function (Rössig et al, 2005; Bandyopadhyay et al, 2012).
Figure 1.
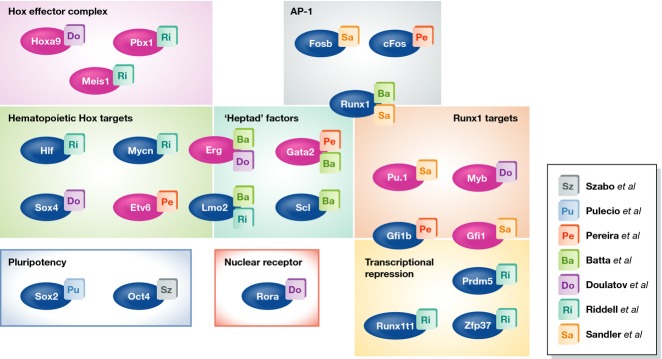
Categorization of transcription factors employed toward de novo HSC generation
TFs employed by Szabo et al (Sz), Pulecio et al (Pu), Pereira et al (Pe), Batta et al (Ba), Doulatov et al (Do), Riddell et al (Ri), and Sandler et al (Sa) are classified into categories based on previously described activities. ‘Heptad’ factors refer to classical hematopoietic TFs known for their physical interactions (Wilson et al, 2010a). Individual loss of function of TFs highlighted in pink has been shown to impair HSC activity.
Species-specific differences may also contribute to the identification of distinct HSPC–inducing TF combinations. It is possible, though not rigorously studied, that divergent gene regulatory networks may govern mouse and human HSCs. For example, although constitutive ectopic expression of Hoxb4 allows the generation of engraftable mouse HSPCs from mouse embryonic stem cells (Wang et al, 2005b; Matsumoto et al, 2009), the same robust effect was not seen using human cells (Wang et al, 2005a). In the erythroid lineage, noticeable transcriptional divergence was found between mouse and human cells isolated from comparative stages of differentiation (Pishesha et al, 2014). Furthermore, overexpression of a dominant-negative isoform of IKAROS (IKZF1) was recently found to impart diametric effects on mouse and human HSPCs (Beer et al, 2014). Whereas IKZF1 overexpression in mouse HSPCs suppressed B but enhanced T lineage outputs, the same manipulation in human cord blood HSPCs significantly increased B lineage cell production without affecting T lineage. Therefore, although the majority of TFs discovered in the studies using murine cells (Pereira et al, 2013; Riddell et al, 2014) are highly homologous between mouse and human, it is uncertain whether the same set of factors capable of inducing mouse HSPCs would also be sufficient for inducing human HSCs.
Another major variable among the studies is the system used for ectopic expression of TFs. An important criterion for complete cell fate conversion is transgene-independent sustenance of destination cell gene regulatory networks. Although transduction with viruses encoding TFs under constitutive promoters may become passively silenced over time, the use of an inducible transgene system (i.e., doxycycline inducible) can give better temporal control over TF expression as well as ensure that the ectopic genes are turned off upon completion of cell fate conversion. Importantly, continued ectopic expression of TFs involved in ‘respecification/reprogramming’ may also interfere with the function of the destination cells. For example, continued ectopic expression of SPI1 in endothelial cell-derived multipotent progenitors (MPPs) was shown to block T-cell differentiation potential (Sandler et al, 2014). In the same study, as only MPPs obtained using constitutive expression vectors were assayed in vivo, it remains to be shown definitively whether the MPPs can stably maintain their identity independent of residual transgene expression.
Since environmental responsiveness and developmental plasticity represent cardinal properties of HSCs, it is conceivable that environmental cues may strongly influence cell fate conversion toward HSCs. Reasoning that reprogramming in the context of the native HSC niche (Morrison & Scadden, 2014) may facilitate the acquisition of HSC identity, Riddell et al (2014) conducted reprogramming experiments in vivo. Similarly, Sandler et al took advantage of an endothelial stromal coculture system developed for ex vivo maintenance of human HSCs to provide an inductive environment for de novo HSC generation (Butler et al, 2012; Sandler et al, 2014). Intriguingly, both of these studies gave rise to serially transplantable cells possessing lympho-myeloid multilineage potential, a feat unattained by any of the other cell fate conversion strategies, which were all conducted in the absence of HSC supportive milieu. Although Riddell et al did not show the absolute requirement of the in vivo environment for HSC induction, an HSC supportive environment was shown to be a necessity in the conversion of endothelial cells to MPPs by Sandler et al Insights from other cell fate conversion systems also point to the importance of the environment. In particular, STAT3, a transcription factor directly activated by growth factor signaling, has been shown to play an central role in embryonic stem cells as well as in induction of pluripotency (Niwa et al, 1998; Raz et al, 1999; Yang et al, 2010; van Oosten et al, 2012), thus emphasizing that cell extrinsic cues can be as important as the intrinsic ones.
Toward understanding the transcriptional regulation of HSC identity
In spite of enormous progress, the era of HSC induction has only dawned. With little consensus on the optimal combination of TFs and/or environmental cues for inducing HSCs, it may be necessary to perform comparative studies or even ‘mix and match’ findings from hitherto studies for even better results. With respect to reevaluating TF combinations for human HSC induction, it may be worth noting the heterogeneity of primary human HSCs. Although immunophenotypically defined single human HSC has been successfully isolated (Notta et al, 2011), a number of studies show that the human HSC compartment can still be fractionated into subpopulations with distinct functional potentials (Anjos-Afonso et al, 2013; Chitteti et al, 2014) similar to the HSC subfractionation that has been demonstrated in the mouse (Dykstra et al, 2007; Beerman et al, 2010; Morita et al, 2010; Babovic & Eaves, 2014). Therefore, TFs enriched in each of these subpopulations may differentially impact HSC induction.
Understanding the function of individual TFs in HSCs constitutes an important step toward elucidating the mechanism of HSC induction. Consistent with the ability to access the HSC gene regulatory program, many of the TFs identified by the studies highlighted in this review have been previously implicated in HSC and progenitor biology. Individual loss of function of Hoxa9 (Lawrence et al, 2005), Gata2 (Lim et al, 2012), Pbx1 (Ficara et al, 2008), Meis1 (Unnisa et al, 2012), Gfi1 (Hock et al, 2004; Zeng et al, 2004), Etv6 (Wang et al, 1998), SPI1 (Pu.1) (Staber et al, 2013), Myb (Lieu & Reddy, 2009), or Erg (Ng et al, 2011) in HSCs has each been shown to compromise their homeostatic maintenance and/or competitive repopulation potential. Although direct loss of function of Lmo2 in HSCs has not been conducted, its critical role in hematopoiesis is underscored by the inability of Lmo2 null ES cells to contribute to postnatal hematopoiesis in chimeric blastocysts (Yamada et al, 1998). Similarly, Runx1 null ES cells cannot contribute to adult hematopoiesis (Okuda et al, 1996) due to impairment of EHT (Chen et al, 2009). Intriguingly, however, conditional loss of Runx1 in the adult hematopoietic system only impairs megakaryocytic and lymphoid differentiation with minimal impact on HSC activity (Ichikawa et al, 2004; Cai et al, 2011). Although the consequence of loss of function of Sox4, Rora, Runx1t1, Zfp37, Prdm5, or Hlf in HSCs has yet to be reported, overexpression and leukemia studies have implicated some of these factors in HSC function. For example, Sox4 has been implicated in self-renewal of leukemic cells (Zhang et al, 2013) and ectopic expression of Hlf, a leukemia-associated TF (Hunger et al, 1992), has been shown to enhance repopulation potential of human HSCs (Shojaei et al, 2005) and allows maintenance of in vitro multilineage differentiation potential and serial plating capacity of murine HSCs and progenitors (Gazit et al, 2013). Also of note, many of these understudied factors, namely Runx1t1 (Lindberg et al, 2003), Zfp37 (Dreyer et al, 1998), and Prdm5 (Duan et al, 2007), identified by Riddell et al (Riddell et al, 2014) function as transcriptional repressors, suggesting that active repression of differentiation-associated genes may be an important component of reprogramming differentiated blood cells back to HSCs. Finally, loss of function of some TFs may not result in HSC defects due to functional redundancies with other TFs. For example, Mycn deletion in HSCs is compensated by the presence of c-Myc (Laurenti et al, 2008). However, combined deletion of Mycn and c-Myc demonstrated the requirement of the Myc genes in the exit from stem cell state as well as preventing apoptosis of HSCs. Similarly, Scl nullizygosity has no effect on HSCs due to its functional redundancy with Lyl1, but their combined ablation leads to loss of HSCs via apoptosis (Souroullas et al, 2009).
Detailing the interactions between the TFs used in HSC induction may provide deeper insights into the gene regulatory network governing HSC identity, which could lend to discoveries of more efficient or alternative TF combinations for inducing HSCs. Many of the reported TFs have already been shown to interact physically and/or at the transcriptional level. As mentioned previously, Hoxa9, Meis1, and Pbx1 directly interact to coregulate transcription (Shen et al, 1997), although they may also function independently as their individual deletions result in differential HSC phenotypes (Lawrence et al, 2005; Ficara et al, 2008; Unnisa et al, 2012). The Hox complex cannot only autoregulate Hox gene expression (Trivedi et al, 2008; Horman et al, 2009) but also enhance the expression of a number of other reprogramming TFs such as Erg, Myb, Sox4, Lmo2, Etv6, Mycn, and Hlf in blood cells (Palmqvist et al, 2007; Nagel et al, 2011; Huang et al, 2012) although there also appears to be redundancies among these targets such as the cross-regulation between Lmo2 and Erg (Oram et al, 2010) and induction of Lmo2 by Hlf (de Boer et al, 2011). However, the diverse and wide-ranging targets of the Hox complex combined with the fact that it regulates distinct processes in non-hematopoietic tissues (i.e., body segmentation, blastema formation, and non-hematopoietic cancers, mentioned previously) suggests that the complex may require interaction with lineage-restricted TFs to exert tissue-specific functions. In support of this, Hoxa9/Meis1 has been shown to specifically bind and activate myeloid enhancers in cooperation with hematopoietic TFs such as SPI1(Pu.1), and Runx1 (Huang et al, 2012). This model implies that HSC specification by the Hox complex requires either a priori patterning of relevant enhancers or coexpression of hematopoietic-specific TFs that can pioneer enhancer establishment. Following this logic, under certain circumstances, overexpression of the Hox complex may impede cell fate conversion by promoting maintenance of enhancers specific to the starting cell type, akin to the antagonism of EHT by Hoxa3 during embryogenesis (Iacovino et al, 2011).
Extensive interactions have also been reported between TFs classically associated with hematopoiesis such as Scl, Runx1, Gata2, Gfi1, and SPI1(Pu.1) that appear in the reprogramming TF cocktails. For example, a heptad of TFs regulating HSPCs, SCL, LYL1, LMO2, GATA2, RUNX1, ERG, and FLI1 have been shown to directly bind each other and cooperatively regulate hematopoietic genes (Wilson et al, 2010a) as well as autoregulate their own expression (Grass et al, 2003; Pimanda et al, 2007; Diffner et al, 2013). While the majority of these TFs is implicated in both hematopoietic and endothelial development (De Val & Black, 2009), Runx1 has received much attention for its specific and critical role in embryonic HSC specification (North et al, 2002). During mid-gestation, Runx1 specifies HSCs by acting as a pioneering TF that initiates expression of hematopoietic genes such as SPI1(Pu.1) in an endothelial-like cell (Huang et al, 2008; Chen et al, 2009; Lichtinger et al, 2012; Tanaka et al, 2012). The activity of Runx1 is modulated not only by its obligate binding partner Cbfb and other members of the heptad TFs but also by the AP1 complex, whose motifs are highly enriched at genomic Runx1 binding peaks (Pencovich et al, 2011; Lie et al, 2014). AP1 complex has been shown to physically interact with Runx1 (Hess et al, 2001; D'Alonzo et al, 2002) and is itself a heterodimer of JUN and FOS family proteins, the latter of which were found to be necessary to induce hematopoiesis from non-hematopoietic cells in the studies by Pereira et al (2013) and Sandler et al (2014). The same studies also identified Gfi1/Gfi1b and SPI1(Pu.1), direct targets of Runx1 that promote the loss of endothelial identity associated with EHT (Lancrin et al, 2012; Pereira et al, 2013; Sandler et al, 2014; Wilkinson et al, 2014). Interestingly, Gfi1 has been shown to directly repress Hoxa9, Pbx1, and Meis1 (Horman et al, 2009), which could support the aforementioned notion that suppression of Hox complex may be important to facilitate lineage switching toward blood. Ultimately, the kinetics and expression levels of TFs may need to be regulated for optimal HSC induction.
TFs mediate cell fate conversion by rewriting epigenetic information, which has been shown to be facilitated by directly modulating nuclear enzymes responsible for chromatin modifications. A number of small molecules that increase chromatin accessibility have been shown to enhance the efficiency of iPSC generation (Federation et al, 2014). One study even demonstrated that a G9a histone methyltransferase inhibitor was able to replace Oct4 in inducing pluripotency from mouse fetal neural progenitors (Shi et al, 2008). Although the mechanisms governing induction of pluripotency or HSCs may differ, barriers to accessing the HSC program may be similarly lowered by the use of such small molecules. Epigenetics has also been shown to underlie functional heterogeneity within the HSC compartment, especially with respect to aging-induced increase in self-renewal and myeloid lineage bias (Beerman et al, 2013; Sun et al, 2014a). Although reprogramming to pluripotency has been demonstrated to be sufficient in reversing epigenetic components of HSC aging (Wahlestedt et al, 2013), other studies have shown functional and molecular heterogeneity of iPSCs that originate from retention of epigenetic information associated with the starting cell type (Hu et al, 2010; Kim et al, 2010; Vaskova et al, 2013). Therefore, an intriguing question would be to determine whether the starting cell age is reset upon direct cell fate conversion to HSCs and whether HSCs induced from committed blood cell lineages exhibit differentiation bias toward the respective lineages. This latter point was not however evident in iHSCs generated in the Riddell et al (2014) study as iHSCs derived from either B-cell progenitors or myeloid progenitor or effector cells appeared comparable in their capacity to give rise to lymphoid and myeloid effector cells in vivo.
Complementary to overexpressing HSC-specific TFs, direct extinction of gene regulatory networks governing starting cell types may further augment cell fate transition. Since deletion of Pax5 has already been shown to be sufficient for liberation from B lineage fate (Nutt et al, 1999), such manipulation may enhance HSC induction from B-cell progenitors. The use of RUNX1 and GFI1 by Sandler et al in inducing hematopoiesis from endothelial cells indeed follows the logic of their developmental role in extinguishing the endothelial program (Iacovino et al, 2011; Lancrin et al, 2012).
Toward human HSCs
Given the rapid progress in the field, derivation of the first human HSCs via cell fate conversion appears increasingly attainable. The diverse strategies presented thus far provide lessons that may help refine the approach to generating human HSCs as well as gain deeper insights into the mechanism of HSC induction.
Though already mentioned earlier, the importance of using inducible expression vectors should be reemphasized. Inducibility is critical to ensure transient ectopic expression of TFs in order to demonstrate that resulting HSCs possess self-sustaining and stable identity. Another advantage of this system is the re-inducibility of the TFs, which can be exploited for secondary reprogramming experiments (Wernig et al, 2008; Riddell et al, 2014).
To accelerate the study of HSC induction, it would be critical to perform experiments in defined media. Although the endothelial stromal system used by Sandler et al appears to provide a powerful inductive milieu, it remains to be seen whether it is generally applicable. Nonetheless, it is also possible that a special environment may not be necessary for certain starting cell types and/or TF combinations. In either case, with an ability to perform the entirety of cell fate conversion in vitro, it should be possible to gain finer control over experimental parameters as well as to obtain kinetic information on the induction of HSC identity.
The crux of HSC induction is the functional evaluation of test cells. Rigorous assays should be employed to confirm the in vivo potential of induced HSCs to self-renew and generate multilineage progeny at the clonal level. Long-term reconstitution and serial transplantation are commonly employed to assess the in vivo self-renewal potential of HSCs. These assays also indirectly provide indications on the ability of test cells to properly home to and engraft in the niches. In vivo functionality of HSCs is contingent upon their successful engraftment. To arrive at bone marrow niches, HSCs must home to the correct vasculature, extravasate, and then migrate to gain contact with the niche components (Lapidot et al, 2005). The surface molecules and signal transduction components necessary for orchestrating this process are thus integral to HSC function. Although it is possible that this facet of HSC identity is within the domains governed by HSC-specific TFs, it could represent a generic functional module shared with other bone marrow resident cell types. In the latter scenario, orthogonally acting TFs or specific environmental signals may be required in addition to ectopic expression of HSC-enriched TFs to induce engraftable HSCs. For instance, it has been shown that expression of Cxcr4, an essential chemokine receptor for HSC migration, is regulated by Hif1a, suggesting that Cxcr4 induction may dependent on hypoxia rather than HSC-specific TFs (Speth et al, 2014).
The prerequisite for homing and engraftment complicates functional assessment of human HSCs in the setting of xenotransplantation experiments. To circumvent graft rejection, immunodeficient mice that lack B, T, and NK cells are used as hosts for human HSCs (Shultz et al, 2012). Even then, the engraftment potential of human HSC in murine hosts is inferior to that of mouse HSCs, likely due to cross-species differences between cytokines and signaling/homing receptors or even due to the incompatibility of human CD47, a ‘don't eat me’ signal, with host phagocytes (Jaiswal et al, 2009; Kwong et al, 2014). To obtain robust chimerism, human HSCs may need to be injected directly into the bone marrow cavity via intrafemoral injection, thus bypassing the complex orchestration of cellular maneuvers leading to engraftment. The caveat, however, is that lowering the hurdle to engraftment or differences in transplantation procedures may result in incongruous definitions for human HSCs. Although intrafemoral injection of induced HSCs could be justified if one were to claim the necessity of the in vivo environment in inducing proper homing/engraftment capacity, this needs to be confirmed with secondary transplantation via intravenous route of injection. The difference in the rigor of in vivo assays used to define HSCs as well as cross-species mismatches in signaling may also underlie molecular differences between mouse and human HSCs, and it is unknown whether experimentally derived cells tested in xenograft models will ultimately function effectively in human patients. An important step, therefore, is to develop better models for native engraftment of human HSCs, such as better humanized mice or reconstituted human bone marrow, so that their functional definition could be refined (Drake et al, 2012; Scotti et al, 2013; Cosgun et al, 2014; Torisawa et al, 2014).
The question of in vivo clonal multilineage differentiation potential, though challenging, can potentially be addressed using two molecular approaches. First, if HSCs can be induced from B or T lineage cells that have undergone receptor rearrangement, it would be possible to use the unique sequences of the recombined loci as a bar code to track the clonal progenitor cell origin of effector cell progeny as was done by Riddell et al (2014). Receptor rearrangement also provides direct evidence for the cell of origin of the reprogrammed cells, an important consideration given that even a single contaminating HSC inadvertently introduced during transplantation experiments has the potential to confound interpretations. The second approach is viral integration analysis that takes advantage of viral insertion sites that can serve as cellular barcodes. Finally, single-cell transplantation experiments can be used to assure clonal multilineage potential in vivo—though this is a very high bar to surmount, especially if cell fate conversion efficacy is low.
Toward clinical translation
Despite the progress, a number of challenges must be met before de novo generated human HSCs can reach clinical translation (Fig2). Despite the diversity of TF combinations hitherto used for hematopoietic induction, a commonality among most of them is their proto-oncogenicity. Although no tumors have thus far been reported by hematopoietic cell fate conversion studies, the potential dangers of these potent TFs combined with the risks of insertional mutagenesis by lentiviral vectors cannot be tolerated for use in generating clinical-grade human HSCs. Therefore, the current lentiviral methods for HSC induction may be most applicable for such uses as disease modeling.
Figure 2.
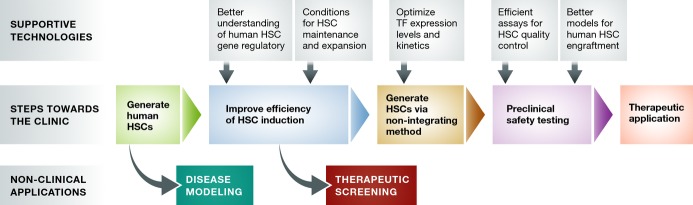
Road map to clinical translation of human induced HSCs
Upon establishment of a method for generating human HSCs from patient-derived cells, a number of critical steps must be taken en route to clinical translation. Although the initial, suboptimal method may be sufficient for initiating patient-specific disease modeling research, substantial improvements in the efficiency of HSC generation must be made to obtain sufficient numbers of HSCs toward therapeutic screening and for use in reconstituting adult patients. Prior to preclinical testing, a non-integrating approach to generating HSCs must be established in order to eliminate the risks associated with insertional mutagenesis and accidental re-induction of reprogramming TFs, many of which are potently oncogenic. During the preclinical phase, quality control and safety testing should be performed at molecular and functional levels. Functional testing should involve the best methods for modeling human HSC engraftment such as humanized immunodeficient animal recipients.
Given the risks associated with lentiviral transduction, non-integrating approaches will need to be considered toward clinical translation. A number of such methods developed for generating integration-free iPSCs including protein transduction, non-integrating viruses, and mRNA based transient protein expression systems could potentially be applied to HSC induction (Fusaki et al, 2009; Warren et al, 2010; Zhang et al, 2012; Mandal & Rossi, 2013; Yoshioka et al, 2013; Elcheva et al, 2014). However, as the non-integrating approaches are highly variable with respect to the duration of ectopic factor expression, the kinetics of respecification/reprogramming will need to be investigated in parallel to optimize the generation of integration-free HSCs. Furthermore, the level of TF induction may also need to be controlled since the stoichiometry of TFs may impact reprogramming efficiency as well as the functionality of reprogrammed cells as has been reported in other systems (Carey et al, 2011).
In addition to guaranteeing safety, it will be necessary to increase the efficiency of HSC induction to obtain sufficient numbers for transplantation. Although the efficiency of generating iPSCs has increased dramatically, their utility is augmented by their limitless self-renewal potential and well-defined culture conditions that support it. Since it is unclear whether HSCs could ever be expanded to the same extent as iPSCs while avoiding functional decline as seen with aging (Rossi et al, 2005; Beerman et al, 2013; Beerman & Rossi, 2014; Sun et al, 2014a), an ideal solution would entail near deterministic induction of HSCs from readily available somatic cells such as peripheral blood or fibroblasts. Until such efficiency can be reached, robust, defined means for ex vivo HSC expansion should be pursued.
Finally, the assays used to confirm HSC identity should be reevaluated. Currently, the only reliable, accepted method for assessing HSC function is competitive transplantation, a resource and time draining procedure incompatible with routine quality control of patient-derived HSCs. Therefore, a bioinformatic metric, like those developed for iPSC quality control (Nestor & Noggle, 2013), predictive of HSC functionality will not only benefit clinical translation but also accelerate the pace of HSC induction research.
Conclusion
“What I cannot create, I do not understand.” Richard Feynman
HSCs have captivated generations of researchers not only because of their clinical importance but also for their intriguing yet elusive biological properties. Arguably, a major contributor to this allure has been the challenges associated with the seemingly simple task of obtaining more HSCs. Even while the biology of HSCs continues to be elucidated in staggering resolution with the advent of increasingly sensitive and high throughput methods (Lu et al, 2011; Gazit et al, 2013; Cabezas-Wallscheid et al, 2014; Lara-Astiaso et al, 2014; Sun et al, 2014a,b; Wu et al, 2014) as well as more accurate description of the HSC niche (Morrison & Scadden, 2014), robust ex vivo expansion or de novo generation of human HSCs still remains elusive. However, major strides made by the recent cohort of fate conversion studies have introduced new hope and perspectives to a field historically reigned by attempts to mimic the natural processes that regulate and specify HSCs. Despite having to induce non-physiologic cell fate transitions, fate conversion toward HSC has begun to yield relevant insights into HSC biology that may synergize with preexisting paradigms to better understand the ontogeny, maintenance, dysregulation, and therapeutic potential of HSCs.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Anjos-Afonso F, Currie E, Palmer HG, Foster KE, Taussig DC, Bonnet D. CD34(−) cells at the apex of the human hematopoietic stem cell hierarchy have distinctive cellular and molecular signatures. Cell Stem Cell. 2013;13:161–174. doi: 10.1016/j.stem.2013.05.025. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Babovic S, Eaves CJ. Hierarchical organization of fetal and adult hematopoietic stem cells. Exp Cell Res. 2014;329:185–191. doi: 10.1016/j.yexcr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Bakanay SM, Demirer T. Novel agents and approaches for stem cell mobilization in normal donors and patients. Bone Marrow Transplant. 2012;47:1154–1163. doi: 10.1038/bmt.2011.170. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Harris DP, Adams GN, Lause GE, McHugh A, Tillmaand EG, Money A, Willard B, Fox PL, Dicorleto PE. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32:1202–1213. doi: 10.1128/MCB.05977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014;9:1871–1884. doi: 10.1016/j.celrep.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer PA, Knapp DJ, Kannan N, Miller PH, Babovic S, Bulaeva E, Aghaeepour N, Rabu G, Rostamirad S, Shih K, Wei L, Eaves CJ. A dominant-negative isoform of IKAROS expands primitive normal human hematopoietic cells. Stem Cell Reports. 2014;3:841–857. doi: 10.1016/j.stemcr.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, Rossi DJ. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, Rossi DJ. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12:413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Beerman I, Rossi DJ. Epigenetic regulation of hematopoietic stem cell aging. Exp Cell Res. 2014;329:192–199. doi: 10.1016/j.yexcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- de Boer J, Yeung J, Ellu J, Ramanujachar R, Bornhauser B, Solarska O, Hubank M, Williams O, Brady HJ. The E2A-HLF oncogenic fusion protein acts through Lmo2 and Bcl-2 to immortalize hematopoietic progenitors. Leukemia. 2011;25:321–330. doi: 10.1038/leu.2010.253. [DOI] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bonzanni N, Garg A, Feenstra KA, Schutte J, Kinston S, Miranda-Saavedra D, Heringa J, Xenarios I, Gottgens B. Hard-wired heterogeneity in blood stem cells revealed using a dynamic regulatory network model. Bioinformatics. 2013;29:i80–i88. doi: 10.1093/bioinformatics/btt243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenkamp N, Ulyanchenko S, O'Neill KE, Manley NR, Vaidya HJ, Blackburn CC. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat Cell Biol. 2014;16:902–908. doi: 10.1038/ncb3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs' eggs. Proc Natl Acad Sci U S A. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Itskovich E, Hu YC, Cheng AW, Ganz K, Sarkar S, Fu D, Welstead GG, Page DC, Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, Kulenkampff E, Faddah DA, Shi L, Gao Q, Sarkar S, Cohen M, Goldmann J, Nery JR, Schultz MD, Ecker JR, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JM, Gars EJ, James DJ, Nolan DJ, Scandura JM, Rafii S. Development of a vascular niche platform for expansion of repopulating human cord blood stem and progenitor cells. Blood. 2012;120:1344–1347. doi: 10.1182/blood-2011-12-398115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, Wünsche P, Zeisberger P, Brocks D, Gu L, Herrmann C, Haas S, Essers MA, Brors B, Eils R, Huber W, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA Methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Cai X, Gaudet JJ, Mangan JK, Chen MJ, De Obaldia ME, Oo Z, Ernst P, Speck NA. Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS ONE. 2011;6:e28430. doi: 10.1371/journal.pone.0028430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, Zappavigna V, Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, Welstead GG, Gao Q, Jaenisch R. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wang IE, Reddien PW. pbx is required for pole and eye regeneration in planarians. Development. 2013;140:719–729. doi: 10.1242/dev.083741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AB, Daley GQ. Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitteti BR, Kobayashi M, Cheng Y, Zhang H, Poteat BA, Broxmeyer HE, Pelus LM, Hanenberg H, Zollman A, Kamocka MM, Carlesso N, Cardoso AA, Kacena MA, Srour EF. CD166 regulates human and murine hematopoietic stem cells and the hematopoietic niche. Blood. 2014;124:519–529. doi: 10.1182/blood-2014-03-565721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474:220–224. doi: 10.1038/nature10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- Cosgun KN, Rahmig S, Mende N, Reinke S, Hauber I, Schäfer C, Petzold A, Weisbach H, Heidkamp G, Purbojo A, Cesnjevar R, Platz A, Bornhäuser M, Schmitz M, Dudziak D, Hauber J, Kirberg J, Waskow C. Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell. 2014;15:227–238. doi: 10.1016/j.stem.2014.06.001. [DOI] [PubMed] [Google Scholar]
- D'Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffner E, Beck D, Gudgin E, Thoms JA, Knezevic K, Pridans C, Foster S, Goode D, Lim WK, Boelen L, Metzeler KH, Micklem G, Bohlander SK, Buske C, Burnett A, Ottersbach K, Vassiliou GS, Olivier J, Wong JW, Göttgens B, et al. Activity of a heptad of transcription factors is associated with stem cell programs and clinical outcome in acute myeloid leukemia. Blood. 2013;121:2289–2300. doi: 10.1182/blood-2012-07-446120. [DOI] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, Daley GQ. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake AC, Chen Q, Chen J. Engineering humanized mice for improved hematopoietic reconstitution. Cell Mol Immunol. 2012;9:215–224. doi: 10.1038/cmi.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer SD, Zhou L, Machado MA, Horton WA, Zabel B, Winterpacht A, Lee B. Cloning, characterization, and chromosomal assignment of the human ortholog of murine Zfp-37, a candidate gene for Nager syndrome. Mamm Genome. 1998;9:458–462. doi: 10.1007/s003359900796. [DOI] [PubMed] [Google Scholar]
- Duan Z, Person RE, Lee HH, Huang S, Donadieu J, Badolato R, Grimes HL, Papayannopoulou T, Horwitz MS. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol Cell Biol. 2007;27:6889–6902. doi: 10.1128/MCB.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee S, Brinkman R, Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Elcheva I, Brok-Volchanskaya V, Kumar A, Liu P, Lee JH, Tong L, Vodyanik M, Swanson S, Stewart R, Kyba M, Yakubov E, Cooke J, Thomson JA, Slukvin I. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espin-Palazon R, Stachura DL, Campbell CA, Garcia-Moreno D, Del Cid N, Kim AD, Candel S, Meseguer J, Mulero V, Traver D. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell. 2014;159:1070–1085. doi: 10.1016/j.cell.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federation AJ, Bradner JE, Meissner A. The use of small molecules in somatic-cell reprogramming. Trends Cell Biol. 2014;24:179–187. doi: 10.1016/j.tcb.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R, Garrison BS, Rao TN, Shay T, Costello JF, Erikson J, Collins JJ, Regev A, Wagers A, Rossi DJ. Transcriptome analysis identifies regulators of hematopoietic stem and progenitor cells. Stem Cell Reports. 2013;1:266–280. doi: 10.1016/j.stemcr.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Keylock S, Medvinsky A. Endothelio-hematopoietic relationship: getting closer to the beginnings. BMC Biol. 2011;9:88. doi: 10.1186/1741-7007-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, McNagny K, Brady G, Frampton J. Chicken “erythroid” cells transformed by the Gag-Myb-Ets-encoding E26 leukemia virus are multipotent. Cell. 1992;70:201–213. doi: 10.1016/0092-8674(92)90096-u. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratwohl A, Baldomero H, Aljurf M, Pasquini MC, Bouzas LF, Yoshimi A, Szer J, Lipton J, Schwendener A, Gratwohl M, Frauendorfer K, Niederwieser D, Horowitz M, Kodera Y Worldwide Network of Blood and Marrow Transplantation. Hematopoietic stem cell transplantation: a global perspective. JAMA. 2010;303:1617–1624. doi: 10.1001/jama.2010.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Han DW, Tapia N, Hermann A, Hemmer K, Höing S, Araúzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, Greber B, Yang JH, Lee HT, Schwamborn JC, Storch A, Schöler HR. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Han JK, Chang SH, Cho HJ, Choi SB, Ahn HS, Lee J, Jeong H, Youn SW, Lee HJ, Kwon YW, Cho HJ, Oh BH, Oettgen P, Park YB, Kim HS. Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation. 2014;130:1168–1178. doi: 10.1161/CIRCULATIONAHA.113.007727. [DOI] [PubMed] [Google Scholar]
- Hess J, Porte D, Munz C, Angel P. AP-1 and Cbfa/runt physically interact and regulate parathyroid hormone-dependent MMP13 expression in osteoblasts through a new osteoblast-specific element 2/AP-1 composite element. J Biol Chem. 2001;276:20029–20038. doi: 10.1074/jbc.M010601200. [DOI] [PubMed] [Google Scholar]
- Heyworth C, Pearson S, May G, Enver T. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 2002;21:3770–3781. doi: 10.1093/emboj/cdf368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, Bilenky M, Cezard T, Tam A, Kamoh B, Birol I, Dai D, Zhao Y, Hirst M, Verchere CB, Helgason CD, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege H, Pfeiffer W, Sie D, Hulsman M, Nicholas TJ, Lee CC, Ross T, Lin J, Miller MA, Ylstra B, Meijers-Heijboer H, Brugman MH, Staal FJ, Holstege G, Reinders MJ, Harkins TT, Levy S, Sistermans EA. Somatic mutations found in the healthy blood compartment of a 115-yr-old woman demonstrate oligoclonal hematopoiesis. Genome Res. 2014;24:733–742. doi: 10.1101/gr.162131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman SR, Velu CS, Chaubey A, Bourdeau T, Zhu J, Paul WE, Gebelein B, Grimes HL. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Friedrich AM, Johnson LV, Clegg DO. Memory in induced pluripotent stem cells: reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28:1981–1991. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Huang H, Yu M, Akie TE, Moran TB, Woo AJ, Tu N, Waldon Z, Lin YY, Steen H, Cantor AB. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Mol Cell Biol. 2009;29:4103–4115. doi: 10.1128/MCB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, Robertson G, MacDonald J, Cezard T, Bilenky M, Thiessen N, Zhao Y, Zeng T, Hirst M, Hero A, Jones S, Hess JL. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Ohyashiki K, Toyama K, Cleary ML. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- Iacovino M, Chong D, Szatmari I, Hartweck L, Rux D, Caprioli A, Cleaver O, Kyba M. HoxA3 is an apical regulator of haemogenic endothelium. Nat Cell Biol. 2011;13:72–78. doi: 10.1038/ncb2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovs A, Rybtsov S, Anderson RA, Turner ML, Medvinsky A. Identification of the niche and phenotype of the first human hematopoietic stem cells. Stem Cell Reports. 2014;2:449–456. doi: 10.1016/j.stemcr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120:593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamao H, Mandai M, Okamoto S, Sakai N, Suga A, Sugita S, Kiryu J, Takahashi M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe H, Kandilci A, Kranenburg TA, Grosveld GC. Overexpression of N-Myc rapidly causes acute myeloid leukemia in mice. Cancer Res. 2007;67:10677–10685. doi: 10.1158/0008-5472.CAN-07-1118. [DOI] [PubMed] [Google Scholar]
- Kazemian M, Pham H, Wolfe SA, Brodsky MH, Sinha S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013;41:8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zuniga-Pflucker JC, Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- Krumsiek J, Marr C, Schroeder T, Theis FJ. Hierarchical differentiation of myeloid progenitors is encoded in the transcription factor network. PLoS ONE. 2011;6:e22649. doi: 10.1371/journal.pone.0022649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong LS, Brown MH, Barclay AN, Hatherley D. Signal-regulatory protein alpha from the NOD mouse binds human CD47 with an exceptionally high affinity– implications for engraftment of human cells. Immunology. 2014;143:61–67. doi: 10.1111/imm.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Mazan M, Stefanska M, Patel R, Lichtinger M, Costa G, Vargel O, Wilson NK, Möröy T, Bonifer C, Göttgens B, Kouskoff V, Lacaud G. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314–322. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, Friedman N, Amit I. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, Bernstein ID, Trumpp A. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Müller D, Clarke D, Assi SA, Lie-A-Ling M, Noailles L, Vijayabaskar MS, Wu M, Tenen DG, Westhead DR, Kouskoff V, Lacaud G, Göttgens B, Bonifer C. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie ALM, Marinopoulou E, Li Y, Patel R, Stefanska M, Bonifer C, Miller C, Kouskoff V, Lacaud G. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood. 2014;124:e11–e20. doi: 10.1182/blood-2014-04-572958. [DOI] [PubMed] [Google Scholar]
- Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci U S A. 2009;106:21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Hosoya T, Brandt W, Ku CJ, Hosoya-Ohmura S, Camper SA, Yamamoto M, Engel JD. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122:3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg SR, Olsson A, Persson AM, Olsson I. Interactions between the leukaemia-associated ETO homologues of nuclear repressor proteins. Eur J Haematol. 2003;71:439–447. doi: 10.1046/j.0902-4441.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- Liu ML, Zang T, Zou Y, Chang JC, Gibson JR, Huber KM, Zhang CL. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Rossi DJ. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Protoc. 2013;8:568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Isagawa T, Nishimura T, Ogaeri T, Eto K, Miyazaki S, Miyazaki J, Aburatani H, Nakauchi H, Ema H. Stepwise development of hematopoietic stem cells from embryonic stem cells. PLoS ONE. 2009;4:e4820. doi: 10.1371/journal.pone.0004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- Mitchell R, Szabo E, Shapovalova Z, Aslostovar L, Makondo K, Bhatia M. Molecular evidence for OCT4-induced plasticity in adult human fibroblasts required for direct cell fate conversion to lineage specific progenitors. Stem Cells. 2014;32:2178–2187. doi: 10.1002/stem.1721. [DOI] [PubMed] [Google Scholar]
- Montserrat N, Nivet E, Sancho-Martinez I, Hishida T, Kumar S, Miquel L, Cortina C, Hishida Y, Xia Y, Esteban CR, Izpisua Belmonte JC. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Morgan R, Pirard PM, Shears L, Sohal J, Pettengell R, Pandha HS. Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer Res. 2007;67:5806–5813. doi: 10.1158/0008-5472.CAN-06-4231. [DOI] [PubMed] [Google Scholar]
- Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel S, Venturini L, Meyer C, Kaufmann M, Scherr M, Drexler HG, Macleod RA. Transcriptional deregulation of oncogenic myocyte enhancer factor 2C in T-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2011;52:290–297. doi: 10.3109/10428194.2010.537003. [DOI] [PubMed] [Google Scholar]
- Najm FJ, Lager AM, Zaremba A, Wyatt K, Caprariello AV, Factor DC, Karl RT, Maeda T, Miller RH, Tesar PJ. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat Biotechnol. 2013;31:426–433. doi: 10.1038/nbt.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Takayama N, Hirata S, Seo H, Endo H, Ochi K, Fujita K, Koike T, Harimoto K, Dohda T, Watanabe A, Okita K, Takahashi N, Sawaguchi A, Yamanaka S, Nakauchi H, Nishimura S, Eto K. Expandable megakaryocyte cell lines enable clinically applicable generation of platelets from human induced pluripotent stem cells. Cell Stem Cell. 2014;14:535–548. doi: 10.1016/j.stem.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- Nestor MW, Noggle SA. Standardization of human stem cell pluripotency using bioinformatics. Stem Cell Res Ther. 2013;4:37. doi: 10.1186/scrt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AP, Loughran SJ, Metcalf D, Hyland CD, de Graaf CA, Hu Y, Smyth GK, Hilton DJ, Kile BT, Alexander WS. Erg is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. Blood. 2011;118:2454–2461. doi: 10.1182/blood-2011-03-344739. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, Dzierzak E, Zon LI. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- van Oosten AL, Costa Y, Smith A, Silva JC. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram SH, Thoms JA, Pridans C, Janes ME, Kinston SJ, Anand S, Landry JR, Lock RB, Jayaraman PS, Huntly BJ, Pimanda JE, Göttgens B. A previously unrecognized promoter of LMO2 forms part of a transcriptional regulatory circuit mediating LMO2 expression in a subset of T-acute lymphoblastic leukaemia patients. Oncogene. 2010;29:5796–5808. doi: 10.1038/onc.2010.320. [DOI] [PubMed] [Google Scholar]
- Orkin SH. Transcription factors and hematopoietic development. J Biol Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- Palmqvist L, Pineault N, Wasslavik C, Humphries RK. Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PLoS ONE. 2007;2:e768. doi: 10.1371/journal.pone.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Ottersbach K, Bollerot K, Orelio C, de Bruijn M, Wijgerde M, Dzierzak E. Ventral embryonic tissues and Hedgehog proteins induce early AGM hematopoietic stem cell development. Development. 2009;136:2613–2621. doi: 10.1242/dev.034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencovich N, Jaschek R, Tanay A, Groner Y. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood. 2011;117:e1–e14. doi: 10.1182/blood-2010-07-295113. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma'ayan A, Schaniel C, Lemischka IR, Moore K. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersdorf EW. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood. 2013;122:1863–1872. doi: 10.1182/blood-2013-05-355982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, Landry JR, Wood AD, Kolb-Kokocinski A, Green AR, Tannahill D, Lacaud G, Kouskoff V, Göttgens B. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M, Kossenkov AV, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC, Weiss MJ. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res. 2014;24:1932–1944. doi: 10.1101/gr.164178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishesha N, Thiru P, Shi J, Eng JC, Sankaran VG, Lodish HF. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc Natl Acad Sci U S A. 2014;111:4103–4108. doi: 10.1073/pnas.1401598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright L, Harrington KJ, Pandha HS, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) Br J Cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Pulecio J, Nivet E, Sancho-Martinez I, Vitaloni M, Guenechea G, Xia Y, Kurian L, Dubova I, Bueren J, Laricchia-Robbio L, Izpisua Belmonte JC. Conversion of human fibroblasts into monocyte-like progenitor cells. Stem Cells. 2014;32:2923–2938. doi: 10.1002/stem.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Hong J, Chaves L, Zhuang Y, Chen Y, Wang D, Chabon J, Graham B, Ohmori K, Li Y, Huang H. Antagonistic regulation by the transcription factors C/EBPalpha and MITF specifies basophil and mast cell fates. Immunity. 2013;39:97–110. doi: 10.1016/j.immuni.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, Rossi DJ. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roensch K, Tazaki A, Chara O, Tanaka EM. Progressive specification rather than intercalation of segments during limb regeneration. Science. 2013;342:1375–1379. doi: 10.1126/science.1241796. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössig L, Urbich C, Brühl T, Dernbach E, Heeschen C, Chavakis E, Sasaki K, Aicher D, Diehl F, Seeger F, Potente M, Aicher A, Zanetta L, Dejana E, Zeiher AM, Dimmeler S. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtsov S, Sobiesiak M, Taoudi S, Souilhol C, Senserrich J, Liakhovitskaia A, Ivanovs A, Frampton J, Zhao S, Medvinsky A. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J Exp Med. 2011;208:1305–1315. doi: 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, Medvinsky A. Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43(−) embryonic precursor. Stem Cell Reports. 2014;3:489–501. doi: 10.1016/j.stemcr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM, Rafii S. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511:312–318. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L, Kelly Modis L, Kastner P, Chan S, Duprez E, Otto C, Sieweke MH. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138:300–313. doi: 10.1016/j.cell.2009.04.057. [DOI] [PubMed] [Google Scholar]
- Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18:1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaniel C, Bruno L, Melchers F, Rolink AG. Multiple hematopoietic cell lineages develop in vivo from transplanted Pax5-deficient pre-B I-cell clones. Blood. 2002;99:472–478. doi: 10.1182/blood.v99.2.472. [DOI] [PubMed] [Google Scholar]
- Scotti C, Piccinini E, Takizawa H, Todorov A, Bourgine P, Papadimitropoulos A, Barbero A, Manz MG, Martin I. Engineering of a functional bone organ through endochondral ossification. Proc Natl Acad Sci U S A. 2013;110:3997–4002. doi: 10.1073/pnas.1220108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Shears L, Plowright L, Harrington K, Pandha HS, Morgan R. Disrupting the interaction between HOX and PBX causes necrotic and apoptotic cell death in the renal cancer lines CaKi-2 and 769-P. J Urol. 2008;180:2196–2201. doi: 10.1016/j.juro.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Shen WF, Montgomery JC, Rozenfeld S, Moskow JJ, Lawrence HJ, Buchberg AM, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Shojaei F, Trowbridge J, Gallacher L, Yuefei L, Goodale D, Karanu F, Levac K, Bhatia M. Hierarchical and ontogenic positions serve to define the molecular basis of human hematopoietic stem cell behavior. Dev Cell. 2005;8:651–663. doi: 10.1016/j.devcel.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Shu J, Wu C, Wu Y, Li Z, Shao S, Zhao W, Tang X, Yang H, Shen L, Zuo X, Yang W, Shi Y, Chi X, Zhang H, Gao G, Shu Y, Yuan K, He W, Tang C, Zhao Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12:786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siena S, Schiavo R, Pedrazzoli P, Carlo-Stella C. Therapeutic relevance of CD34 cell dose in blood cell transplantation for cancer therapy. J Clin Oncol. 2000;18:1360–1377. doi: 10.1200/JCO.2000.18.6.1360. [DOI] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth JM, Hoggatt J, Singh P, Pelus LM. Pharmacologic increase in HIF1alpha enhances hematopoietic stem and progenitor homing and engraftment. Blood. 2014;123:203–207. doi: 10.1182/blood-2013-07-516336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber PB, Zhang P, Ye M, Welner RS, Nombela-Arrieta C, Bach C, Kerenyi M, Bartholdy BA, Zhang H, Alberich-Jordà M, Lee S, Yang H, Ng F, Zhang J, Leddin M, Silberstein LE, Hoefler G, Orkin SH, Göttgens B, Rosenbauer F, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013;49:934–946. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MH. Pluripotency and targeted reprogramming: strategies, disease modeling and drug screening. Curr Drug Deliv. 2014;11:592–604. doi: 10.2174/156720181105140922123751. [DOI] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat Biotechnol. 2013;31:416–418. doi: 10.1038/nbt.2571. [DOI] [PubMed] [Google Scholar]
- Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, Rosner MR. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, Gu H, Bock C, Meissner A, Göttgens B, Darlington GJ, Li W, Goodell MA. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014a;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD. Clonal dynamics of native haematopoiesis. Nature. 2014b;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Joshi A, Wilson NK, Kinston S, Nishikawa S, Gottgens B. The transcriptional programme controlled by Runx1 during early embryonic blood development. Dev Biol. 2012;366:404–419. doi: 10.1016/j.ydbio.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tober J, Yzaguirre AD, Piwarzyk E, Speck NA. Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development. 2013;140:3765–3776. doi: 10.1242/dev.094961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisawa YS, Spina CS, Mammoto T, Mammoto A, Weaver JC, Tat T, Collins JJ, Ingber DE. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods. 2014;11:663–669. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Patel RC, Patel CV. Differential regulation of HOXA9 expression by nuclear factor kappa B (NF-kappaB) and HOXA9. Gene. 2008;408:187–195. doi: 10.1016/j.gene.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG, Jenkins NA, Grimes HL, Kumar AR. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood. 2012;120:4973–4981. doi: 10.1182/blood-2012-06-435800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Vaskova EA, Stekleneva AE, Medvedev SP, Zakian SM. “Epigenetic memory” phenomenon in induced pluripotent stem cells. Acta Naturae. 2013;5:15–21. [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt M, Norddahl GL, Sten G, Ugale A, Frisk MA, Mattsson R, Deierborg T, Sigvardsson M, Bryder D. An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood. 2013;121:4257–4264. doi: 10.1182/blood-2012-11-469080. [DOI] [PubMed] [Google Scholar]
- Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, Berry JD, Brown RH, Jr, Cudkowicz ME, Bean BP, Eggan K, Woolf CJ. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Swat W, Fujiwara Y, Davidson L, Visvader J, Kuo F, Alt FW, Gilliland DG, Golub TR, Orkin SH. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–2402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Menendez P, Shojaei F, Li L, Mazurier F, Dick JE, Cerdan C, Levac K, Bhatia M. Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. J Exp Med. 2005a;201:1603–1614. doi: 10.1084/jem.20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yates F, Naveiras O, Ernst P, Daley GQ. Embryonic stem cell-derived hematopoietic stem cells. Proc Natl Acad Sci U S A. 2005b;102:19081–19086. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RN, Pouget C, Gering M, Russell AJ, Davies SG, Kimelman D, Patient R. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev Cell. 2009;16:909–916. doi: 10.1016/j.devcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson AC, Kawata VK, Schütte J, Gao X, Antoniou S, Baumann C, Woodhouse S, Hannah R, Tanaka Y, Swiers G, Moignard V, Fisher J, Hidetoshi S, Tijssen MR, de Bruijn MF, Liu P, Göttgens B. Single-cell analyses of regulatory network perturbations using enhancer-targeting TALEs suggest novel roles for PU.1 during haematopoietic specification. Development. 2014;141:4018–4030. doi: 10.1242/dev.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn MF, Göttgens B. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010a;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Timms RT, Kinston SJ, Cheng YH, Oram SH, Landry JR, Mullender J, Ottersbach K, Gottgens B. Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol Cell Biol. 2010b;30:3853–3863. doi: 10.1128/MCB.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Runnels JM, Lin CP. Intravital imaging of hematopoietic stem cells in the mouse skull. Methods Mol Biol. 2014;1185:247–265. doi: 10.1007/978-1-4939-1133-2_17. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Porayette P, Glosson NL, Conway SJ, Carlesso N, Cardoso AA, Kaplan MH, Yoder MC. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 2012;119:5706–5714. doi: 10.1182/blood-2011-12-397489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka N, Gros E, Li HR, Kumar S, Deacon DC, Maron C, Muotri AR, Chi NC, Fu XD, Yu BD, Dowdy SF. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13:246–254. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- Zhang H, Ma Y, Gu J, Liao B, Li J, Wong J, Jin Y. Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials. 2012;33:5047–5055. doi: 10.1016/j.biomaterials.2012.03.061. [DOI] [PubMed] [Google Scholar]
- Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio A, Welner RS, Ebralidze A, Zhang J, Levantini E, Lefebvre V, Valk PJ, Delwel R, Hoogenkamp M, Nerlov C, Cammenga J, Saez B, Scadden DT, Bonifer C, Ye M, et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell. 2013;24:575–588. doi: 10.1016/j.ccr.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, Tallquist MD, Iruela-Arispe ML. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


