Abstract
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is a global epidemic threat. The aim of this study was to determine which globally known MRSA lineages are currently present at our tertiary care hospital in Switzerland, a hospital with low MRSA prevalence. In light of the increasing prevalence of multi drug resistance including vancomycin resistance we also assessed antibiotic susceptibilities.
Methods
The 146 MRSA strains collected over two years (March 2012 until February 2014) at the University Hospital Zurich, Switzerland, were analyzed by PFGE analysis of SmaI digests in combination with spa-typing. In addition, representative isolates were analyzed by multi locus sequence typing (MLST). Susceptibilities to eight antibiotics were assessed using the Kirby-Bauer disc diffusion method.
Results
Isolates showed resistance to erythromycin (48%), ciprofloxacin (43%), clindamycin (31%), tetracycline (22%), and gentamicin (16%). All isolates were susceptible to vancomycin, 95% were susceptible to sulfamethoxazole/trimethoprim and rifampicin, respectively. PFGE analysis revealed 22 different patterns, with four major patterns that accounted for 53.4% of all MRSA isolates, and seven sporadic patterns. Spa typing revealed 50 different spa types with the predominant types being t008 (14%), t002 (10%), and t127 (9%). 82% of the MRSA isolates could be assigned to six clonal complexes (CCs) namely CC1 (10%), CC5 (23%), CC8 (18%), CC22 (17%), CC30 (11%), and CC45 (3%) based on spa-types, PFGE patterns, and MLST. Two isolates could not be typed by either PFGE analysis or spa-typing and three isolates had spa-types that have not yet been described.
Conclusions
The combination of the two typing methods was more discriminatory as compared to the use of a single method. Several of the lineages that are predominant in Europe are present in our hospital. Resistances to antibiotics have decreased in comparison to a study conducted between 2004 and 2006.
Electronic supplementary material
The online version of this article (doi:10.1186/s12941-015-0075-3) contains supplementary material, which is available to authorized users.
Keywords: MRSA, Epidemiology, Antibiotic susceptibility, Molecular typing
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the major causes of healthcare-associated infections worldwide [1]. In addition, the increasing prevalence of multi drug resistance including vancomycin resistance emphasizes the importance of infection control measures such as MRSA typing. There are considerable variations in the prevalence of MRSA according to geographic area and rates reach over 50% in some regions of the world (Reviewed by Stefani et al. 2012 [2]). Most MRSA strains belong to a few distinct pandemic lineages. The current terminology to describe S. aureus lineages is based on the clonal complexes (CCs) identified by multilocus sequence typing (MLST). MLST involves the sequencing of seven housekeeping genes and each unique allelic profile is assigned a sequence type (ST) [3]. Clonal complexes are defined as groups of STs in which every ST shares at least five of seven identical alleles with at least one other ST in the group [4]. Even though numerous studies have used whole genome sequencing to explore the local and global dissemination of distinct lineages recently (for example [5]), MLST is still considered the “gold standard” of typing. But since for MLST seven house-keeping genes have to be sequenced it is expensive and time-consuming. Therefore, many epidemiological studies have used spa-typing, which includes the sequencing of the polymorphic repeat region of protein A [6]. In the meantime, most CCs can be predicted from spa-types using previous publications (for example [7]) or the ridom spa-server (http://spa.ridom.de/spatypes.shtml [8]). Other methods currently used to type MRSA include SCCmec typing, or multilocus variable-number tandem repeats analysis (MLVA), and were recently reviewed [2]. SCCmec typing uses a defined nomenclature, but there are several typing and subtyping schemes that are not harmonized, and the discriminatory power of this method is limited. Even though MLVA is rapid, high-throughput and has high discriminatory power, no standard methodology or nomenclature has been defined, which makes this method less suitable for assessing long-term and global epidemiology [9].
Especially before the introduction of sequence-based approaches pulsed field gel electrophoresis (PFGE) used to be the “gold standard” of MRSA typing [10]. PFGE analysis is very convenient and has high discriminatory power. PFGE is still widely used for short-term, local epidemiology, and to identify outbreaks. However, it is less suited for studying long-term and global epidemiology since it does not permit to compare between centers. This is reflected by the fact that PFGE patterns (e.g. numbers) used at our hospital are only valid for strains typed at our hospital. In addition, it is challenging to compare PFGE data over long time periods, since several variables (i.e. devices and personnel) might change over time and since PFGE analysis is, at least to a certain degree, a matter of subjective interpretation, i.e. whether a weak band is assessed or not.
All MRSA isolates collected at our hospital, a tertiary care hospital in Switzerland with low MRSA prevalence (3-6%, [11]), have been analyzed by PFGE since 1992 within the scope of our local isolation management. The objectives of the current study were to combine the PFGE data with an additional typing method (spa-typing), and to determine which of the globally known clones were found at our hospital during a two-year study period. MLST served as reference. In addition, antibiotic susceptibilities of the MRSA strains were assessed.
Methods
Bacterial strains
All consecutive, non-duplicate MRSA strains collected at the University Hospital Zurich, Switzerland between March 2012 and February 2014 were included in the study. Isolates included MRSA from patients with infections (including three bacteremias) as well as from colonized individuals. Several reference strains were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) and included USA300-114 (ST8), USA100 (ST5), USA1100 (ST30), USA400 (ST1), USA700 (ST72), and USA500 (ST8). EMRSA-15 (ST22) was a kind gift of Dr. Patrice François (Geneva, Switzerland). Strain CHE482 (ST45) has been previously described [12].
Methicillin resistance detection
Isolates were identified as S. aureus by StaphAureux tests (Remel, Kent, UK). Methicillin resistance was detected by testing cefoxitin resistance using the Kirby-Bauer disc diffusion method [13] and/or by detection of the penicillin binding protein 2a (PBP2a) using the MRSA Screen kit (Denka Seiken Co., Ltd., Tokyo, Japan).
Susceptibility testing
Resistance levels for ciprofloxacin, clindamycin, erythromycin, rifampicin, tetracycline, gentamicin, sulfamethoxazole/trimethoprim, and vancomycin were determined by disk diffusion according to EUCAST guidelines [13]. To compare susceptibility patterns of the current strain collection with a collection from a previous study that has been conducted in 2007 [11], susceptibilities were also interpreted using CLSI guidelines [14]. Intermediate resistance and inducible resistance were assumed to be resistant.
Molecular typing
PFGE typing was done with SmaI digests according to standard protocols and analyzed using the BioNumerics software package (Applied Maths, Belgium). The dice coefficient was used with 1.25% optimization and 1% tolerance to calculate similarities between PFGE patterns. Isolates with >80% identity were considered identical. Isolates with similar PFGE patterns (70% identity) were assigned with letters (e.g. 21a). Numbers were assigned to isolates that occurred at least three times in our laboratory including isolates from other hospitals and isolates that were collected before 2012. Therefore, some of the PFGE patterns only occurred once in this study but have a number. Spa sequence typing of the polymorphic repeat regions of protein A [6] was performed using the Ridom StaphType spa-sequencing protocol and the spaTyper (http://spatyper.fortinbras.us/). Multi-locus sequence typing (MLST) of representative isolates was performed as described [3] and sequence types (STs) were derived using the MLST database (http://www.mlst.net).
Results and discussion
Resistance patterns
Of the 146 isolates 48% showed resistance (or intermediate resistance) to erythromycin, 43% to ciprofloxacin, 31% to clindamycin (of which 14% were inducible), 22% to tetracycline, and 16% to gentamicin (Table 1). All of the 146 isolates were susceptible to vancomycin, and 95% of isolates were susceptible to sulfamethoxazole/trimethoprim and rifampicin, respectively (Table 1). Thirty seven isolates (25%) were susceptible to all antibiotics other than oxacillin. There was a decrease in resistances to ciprofloxacin, erythromycin, gentamicin, rifampicin, and tetracycline as compared to MRSA strains collected between 2004 and 2006 at our hospital [11]. In this previous study, 67% of isolates were resistant to ciprofloxacin (25% more than in the current study), 57% to erythromycin (9% more), 34% to tetracycline (12% more), 22% to gentamicin (7% more), and 11% to rifampicin (6% more). No information was available about the resistance towards sulfamethoxazole/trimethoprim or whether the clindamycin-resistant strains exhibited inducible resistance. Because some of the changes in resistances might be attributable to the change in guidelines from CLSI to EUCAST we also analyzed antibiotic susceptibilities of the current collection using the CLSI guidelines which had been used in the previous study [14]. Decreases were identical for ciprofloxacin, erythromycin, and tetracycline when applying CLSI guidelines. Decreases in resistance to gentamicin and rifampicin were slightly higher (9% and 7% less resistant strains in the current vs. the previous study, respectively).
Table 1.
Resistance phenotypes of MRSA
| Resistance profile | No. (%) of isolates |
|---|---|
| Individual antibiotics | |
| Erythromycin | 70 (47.9) |
| Ciprofloxacin | 62 (42.5) |
| Clindamycin (including inducible resistance) | 45 (30.8) |
| (Inducible Clindamycin resistance) | 20 (13.7)) |
| Tetracycline | 32 (21.9) |
| Gentamicin | 23 (15.6) |
| Sulfamethoxazole/trimethoprim | 8 (5.4) |
| Rifampicin | 7 (4.8) |
| Vancomycin | 0 (0) |
| Multiresistance | |
| Oxacillin + 1 antibiotic | 42 (28.8) |
| Ciprofloxacin | 16 (11.0) |
| Erythromycin | 10 (6.8) |
| Tetracycline | 9 (6.1) |
| Gentamycin | 6 (4.1) |
| Sulfamethoxazole/trimethoprim | 1 (0.7) |
| Oxacillin + 2 antibiotics | 22 (15.1) |
| Ciprofloxacin + erythromycin | 7 (4.8) |
| Clindamycin + erythromycin | 6 (4.1) |
| Erythromycin + tetracycline | 3 (2.1) |
| Other combinations* | 6 (4.1) |
| Oxacillin + 3 antibiotics | 29 (19.7) |
| Ciprofloxacin + clindamycin + erythromycin | 16 (11.0) |
| Ciprofloxacin + erythromycin + tetracyclin | 7 (4.8) |
| Other combinations* | 6 (4.1) |
| Oxacillin + 4 antibiotics | 11 (7.5) |
| Ciprofloxacin + clindamycin + erythromycin + gentamicin | 6 (4.1) |
| Other combinations* | 5 (3.5) |
| Oxacillin + 5 antibiotics* | 1 (0.7) |
| Oxacillin + 6 antibiotics* | 4 (2.7) |
| Ciprofloxacin + clindamycin + erythromycin + tetracyclin + rifampicin + gentamicin | 3 (2.1) |
*Combinations that occurred less than three times are not specified. Bold numbers indicate the total number of each subgroup.
Multiresistance to one, two, three, four, five, and six antibiotics in addition to oxacillin was observed for 29%, 15%, 20%, 8%, 1%, and 3%, of the isolates, respectively (Table 1). The most prominent combinations of resistances are shown in Table 1. Together, ~30% of the MRSA strains isolated were resistant to at least four antimicrobial agents (including oxacillin), emphasizing that antibiotic resistance remains a problem. These findings underline the importance of infection control measures.
PFGE analyses
Of the 146 isolates 136 isolates were grouped into 22 PFGE patterns. The four most frequent PFGE patterns were PFGE-21 (n = 27, 19%), PFGE-20 (n = 20, 14%), PFGE-08 (n = 16, 11%) and PFGE-41 (n = 15, 10%), and together made up more than 50% of all isolates (Figure 1a). Nine isolates had a pattern that had never occurred in our laboratory and were therefore not assigned a number. One isolate (MRSA3687) could not be typed by PFGE analysis (Additional file 1: Table S1).
Figure 1.
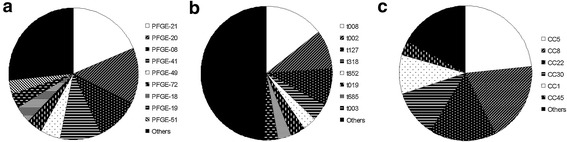
Genetic background of MRSA. Frequencies of PFGE types (a), spa-types (b), and predicted clonal complexes (c). CC, clonal complex.
Spa-types
The 146 isolates had 50 distinct spa-types. The three most common spa-types were t008 (n = 21, 14%), t002 (n = 14, 10%), and t127 (n = 13, 9%) and accounted for 33% of all isolates (Figure 1b). All other spa-types occurred less than ten times and 47% of all isolates had a spa-type that occurred less than three times (Figure 1b). One of the 146 isolates (MRSA3984) was not spa-typable (no PCR product for spa). Three isolates had spa-types not described so far: MRSA3903 (T1-J1-J1-*-J1-N1-F2-M1-N1-F2-M1-O1-O1-K1-R1) had a repeat that differed in one nucleotide from repeat r13 (E1). MRSA 3771 had a new combination of spa-repeats (T1-N1-F2-M1-O1-M1-O1-O1-R1) as well as MRSA3670 (U1-J2-G1-M1-K1-M1-K1-K1-P1-N1-S1-G1, Additional file 1: Table S1).
The two most common spa-types found in this study (t008 and t002) were among the most frequent spa-types in MRSA blood stream isolates collected all over Europe from 2006 to 2007 [15]. In contrast to this previous study, our MRSA strains were derived from various infections including three blood stream isolates (one of which was of t008) as well as from colonized patients. We thus provide information on both invasive and colonizing MRSA strains. In contrast, we have no information whether the European blood stream isolates represent the MRSA circulating in the population. Spa-type t127, our third most common spa-type, was only sporadically identified in the European blood stream isolates [15]. However, this spa-type has been identified as a common MRSA lineage in pigs in Europe, mainly in Italy [16]. Several common spa-types found in European blood stream isolates e.g. t032 (14.5%) or t041 (7.4%) only occurred sporadically in our hospital (t032 (5%)) or were absent (t041). In contrast, t041 was the local epidemic spa-type of MRSA isolates (including invasive and colonizing isolates) at the University Hospital Basel, Switzerland between 2000 and 2005 [17], another low-prevalence hospital less than 100 kilometers away from our hospital.
Correlation between PFGE analyses and spa-typing
Results of PFGE analyses and spa-typing are shown in Figure 2. All isolates were typable by at least one of the two methods. Two isolates could not be typed by either PFGE analysis or spa-typing. Both phenomena have been previously described and were reported to be due to methylation of the SmaI restriction site as well as due to mutations of the protein A gene spa, respectively [18,19].
Figure 2.
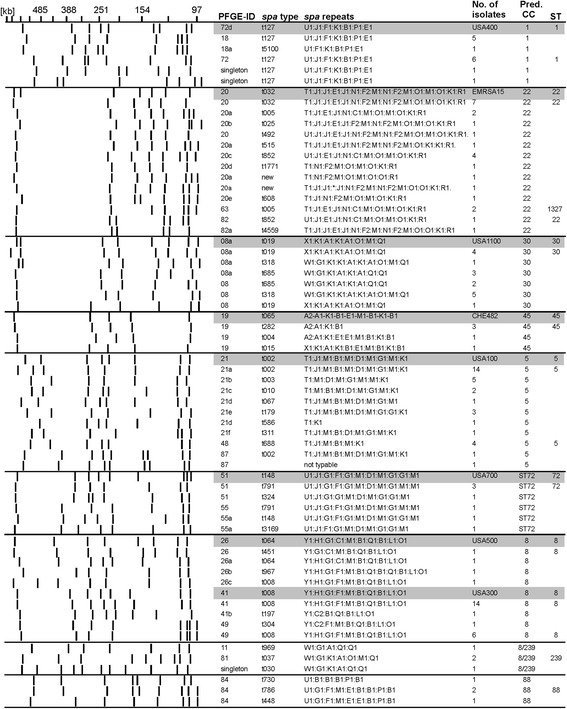
Correlation between PFGE analyses and spa -typing. Genetic relationships between the 146 MRSA isolates, correlations between PFGE typing and spa-typing (spa). Strains presented were sorted according to PFGE patterns and similarities of spa-types. Shown are strains of clonal complexes (CC) that occurred at least four times. Reference strains are marked with grey boxes. ST, Sequence type.
In most of the cases, isolates with identical PFGE patterns had identical or related spa-types. Only two isolates (MRSA3548 and MRSA3619) which were both of spa-type t127 were both singletons with low similarity (~50%) to the other t127 isolates (Figure 2).
Different PFGE patterns shared common spa-types (e.g. PFGE-41 and 49 both included spa-type t008). Vice versa, several strains with different spa-types shared the same PFGE patterns (e.g. PFGE-21 included seven different spa-types, Figure 2). These findings emphasize that the combination of both methods is more discriminatory as compared to using one method only as has been described previously [20].
Clonal complex 5
Thirty-four isolates were of spa-types t002, t003, t010, t067, t179 or t688 that corresponded to CC5 [7,8,21,22]. These 34 isolates were grouped into three related PFGE-patterns (PFGE-21, 48, and 87 (Figure 2)), one of which (PFGE-21) was identical to USA100 (ST5, CC5). MLST of representative PFGE-21/t002 and PFGE-48/t688 isolates confirmed that both were of ST5, CC5. ST5 was one of the predominant MRSA clones among blood culture isolates in Europe [15].
Clonal complex 8 and related lineages
Twenty-eight isolates were grouped into CC8 based on their spa-types, which were t008, t064, t304, t451, t967 [22,23]. These isolates displayed the PFGE patterns 41, 49, and 26 (Figure 2). PFGE-41 had previously been identified as USA300 (ST8, CC8) [24]. This lineage is the most prevalent MRSA in the United States of America (USA) and a global epidemic threat (Reviewed by Nimmo 2012 [25]). Special care must be taken to avoid the spread of this particular clone. The PFGE-49 pattern has been previously shown to be ST8 [24]. PFGE-26 was identical to USA500 (ST8, CC8), and we confirmed ST8 by MLST.
Isolates that were related to CC8, but were grouped separately as previously suggested [26], included four isolates that were assigned to ST239 (CC8/239) based on their spa-types t030, t037 and t969 [26]. MLST of one of these isolates confirmed that they were of ST239. Seven isolates were of spa-types t148, t324 and t791, t3169, which are predictive of ST72 and also related to CC8 [26]. These isolates belonged to PFGE-51 (n = 4) or PFGE-55 (n = 3). PFGE-51 exhibited >70% identity with USA700 (ST72) and MLST confirmed that PFGE-51 was ST72.
Clonal complex 22
Twenty-four isolates were of spa-types t005, t032, t492, t515, t852, t1771, t4325, t4559 (Table 1), which was predictive of CC22 [8,27]. Twenty of these isolates (PFGE-20) were identical to EMRSA-15 (ST-22, CC22) in terms of PFGE-restriction patterns. MLST of a representative isolate confirmed that these isolates were of ST22. EMRSA-15 (ST22) was also widely distributed among MRSA blood stream isolates in Europe [15]. The other four isolates were of PFGE-82, which was also of ST22 and PFGE-63, which was of ST1327, a sequence type that differs in one allele (pta) from ST22 (Figure 2).
Clonal complex 30
Sixteen isolates had spa-types t019, t318 and t685, which are indicative of CC30 [22]. All of these isolates had the PFGE-08 pattern, which exhibited at least 80% identity to USA1100 (ST30, CC30). MLST of a representative PFGE-08 isolate confirmed that it was of ST30.
Clonal complex 1
Fourteen isolates were of spa-type t127 and the related spa-type t5100 which was assigned to CC1 [22]. These isolates had PFGE-18 (n = 6) and PFGE-72 (n = 6) patterns and two isolates were singletons. PFGE-72 exhibited >80% similarity to USA400 (ST1, CC1). MLST of a PFGE-72/t127 isolate confirmed that it was of ST1, confirming its grouping into CC1 (Figure 2).
Clonal complex 45
Five isolates were of spa-types t004, t015 or t282, which are predictive of CC45 [8], a lineage that is common in Europe [2]. These isolates were of PFGE-19, which is identical to the Swiss drug clone CHE482-ST45 [12]. MLST of a representative PFGE-19 isolate confirmed that it was ST45. The Swiss drug clone is a community acquired MRSA (CA-MRSA) clone, which was spread among injection drug users, was predominant at our hospital between 1998 and 2004 [11], and still occurs sporadically.
Summary of clonal complexes
Four isolates were of spa-types t448, t730 and t784, being indicative of CC88 [28]. MLST of one of these isolates confirmed that it was ST88. The remaining thirteen isolates were of diverse genetic backgrounds (Additional file 1: Table S1). Together, 82% of the MRSA isolates could be grouped into the six clonal complexes (CCs): CC1 (10%), CC5 (23%), CC8 (18%), CC22 (17%), CC30 (11%), and CC45 (3%) based on their PFGE pattern, spa types, MLST and previously published literature (Figure 1c).
Correlation between genetic background and antibiotic sucseptibility
Additional file 1: Table S1 shows the antibiograms of the individual MRSA strains. While some clones, such as PFGE-21a/t002 or PFGE-41/t008 showed high heterogeneity in their antibiograms, other clones had homogenous resistance patterns. Spa/PFGE-types that shared identical antibiograms included t003 isolates (n = 5, PFGE-21b, CC5), t688 isolates (n = 4, PFGE-48, CC5), t685 isolates (n = 5, PFGE-08 and PFGE-08a, CC30), t005 (n = 4, PFGE-20 and PFGE-63, CC22), t282 isolates (n = 3, PFGE-19, CC45), and PFGE-51 isolates (n = 4, t791 or t324, ST72) (Additional file 1: Table S1). Interestingly, all four ST239 isolates were resistant to six antibiotics in addition to oxacillin, which is in accordance with previous findings, in which ST239 has been described as multi resistant [5].
Conclusions
In summary, we have demonstrated that i) several of the lineages that are predominant in Europe are present in our hospital, ii) the combination of PFGE and spa-typing is more discriminatory as compared to using a single method only and will allow to better monitor and recognize changes in MRSA epidemiology over time, iii) clonal complex predictions based on PFGE and spa-typing could be confirmed by MLST, and iv) there was a decrease in resistances to ciprofloxacin, erythromycin, gentamicin, rifampicin, and tetracycline as compared to MRSA strains collected in an earlier period.
Acknowledgements
This work was supported by Grants of the Foundation for Research at the Medical Faculty, University of Zurich, Switzerland (to ASZ and KS), and a research Grant 2013 by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) (to KS).
We thank Dr. Vera Bruderer-Rüegger (Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland) for searching zone diameters of all isolates for the reinterpretation of breakpoints according to CLSI guidelines.
Additional file
Genetic background and antibiogramms of all 146 MRSA isolates.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KS conceived the study, carried out spa-typing, PFGE analyses, and MLST, analyzed the data and wrote the manuscript. NL carried out spa-typing, MLST, and wrote the manuscript. MPM and AF carried out spa-typing. AHB and GS carried out PFGE analyses. RZ was involved in the identification of the strains and susceptibility testing. AZ conceived the study, participated in its design and coordination and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Kati Seidl, Email: kati.seidlspirig@usz.ch.
Nadja Leimer, Email: nadja.leimer@usz.ch.
Miguel Palheiros Marques, Email: miguel.palheirosmarques@usz.ch.
Alexandra Furrer, Email: alexandra.furrer@access.uzh.ch.
Anne Holzmann-Bürgel, Email: anne.holzmannbuergel@usz.ch.
Gabriela Senn, Email: gabrielasennhunn@gmail.com.
Reinhard Zbinden, Email: rzbinden@imm.uzh.ch.
Annelies S Zinkernagel, Email: annelies.zinkernagel@usz.ch.
References
- 1.Köck R, Becker K, Cookson B, van Gemert-Pijne NJ, Harbarth S, Kluytmans J, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010;15(41):19688. doi: 10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- 2.Stefani S, Chung D, Lindsay J, Friedrich A, Kearns A, Westh H, et al. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents. 2012;39(4):273–82. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99(11):7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37(11):3556–63. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCalla C, Smyth DS, Robinson DA, Steenbergen J, Luperchio SA, Moise PA, et al. Microbiological and genotypic analysis of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother. 2008;52(9):3441–3. doi: 10.1128/AAC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–8. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, et al. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus. Comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One. 2009;4(4):e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41(11):5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ender M, Burger S, Sokoli A, Zbinden R, Berger-Bächi B, Heusser R, et al. Variability of SCCmec in the Zurich area. Eur J Clin Microbiol Infect Dis. 2009;28(6):647–53. doi: 10.1007/s10096-008-0663-7. [DOI] [PubMed] [Google Scholar]
- 12.Ender M, Berger-Bächi B, McCallum N. Variability in SCCmecN1 spreading among injection drug users in Zurich. Switzerland BMC Microbiol. 2007;7(1):62. doi: 10.1186/1471-2180-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.EUCAST. Staphylococcus aureus - Calibration of zone diameter breakpoints to MIC values and/or resistance mechanisms. Breakpoint tables for interpretation of MICs and zone diameters. The European Committee on Antimicrobial Susceptibility Testing; 2012 Version 2.1. http://www.eucast.org/.
- 14.CLSI . Performance standards for antimicrobial susceptibility testing. 17th informational supplement. Wayne, Pennsylvania, USA: Clinical Laboratory Standards Institute; 2007. [Google Scholar]
- 15.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, the European Staphylococcal Reference Laboratory Working G et al. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7(1):e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco A, Hasman H, Iurescia M, Lorenzetti R, Stegger M, Pantosti A, et al. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J Antimicrob Chemother. 2011;66(6):1231–5. doi: 10.1093/jac/dkr115. [DOI] [PubMed] [Google Scholar]
- 17.Fenner L, Widmer AF, Dangel M, Frei R. Distribution of spa types among meticillin-resistant Staphylococcus aureus isolates during a 6 year period at a low-prevalence university hospital. J Med Microbiol. 2008;57(5):612–6. doi: 10.1099/jmm.0.47757-0. [DOI] [PubMed] [Google Scholar]
- 18.Bens CCPM, Voss A, Klaassen CHW. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J Clin Microbiol. 2006;44(5):1875–6. doi: 10.1128/JCM.44.5.1875-1876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baum C, Haslinger-Loffler B, Westh H, Boye K, Peters G, Neumann C, et al. Non-spa-typeable clinical Staphylococcus aureus strains are naturally occurring protein A mutants. J Clin Microbiol. 2009;47(11):3624–9. doi: 10.1128/JCM.00941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fossum Moen AE, Holberg-Petersen M, Andresen LL, Blomfeldt A. Spa typing alone is not sufficient to demonstrate endemic establishment of meticillin-resistant Staphylococcus aureus in a low-prevalence country. J Hosp Infect. 2014, in press (0). [DOI] [PubMed]
- 21.Cuny C, Friedrich AW, Witte W. Absence of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex CC398 as a nasal colonizer of pigs raised in an alternative system. Appl Environ Microbiol. 2012;78(4):1296–7. doi: 10.1128/AEM.07260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strommenger B, Braulke C, Pasemann B, Schmidt C, Witte W. Multiplex PCR for rapid detection of Staphylococcus aureus isolates suspected to represent community-acquired strains. J Clin Microbiol. 2008;46(2):582–7. doi: 10.1128/JCM.01600-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlemann A-C, Dumortier C, Hafer C, Taylor BS, Sánchez EJ, Rodriguez-Taveras C, et al. Molecular characterization of Staphylococcus aureus from outpatients in the Caribbean reveals the presence of pandemic clones. Eur J Clin Microbiol Infect Dis. 2012;31(4):505–11. doi: 10.1007/s10096-011-1339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seidl K, Leimer N, Furrer A, Senn G, Holzmann-Bürgel A, Palheiros Marques M, et al. USA300 methicillin-resistant Staphylococcus aureus in Zurich, Switzerland between 2001 and 2013. Intern J Med Microbiol. 2014;304(8):1118–22. doi: 10.1016/j.ijmm.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Nimmo GR. USA300 abroad: global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2012;18(8):725–34. doi: 10.1111/j.1469-0691.2012.03822.x. [DOI] [PubMed] [Google Scholar]
- 26.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(4):e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen AR, Stegger M, Böcher S, Sørum M, Monnet DL, Skov RL. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999 to 2006. J Clin Microbiol. 2009;47(1):73–8. doi: 10.1128/JCM.01557-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breurec S, Fall C, Pouillot R, Boisier P, Brisse S, Diene-Sarr F, et al. Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton–Valentine leukocidin genes. Clin Microbiol Infect. 2011;17(4):633–9. doi: 10.1111/j.1469-0691.2010.03320.x. [DOI] [PubMed] [Google Scholar]


