Abstract
Background
Cardiovascular disease (CVD) reflects a highly coordinated complex of traits. Although genome-wide association studies have reported numerous single nucleotide polymorphisms (SNPs) to be associated with CVD, the role of most of these variants in disease processes remains unknown.
Methods and Results
We built a CVD network using 1512 SNPs associated with 21 CVD traits in genome-wide association studies (at P≤5×10−8) and cross-linked different traits by virtue of their shared SNP associations. We then explored whole blood gene expression in relation to these SNPs in 5257 participants in the Framingham Heart Study. At a false discovery rate <0.05, we identified 370 cis-expression quantitative trait loci (eQTLs; SNPs associated with altered expression of nearby genes) and 44 trans-eQTLs (SNPs associated with altered expression of remote genes). The eQTL network revealed 13 CVD-related modules. Searching for association of eQTL genes with CVD risk factors (lipids, blood pressure, fasting blood glucose, and body mass index) in the same individuals, we found examples in which the expression of eQTL genes was significantly associated with these CVD phenotypes. In addition, mediation tests suggested that a subset of SNPs previously associated with CVD phenotypes in genome-wide association studies may exert their function by altering expression of eQTL genes (eg, LDLR and PCSK7), which in turn may promote interindividual variation in phenotypes.
Conclusions
Using a network approach to analyze CVD traits, we identified complex networks of SNP-phenotype and SNP-transcript connections. Integrating the CVD network with phenotypic data, we identified biological pathways that may provide insights into potential drug targets for treatment or prevention of CVD.
Keywords: cardiovascular disease, gene expression/regulation network, genetic variation
Cardiovascular disease (CVD) is a group of disorders affecting the heart and blood vessels including coronary heart disease, stroke, hypertension, and peripheral arterial disease. CVD is caused by interactions of genetic, environmental, and lifestyle factors.1 During the past half century, prevention and treatment efforts have focused on modifiable CVD risk factors such as elevated blood cholesterol level, hypertension, type 2 diabetes mellitus, and tobacco use. Although these targeted efforts have contributed to steady declines in CVD mortality over this time period, CVD remains the leading cause of death across the globe.2
Genome-wide association studies (GWAS) have successfully identified thousands of single nucleotide polymorphisms (SNPs) that underlie CVD and its major risk factors.3 Many genetic loci appear to affect multiple phenotypes. One example is the SH2B3 gene region on chromosome 12, which harbors variants that are associated with myocardial infarction4 and blood pressure5 and also with rheumatoid arthritis6 and type 1 diabetes mellitus.7 Several common genetic variants associated with coronary artery disease (CAD) or myocardial infarction in GWAS also reveal associations with CVD risk factors and other complex traits,8 suggesting that these common genetic variants have multiple molecular functions or that they have a single molecular function with multiple downstream consequences. Although pleiotropic effects have been widely observed, their presence in relation to CVD and their downstream effects have not been evaluated systematically.
Despite the identification of thousands of common SNPs that are associated with an increased propensity toward CVD, the variants identified thus far explain only a small fraction of the overall genetic contribution to disease risk.9 It is likely that disease-promoting SNPs act by affecting the amino acid sequences of the corresponding coded proteins (ie, nonsynonymous SNPs) or by altering mRNA expression levels (ie, expression quantitative trait loci [eQTLs]).10 A growing number of eQTLs have been found to be associated with human diseases.11 For example, multiple SNPs that were associated with blood lipid levels in GWAS were also found to be eQTLs for nearby genes (eg, in SORT1, PPP1R3B, and TTC39B),12 suggesting that eQTLs play an important functional role.
We hypothesized that genetic variants influence CVD phenotypes by altering expression of genes and that systematic analysis of multiple traits might reveal high-order interactions of CVD and its risk factors.13,14 To that end, we built a CVD network using SNP-CVD phenotype associations and dissected the relationships between genetic variants, gene expression, and CVD phenotypes. By integrating these 3 layers of information from >5000 Framingham Heart Study (FHS) participants with deep phenotyping for CVD and extensive genotyping and gene expression profiling, we were able to study the role of genetic variation in relation to gene expression and to integrate this information across multiple complex CVD phenotypes. Our results revealed a dense network in which genetic variation was linked to gene expression and CVD phenotypes. We identified several modules that support the existence of pathways affected by genetic variants. We highlighted examples in which genetic variants may play a causal role in CVD and hypothesized that they affect CVD phenotypes by regulating (cis and trans) gene expression. Identifying these genetic variants that mediate gene expression may aid in understanding biological mechanisms underlying CVD and in targeting therapies for its treatment and prevention.
Methods
Study Sample
Beginning in 1948, the FHS recruited participants from Framingham, MA, to undergo biennial examinations to investigate CVD and its risk factors.15 In 1971 and 2002, offspring (and their spouses) and adult grandchildren of the original cohort participants were recruited into the second- and third-generation cohorts, respectively. Collection of blood samples and RNA preparation were described previously.16 A total of 5257 participants from the offspring cohort (at examination 8) and third-generation cohort (at examination 2) who had both genome-wide genotyping (institutional review board approval No. H-26671) and gene expression profiling (institutional review board approval No. H-27984) were included in this study (Figure 1).
Figure 1.
Flowchart of integromic analysis. A total of 1512 single nucleotide polymorphisms (SNPs) associated with 21 cardiovascular disease (CVD) traits (at P ≤ 5×10−8) were derived from database of Genotypes and Phenotypes and the National Human Genome Research Institute genome-wide association studies (GWAS) catalog. We built a CVD phenotype network by connecting 2 traits if they share the same GWAS SNP. Whole blood samples were collected from 5257 FHS participants. Genome-wide genotyping and mRNA expression levels were assayed. We correlated 1077 SNPs (after genotyping quality control of 1512 SNPs) with 17873 gene expression values to assess expression quantitative trait loci (eQTLs). We replicated these eQTLs in 2 large databases. We then built an eQTL network by connecting eQTLs to their associated genes and traits. We identified modules associated with different CVD traits within the network. Finally, we conducted mediation analyses to test whether the genetic effect appears to influence the CVD phenotype through effects of the eQTL (ie, GWAS SNP) on gene expression. BMI indicates body mass index; FHS, Framingham Heart Study; HDL-C, high-density lipoprotein cholesterol; and LDL-C, low-density lipoprotein cholesterol.
Trait-Associated SNP
A total of 1512 SNPs associated in GWAS with 21 cardiovascular traits (Table 1) with the use of data from the database of Genotypes and Phenotypes (dbGaP)17 and the National Human Genome Research Institute GWAS catalog3 (at P≤5×10−8, downloaded in January 2014) were curated and matched with Framingham Affymetrix 550K array genotype data.18 The dbGaP resource lists results of GWAS whether published or not. The National Human Genome Research Institute GWAS catalog lists only published GWAS studies. Genotyping and quality control methods in the FHS have been described previously.18 Briefly, SNPs were inputted to Minimac,19 an implementation of genotype imputation software. SNP imputation combined genotype data with the HapMap CEU samples and then inferred genotypes probabilistically on the basis of shared haplotype stretches between study samples and HapMap release 22 build 36. Imputation results were summarized as an “allele dosage,” defined as the expected number of copies of the minor allele at that SNP (a fractional value between 0 and 2) for each genotype. SNPs with imputed quality score (r2) <0.3 or minor allele frequency <0.01 were filtered out, resulting in 1077 SNPs for eQTL analysis.
Table 1.
Cardiovascular Disease Phenotypes Included in Analyses
| Cardiovascular Disease Phenotypes (Named by MeSH Terms) |
Cardiovascular Disease Risk Factors (Named by MeSH Terms) |
|---|---|
|
|
HDL indicates high-density lipoprotein; LDL, low-density lipoprotein; MeSH, Medical Subject Headings; VLDL-C, very low-density lipoprotein; and /, similar traits that were merged.
Gene Expression
Whole blood was collected in PAXgene tubes (PreAnalytiX, Hombrechtikon, Switzerland) and frozen at −80°C. RNA was extracted from whole blood with the use of the RNA System Kit (Qiagen, Venlo, Netherlands), and mRNA expression profiling was assessed with the use of the Affymetrix Human Exon 1.0 ST GeneChip platform (Affymetrix Inc, Santa Clara, CA), which contains >5.5 million probes targeting the expression of 17873 genes. The Robust Multi-Array Average package20 was used to normalize the gene expression values and remove any technical or spurious background variation. Linear regression models were used to adjust for technical covariates (batch, first principal component, and residual of all probeset mean values).
Statistical Analysis
eQTL analysis was conducted with the use of the pedigreemm21 package in R with gene expression as the dependent variable and genotype, sex, and age as independent variables. Technical covariates and imputed whole blood cell counts (or proportions) were adjusted for with the use of a linear mixed effects model. Familial relatedness was modeled as a random effect. The cis effect for a given expression trait was defined by testing all SNPs located within 1 Mb upstream or downstream of the transcription start site of a gene (cis-eQTL). SNPs that were mapped to different chromosomes from their associated gene transcripts were defined as trans-eQTLs. The false discovery rate22 was applied to account for multiple testing. SNPs at false discovery rate <0.05 were selected as significant eQTLs. For trans-eQTLs that were also cis-eQTLs, we examined whether the genes regulated in cis play a role in the regulation of the trans genes by conditioning on expression of the cis gene in the linear regression model. Mediation analysis was conducted with the use of the mediation package23 in R with SNP as the “exposure,” gene expression as the “mediator,” and phenotype as the “outcome.” A 100% proportion of mediation effect indicates that the entire association between a SNP and a phenotype (direct effect) is explained by changes in gene expression. The significant mediation effects were selected at a permutation P<0.0005 (based on 10000 permutations).
Annotation and Enrichment Analysis of eQTLs With Encyclopedia of DNA Elements Data
The Encyclopedia of DNA Elements (ENCODE)24 cataloged many regulatory elements including DNase I hypersensitive regions profiled in 82 cell lines, 149 transcription factor (TF) binding regions profiled in different cell lines resulting in a total of 406 different cell line–TF pairs, and 162 histone modification–cell line pairs (ENCODE January 21,2011 freeze). We used GLANET (publication in preparation, software available at https://github.com/burcakotlu/GLANET and documentation at https://glanet.readthedocs.org/en/latest/) to annotate our list of eQTLs by overlapping them with the ENCODE peak lists. We then evaluated the significance of the overlap using GLANET’s resampling-based enrichment analysis. Specifically, we sampled multiple (n=100000) random SNP sets matching in size and numbers per chromosome to the original eQTL SNP set and computed the size of the overlap for each random set. To account for systematic biases, our random sampling scheme took into account the “mappability” and guanine-cytosine content of the SNPs and matched the random SNP sets to the actual SNP set in terms of mappability and guanine-cytosine content. The collection of overlap statistics across multiple random samplings was then used to estimate an empirical null distribution for the overlap statistic. The resulting P values were adjusted for multiple testing using both the Benjamini Hochberg22 and Bonferroni correction methods. We used the FIMO tool from the MEME suite25 to assess whether the eQTLs disrupted the binding sites of the TFs that they were bound by in the ENCODE data.
In Silico Validation of eQTLs
Whole blood eQTLs were downloaded from the Blood eQTL Browser.11 This resource contains the results of an eQTL meta-analysis from 5311 peripheral blood samples from 7 studies. To explore tissue-specific effects, we also collected and analyzed results from 53 eQTL population data sets (Table I in the online-only Data Supplement). These 53 data sets represent analyses from 24 published manuscripts and 13 unpublished data sets reflecting >27 cell and tissue types.26 Cis- and trans-eQTLs from the present study were cross-referenced with significant eQTLs reported in the aforementioned data sets directly by matching SNP identifiers.
Network Construction and Modules Identification
On the basis of the SNP–trait relationships, we constructed a CVD network. In the network, each node corresponds to a CVD trait, and 2 traits were connected to each other if they shared at least 1 SNP in GWAS. The width of each edge was weighted by the proportion of shared SNPs between traits. To explore relations between CVD traits and other complex traits (GWAS SNP P<5×10−8), we expanded the connections if SNPs associated with CVD traits were also found to be associated with other diseases in GWAS. Networks were visualized with the use of Cytoscape software.27
On the basis of SNP–gene expression associations, we constructed an eQTL network. The TFit (iterated Transfer-Fusion) algorithm with default parameters in the Clust&see28 plugin of Cytoscape was used to search for modules within this network. The TFit algorithm29 is based on modularity optimization, which uses a vertex transfer procedure at every level. Level 1 is the entire network; each node is assigned to its best adjacent cluster, as long as modularity increases. Then classic transfers were performed, and vertices belonging to the same cluster were merged.
Results
Genetic Variation Network for Complex CVD Traits
We restricted our analysis to 1512 significant GWAS SNPs associated (at P≤5×10−8) with 21 CVD traits listed in Table 1. Fifteen of the 21 CVD traits shared at least 1 SNP with another trait (Figure 2 and Table II in the online-only Data Supplement). Among the CVD traits, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides, body mass index, type 2 diabetes mellitus, and blood pressure served as “hub” phenotypes that connected multiple CVD traits, mirroring epidemiological observations about the clustering of metabolic risk factors.30 We found that C-reactive protein and LDL-C had a strong genetic connection via 6 shared SNPs (rs1260326 of GCKR, rs1800961 of HNF4A, rs2075650 of TOMM40, rs2650000 of RPL12P33-NCRNA00262, rs4420638 of APOC1, and rs9987289 of PPP1R3B–TNKS). CAD and LDL-C had a strong genetic connection through 5 shared SNPs (rs11206510 of BSND-PCSK9, rs599839 of PSRC1, rs12740374 and rs646776 of CELSR2, and rs964184 of ZNF259). We also identified some hub SNPs; for example, rs964184, an intronic variant in ZNF259, was associated in GWAS with HDL-C,31 LDL-C,12 triglycerides,12 and CAD.4 rs1260326 (GCKR) was associated in GWAS with triglycerides,12 total cholesterol,12 and C-reactive protein32; rs13107325 (SLC39A8) was associated in GWAS with blood pressure,5 body mass index,33 and HDL-C.12 We further considered SNPs in linkage disequilibrium with an index SNP. Two traits were connected if they shared the same GWAS SNP or proxy SNPs that are in high linkage disequilibrium (r2>0.8) with the index SNP. When modified through the inclusion of proxy SNPs, the CVD phenotype network encompassed 19 of the 21 CVD traits (Figure I in the online-only Data Supplement). Four traits (coagulation, venous thrombosis, sudden cardiac death, and abdominal aortic aneurysm) with no connections by virtue of directly shared SNPs all had proxy SNPs in perfect linkage disequilibrium (r2=1) with the index SNPs, and the combination of index and proxy SNPs identified new trait connections: coagulation and venous thrombosis; sudden cardiac death and HDL cholesterol; and abdominal aortic aneurysm and CAD.
Figure 2.
Cardiovascular disease phenotype network by virtue of shared genome-wide association study single nucleotide polymorphisms. Each node represents a cardiovascular disease trait, and 2 traits are connected if they share at least 1 single nucleotide polymorphism in genome-wide association studies. The width of each line is weighted by the proportion of shared single nucleotide polymorphisms between 2 connected traits. HDL indicates high-density lipoprotein; and LDL, low-density lipoprotein.
Expanding the connections across all 409 complex traits containing genome-wide significant SNPs within dbGaP and the National Human Genome Research Institute GWAS catalog, we found that CVD-associated SNPs from GWAS were strongly linked with many other complex traits (Figure II in the online-only Data Supplement). These associations include HDL-C and LDL-C with alcohol consumption, Alzheimer disease (Figure III in the online-only Data Supplement) and blood pressure with CD40 ligand, and resistin with vitamin K levels (Table III in the online-only Data Supplement). Using this approach, we found that the phenotype network linked by common SNPs may reveal unexpected genetic connections with numerous non-CVD traits.
Regulation of the Genetic Variation Network
At a minor allele frequency >0.01 and imputation r2>0.3, 1077 genome-wide significant (P<5×10−8) SNPs from GWAS were available for analysis. At false discovery rate <0.05, we identified 370 cis-eQTLs (associated with expression of 400 genes at P<10−4) and 44 trans-eQTLs (associated with expression of 76 genes at P<10−6; Table IV in the online-only Data Supplement). For 696 SNPs (65%) not associated with expression traits, we further tested the association between their perfect proxy SNPs (linkage disequilibrium r2=1 in SNAP)34 and gene expression levels. Using proxy SNPs, we identified an additional 54 cis-eQTLs for 6 CVD trait–associated SNPs (Table V in the online-only Data Supplement).
To assess whether the eQTLs significantly overlap with regulatory regions, we performed annotation and enrichment analysis with the DNase, histone modification, and TF peaks from the ENCODE project (see Methods for details). We first annotated each eQTL by intersecting the SNP locus with ENCODE peaks and then evaluated the significance of overlap with functional elements using GLANET. This analysis revealed that the eQTLs are significantly enriched for DNase I hypersensitive regions in 16 cell lines and 133 histone modification–cell line pairs (Table VI in the online-only Data Supplement). Thirty of our eQTLs are located within 10 kb upstream of the transcription start site of the expressed gene associated with the corresponding SNP (Table VI in the online-only Data Supplement). Our annotation analysis indicated that all of these promoter eQTLs are within 1 or more histone modification region, and 10 of them overlap with a TF peak. Notably, rs7528684, which is a cis-eQTL associated with expression of FCRL3, resides 2 kb upstream of the transcription start site of FCRL3 and is bound by Nfkb in the Gm12891 cell line. Our sequence analysis revealed that this SNP is an eQTL that regulates expression of FCRL3 by disrupting the Nfkb binding site (Figure 3).
Figure 3.
Reference and single nucleotide polymorphism (rs7528684) allele matches to the Nfkb sequence logo (Encyclopedia of DNA Elements [ENCODE] motif logo NFKB_disci from http://compbio.mit.edu/encode-motifs/).
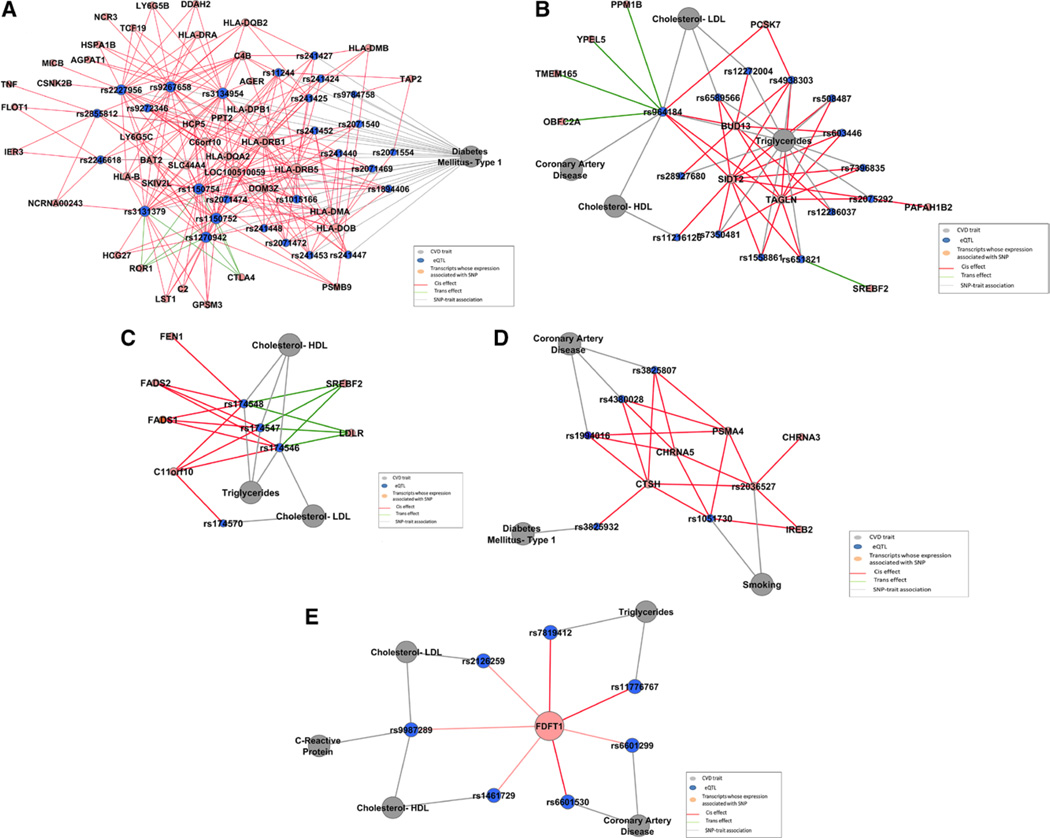
By connecting eQTLs and their associated genes, we built a SNP-gene association network (Figure IV in the online-only Data Supplement). Using the TFit algorithm,28 we identified 13 modules containing >10 nodes (Table VII in the online-only Data Supplement). These modules may reveal genetic pathways affecting CVD phenotypes. For example, SNPs associated with type 1 diabetes mellitus displayed cis associations with genes in 6p21 and trans associations with ROR1 and CTLA4 (Figure 4A). Using gene set enrichment analysis, we found that these genes were significantly enriched for the KEGG type 1 diabetes mellitus pathway (P<10−6). Of note, GWAS and gene expression studies have identified association between CTLA4 (DNA and mRNA level) and type 1 diabetes mellitus.35 In another module, rs964184 in ZNF259, which was associated in GWAS with HDL-C, LDL-C, triglycerides,12,31 and CAD,4 was found to have cis associations with PCSK7, SIDT2, TAGLN, and BUD13 and trans associations with TMEM165, YPEL5, PPM1B, and OBFC2A (Figure 4B). Three linked SNPs in FADS1 (rs174546, rs174547, and rs174548; pairwise R2=0.80–0.97) were associated in GWAS with multiple lipids traits36; we found that these SNPs have cis associations with C11orf10, FADS1, FADS2, and FEN1 and trans associations with LDLR and SREBF2 (Figure 4C). Using gene set enrichment analysis, we found that genes in these 2 modules are significantly enriched for lipid metabolic processes (P<10−6). rs1994016, rs3825807, and rs4380028 (pairwise r2=0.52–0.87) in ADAMTS7 were associated in GWAS with CAD,4 whereas rs1051730 and rs2036527 (pair-wise r2=0.90) in CHRNA3 were associated in GWAS with smoking.37,38 We discovered that these SNPs all displayed cis association with 3 genes (PSMA4, CHRNA5, CTSH). Variants in PSMA4 and CHRNA5 were found to be associated with chronic obstructive pulmonary disease and lung function.39 The CHRNA5 variants were also found to be associated with nicotine and alcohol dependence.40 Expression levels of PSMA4 and CTSH were found to regulate immune function in type 1 diabetes mellitus.41 Therefore, the clustering of these 3 genes with multiple disease-associated SNPs may explain in part the concurrence of CAD and chronic obstructive pulmonary disease and the strong association between smoking, CAD, and diabetes mellitus (Figure 4D).
Figure 4.
Modules in the cardiovascular disease (CVD) expression quantitative trait loci (eQTL) network. Gray nodes represent CVD traits. Blue nodes represent single nucleotide polymorphisms (SNPs) associated with CVD traits in genome-wide association studies. Orange nodes represent genes whose expression is associated with SNPs in Framingham Heart Study participants. Gray edges represent SNP-trait associations. Red edges represent cis associations between SNPs and gene expression. Green edges represent trans associations between SNPs and gene expressions. A, Type 1 diabetes mellitus eQTL module. B, rs964184 pleiotropic eQTL module. C, Lipids eQTL module. D, Coronary artery disease and smoking eQTL module. E, eQTLs associated with FDFT1. HDL indicates high-density lipoprotein; LDL, low-density lipoprotein; and LDLR, low-density lipoprotein receptor.
Reproducibility of eQTLs
To validate the eQTLs detected above, we first queried the Blood eQTL Browser11 meta-analysis of eQTL associations in nontransformed peripheral blood samples from 5311 individuals. A total of 240 cis-eQTLs and 25 trans-eQTLs from our data set were also detected as eQTLs in the Blood eQTL Browser database. Among them, 165 cis-eQTLs (69%) and 25 trans-eQTLs (100%) were associated with expression of the same genes and showed the same directions of association as our eQTL findings. In addition, we found 7 cis-eQTLs from our results that were perfect proxies (r2=1) of eQTLs in the Blood eQTL Browser (Table VIII in the online-only Data Supplement). Because eQTLs are highly tissue specific,42 we further queried our multitissue eQTL databases, which integrated 53 data sets from multiple tissues (see Methods for details). One hundred sixty-one cis-eQTLs from our data also were detected as eQTLs in this database (no trans-eQTLs were found). Among them, 116 cis-eQTLs (72%) were associated with the same genes across eQTL databases (Table IX in the online-only Data Supplement). rs17030613 in CAPZA1, associated with blood pressure in GWAS,43 was associated with the expression of ST7L in our data and in 2 other tissues (brain and CD4+ lymph). Lower ST7L transcript levels were found to be associated with lower blood pressure in East Asian populations.43 In the FHS samples, we found that ST7L transcript levels were associated with diastolic blood pressure (P=0.023). rs1412444 in LIPA, associated with CAD in GWAS,44 was associated with expression of LIPA in our data and in 2 other tissues (blood and liver). rs2531995 in ADCY9 was associated with obesity in GWAS45 and with expression of ADCY9 in our data and in 4 other tissues (brain, blood, liver, and omentum).
SNP Effects on Gene Expression May Mediate Phenotype Variation
To test whether expression levels of genes regulated by eQTLs might explain the observed associations between eQTLs and phenotypes, we tested the association between expression of eQTL genes and 7 metabolic CVD phenotypes (body mass index, LDL-C, HDL-C, triglycerides [log-transformed], fasting blood glucose, and systolic and diastolic blood pressure; Table 2) in 5257 FHS participants. We found several examples in which the expression level of the eQTL-associated gene was significantly associated with the same trait that was associated in GWAS with the index SNP (hypergeometric test P<0.001; Table 3). For 7 continuous CVD phenotypes that were available for analysis in the FHS, the eQTLs explained 0.5% to 5% of interindividual phenotype variation; in contrast, expression levels of the eQTL genes explained 4% to 13%of interindividual phenotype variation (Table 3). These results are consistent with the hypothesis that genetic variation affects phenotypes via effects on gene expression (see Figure 5 for an example).
Table 2.
Clinical Characteristics of Framingham Heart Study Participants
| Age, y | 51.4 (15.7) |
| Male sex, % | 46 |
| Fasting blood glucose, mg/dL | 100 (21.5) |
| Body mass index, kg/m2 | 27.5 (5.5) |
| Systolic blood pressure, mm Hg | 121.7 (16.6) |
| Diastolic blood pressure, mm Hg | 74.4 (9.9) |
| Total cholesterol, mg/dL | 187.7 (36.3) |
| Triglycerides, mg/dL | 116.4 (83.5) |
| HDL cholesterol, mg/dL | 55.8 (17.0) |
| Hypertension,* % | 40 |
| Diabetes mellitus,* % | 8.4 |
| Lipid treatment, % | 27.8 |
Values are mean (SD) unless indicated otherwise. HDL indicates high-density lipoprotein.
Hypertension: systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg or currently taking medication to treat elevated blood pressure. Diabetes mellitus: participants with fasting blood glucose ≥126 mg/dL or currently taking medication to treat an elevated blood glucose level.
Table 3.
Cardiovascular Phenotypes and Proportion of Interindividual Variation Explained by Associated eQTLs and eQTL Genes in Framingham Heart Study Participants
| Phenotype | Interindividual Variation Explained by eQTLs, % (No. of eQTLs) |
Interindividual Variation Explained by Expression of eQTL Genes, % (No. of eQTL Genes) |
No. of eQTL Genes Also Associated With CVD Phenotype |
|---|---|---|---|
| Body mass index | 3 (39) | 6 (59) | 29 |
| Blood pressure | 0.5 (DBP) | 4 (DBP) | 35 (27 for DBP |
| (SBP and DBP) | 0.7 (SBP)(22) | 4 (SBP) (47) | and 21 for SBP) |
| HDL cholesterol | 4 (60) | 13 (89) | 44 |
| LDL cholesterol | 3 (57) | 13 (101) | 30 |
| Triglycerides | 6 (50) | 13 (60) | 40 |
| Fasting blood | 2 (89) | 9 (183) | 56 |
| glucose |
CVD indicates cardiovascular disease; DBP, diastolic blood pressure; eQTL, expression quantitative trait loci; HDL, high-density lipoprotein; LDL, low-density lipoprotein; and SBP, systolic blood pressure.
Figure 5.
Example of triangular relations among phenotype, single nucleotide polymorphism, and gene expression. rs174546 (in FADS1) was associated with high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides in genome-wide association studies (GWAS). This single nucleotide polymorphism was significantly associated with expression of LDLR in Framingham Heart Study participants (P = 2.9×10−7). The expression of LDLR was also significantly associated with HDL-C, LDL-C, and triglyceride levels in Framingham Heart Study participants. eQTL indicates expression quantitative trait loci.
To test whether the association of a SNP with a phenotype was potentially mediated via its effect on gene expression, we conducted mediation analysis to identify the proportion of the association between a SNP and its corresponding phenotype that was attributable to SNP-related changes in gene expression and subsequent differences in phenotype levels. At P<0.0005 for average causal mediation effects, we identified several potential mediation effects for HDL-C, LDL-C, and triglycerides (Table 4; no significant results were obtained for body mass index, fasting blood glucose, or blood pressure). For example, rs174546, rs174547, and rs174548 (intronic to FADS1) were found to be associated in GWAS with multiple metabolic traits (HDL-C, triglycerides, and phospholipids).31 For these SNPs, we found that 46% of their genetic effect on HDL-C, 59% of their genetic effect on LDL-C, and 47% of their genetic effect on triglycerides were mediated through FADS1 expression (Table 4). In addition, as shown in Figure 4C, these 3 SNPs have trans associations with LDLR and SREBF2, which also demonstrate strong mediation effects on HDL-C, LDL-C, and triglycerides: LDLR (19% mediation for HDL-C, 29% mediation for LDL-C, and 15% mediation for triglycerides) and SREBF2 (19% mediation for HDL-C, 28% mediation for triglycerides). rs964184 was reported to be associated in GWAS with LDL-C, HDL-C, and CAD.4,12 Mediation analyses revealed that a substantial proportion of its genetic effect on lipids is mediated through its trans association with expression of PPM1B (4% mediation of HDL-C and triglycerides) and YPEL5 (7% mediation of HDL-C and 6% mediation of triglycerides). Because the expression levels of all of these genes were associated with HDL-C, LDL-C, or triglyceride levels, the module they constitute may represent an important target for lipid treatment.
Table 4.
Mediation Test Results
| GWAS- Associated Phenotype |
Phenotype-Associated SNP From GWAS (Mapped Gene) |
Expressed Gene Associated With SNP (cis, trans) |
SNP-Gene R2 |
SNP-Gene P Value |
Gene- Phenotype R2 |
Gene- Phenotype P Value |
SNP-Phenotype β (Controlling for Expression) |
Proportion of Mediation of SNP-Phenotype Association by Expression, % |
|---|---|---|---|---|---|---|---|---|
| HDL-C | rs7120118 (NR1H3) |
DDB2 (cis) |
0.0069 | 1.7×10−9 | 0.0033 | 9.1×10−7 | 0.17 | 18.0 |
|
MADD (cis) |
0.015 | 2.6×10−19 | 0.0055 | 1.9×10−5 | 0.22 | 24.0 | ||
| rs174546/rs174547/ rs174548 |
FADS1 (cis) |
0.046 | 9.4×10−56 | 0.0037 | 3.1 ×10−8 | −0.45 | 45.8 | |
| (FADS1) |
SREBF2 (trans) |
0.0054 | 8.9×10−8 | 0.015 | 2.2×10−24 | −0.26 | 21.1 | |
|
LDLR (trans) |
0.005 | 2.9×10−7 | 0.0084 | 1.1×1012 | −0.21 | 18.6 | ||
| rs964184 (ZNF259) |
PPM1B (trans) |
0.0051 | 2.3×10−7 | 0.0017 | 6.1 ×10−5 | −0.073 | 3.7 | |
|
YPEL5 (trans) |
0.0056 | 6.1 ×10−8 | 0.0085 | 1.5×10−12 | −0.13 | 7.0 | ||
| rs4759375 (SBN01) |
CDK2AP1 (cis) |
0.0047 | 6.1 ×10−7 | 0.0074 | 1.6×10−6 | 0.26 | 10.1 | |
| rs3136441 (F2) |
DDB2 (cis) |
0.012 | 1.1×10−15 | 0.0033 | 9.1 ×10−7 | 0.30 | 32.4 | |
| s2271293/rs16942887 (NUTF2/PSKH1) |
DPEP2 (cis) |
0.0061 | 1.3×10−8 | 0.015 | 5.9×10−16 | 0.41 | 23.2 | |
|
SLC12A4 (cis) |
0.0046 | 9.2×10−8 | 0.0035 | 4.6×10−4 | 0.14 | 8.3 | ||
| HDL-C | rs255049 (DPEP3) |
DPEP2 (cis) |
0.0084 | 2.5×10−11 | 0.015 | 5.9×10−16 | 0.38 | 43.7 |
|
SLC12A4 (cis) |
0.0045 | 1.2×10−6 | 0.0035 | 4.6×10−4 | 0.11 | 13.3 | ||
| LDL-C | rs964184 (ZNF259) |
PCSK7 (cis) |
0.0038 | 7.0×10−6 | 0.0040 | 3.3×10−4 | 0.15 | 7.5 |
| rs174546 (FADS1) |
LDLR (trans) |
0.005 | 2.9×10−7 | 0.0034 | 9.6×10−13 | −0.35 | 28.5 | |
|
FADS1 (cis) |
0.046 | 9.4×10−56 | 0.0018 | 4.4×10−7 | −0.72 | 58.6 | ||
| Triglycerides | rs10761731 (JMJD1C) |
CXCL5 (trans) |
0.0061 | 1.4×10−8 | 0.012 | 2.9×10−15 | −0.0063 | 17.0 |
|
ITGB3 (trans) |
0.0062 | 1.2×10−8 | 0.011 | 1.3×10−11 | −0.0042 | 11.8 | ||
|
AQ10 (trans) |
0.0067 | 2.9×10−9 | 0.0044 | 3.3×10−5 | −0.0046 | 12.8 | ||
|
ITGA2B (trans) |
0.0051 | 2.4×10−7 | 0.02 | 7.5×10−6 | −0.0063 | 16.6 | ||
|
CLU (trans) |
0.005 | 2.7×10−7 | 0.022 | 9.9×10−23 | −0.0066 | 17.6 | ||
| rs174546/rs174558 (FADS1) |
FADS1 (cis) |
0.046 | 9.4×10−56 | 0.0017 | 2.7×10−6 | 0.011 | 47.4 | |
| Triglycerides | rs174546/rs174558 (FADS1) |
SREBF2 (trans) |
0.0054 | 8.9×10−8 | 0.017 | 1.1×10−26 | 0.0074 | 27.6 |
|
LDLR (trans) |
0.005 | 2.9×10−7 | 0.0024 | 2.8×10−7 | 0.0038 | 15.1 | ||
| rs4938303 (RPL15P15, BUD13) |
PCSK7 (cis) |
0.0041 | 3.64×10−6 | 0.0047 | 2.6×10−11 | 0.0035 | 5.0 | |
| rs651821 (APOA5) |
SREBF2 (trans) |
0.0047 | 6.4×10−7 | 0.017 | 1.1×10−26 | 0.014 | 9.8 | |
| rs964184 (ZNF259) |
PPM1B (trans) |
0.0051 | 2.26×10−7 | 0.0087 | 3.0×10−19 | 0.0055 | 4.4 | |
|
YPEL5 (trans) |
0.0056 | 6.11×10−8 | 0.027 | 1.3×10−38 | 0.0071 | 5.7 |
GWAS indicates genome-wide association studies; HDL-C, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; R2, percent variance explained; SNP, single nucleotide polymorphism; and /, SNP in linkage disequilibrium (R2 > 0.8).
Metabolite SNPs and CVD Traits
Metabolomic findings can be used to unravel novel biochemical mechanisms involved in a variety of disease processes, including atherogenesis. To identify genetic and biochemical underpinnings of our CVD network and pathways, we incorporated 170 genome-wide significant SNPs from 2 recently published metabolomic GWAS.36,46 We found 13 SNPs that were shared between metabolites and the 21 CVD phenotypes in our network. As shown in Figure 6, several metabolites are associated with the 21 CVD traits in our network by virtue of shared GWAS SNPs. This was especially notable for lipid traits. For 13 SNPs that were shared between metabolites and the CVD traits in our network (Figure 6), 6 of them were also associated with expression levels of genes (Table 5), including rs174548 (FADS1), which was associated in GWAS with arachidonic acid (C20:4), a product regulated by FADS1, and with its, substrate, dihomolinolenate. These eQTLs belong to 3 eQTL subnetworks (Figure 4A through 4C), suggesting genetic regulation of intermediate metabolites or the lipid end-products in our pathways. When we further included perfect proxy SNPs (r2=1) for the index GWAS SNPs associated with metabolites and CVD traits, we identified 8 eQTLs for 3 metabolite SNPs (Table X in the online-only Data Supplement) that were associated with additional CVD traits, including variants in ABO associated with venous thrombosis, CAD, and LDL-C.
Figure 6.
Cardiovascular disease phenotype and metabolite network by virtue of shared genome-wide association study single nucleotide polymorphisms. Gray nodes represent cardiovascular disease traits. Red nodes represent metabolites. Two traits are connected if they share at least 1 single nucleotide polymorphism in genome-wide association studies. HDL-C indicates high-density lipoprotein cholesterol; and LDL, low-density lipoprotein.
Table 5.
eQTLs Among Metabolite-Associated GWAS Single Nucleotide Polymorphisms
| eQTL | Gene Symbol and Locus |
Metabolite Associated With eQTL |
Traits Associated With eQTL in GWAS |
Expressed Gene Associated With eQTL |
|---|---|---|---|---|
| rs1260326 |
GCKR (2p23.3) |
Glucose/mannose | C-reactive protein; triglycerides; LDL cholesterol |
NRBP1* |
| rs174547/ | FADS1 | Arachidonate (20:4n6)/ | HDL cholesterol; | C11orf10*; FADS2*; FADS1*; FEN1*; |
| rs174548 | (11q12.2) | dihomo-linolenate (20:3n3 or n6) | triglycerides | LDLR†; SREBF2† |
| rs3184504 | SW2B3(12q24.12) | Kynurenine | Blood pressure; type 1 diabetes mellitus |
TRAFD1*; ALDH2*; HVCN1*; TCTN1*; ANKRD22†; ARHGEF40†; CD274†; FCGRIA†; GBP1†; GBP4†; GBP5†; GBP7†; IDS†; IFIT3†; IRF9†; MYADM†; PARP14†; PSMB9†; PSTPIP2†; RFX2†; RNF31†; SAMD9L†; SERPINGI†; SRBDI†; STATI†; TRIM22†; UBE2L6†; WDFY2† |
| rs7570971 | RAB3GAP1 (2q21.3) | 1,5-Anhydroglucitol (1,5-AG) | LDL cholesterol | MCM6*; R3HDM1*; IRF8†; TNFRSF21†; ULRA4†; SERPINF1†; DARS† |
| rs964184 | ZNF259 (11q23.3) | DAG 36:2/ TAG 56:3 /X-03094 |
HDL cholesterol | BUD13*; PCSK7*; SIDT2*; TAGLN*; 0BFC2A†; TMEM165†; PPM1B†; YPEL5† |
| rs651821 |
AP0A5 (11q23.3) |
Valine | Triglycerides | TAGLN*; SIDT2*; SREBF2† |
eQTL indicates expression quantitative trait loci; GWAS, genome-wide association studies; HDL, high-density lipoprotein; and LDL, low-density lipoprotein.
Denotes cis association with eQTL.
Denotes trans association with eQTL.
Discussion
CVD is the consequence of the intricate interplay between multiple genetic variants, clinical risk factors, and environmental factors. Our phenotype network, composed of pleiotropic SNPs, provided evidence of the shared genetic underpinnings of CVD and its risk factors. Our eQTL network, which integrated SNPs, gene expression, and phenotype, identified several pathways affected by genetic variants associated with CVD and its major risk factors.
With the use of GWAS results alone, it is not possible to identify the causal variant, the causal gene, or the mechanism by which a SNP or nearby gene affects the phenotype. By integrating multidimensional data (ie, GWAS SNPs and gene expression analyses), we provide evidence that GWAS loci have strong associations (cis or trans) with expression levels of genes.47 We replicated our eQTL results in 2 large databases. The relatively low replication of some eQTLs from our data set in other databases may be attributable to the different genotyping and gene expression platforms (the Blood eQTL Browser used iIllumina arrays for SNPs and gene expression, whereas we used an Affymetrix array) or from the fact that our data set arose from a larger single cohort with uniform data collection techniques, whereas the Blood eQTL Browser relied on meta-analysis of many separate data collection efforts. On the other hand, for the eQTLs identified both in our data and in other databases, we found a high concordance of SNP-gene associations, further indicating that these eQTLs are replicable. Many of the genes associated with CVD SNPs were previously reported to be associated with CVD or its risk factors, including FADS1, HMGCR, LPL, LDLR, and SREBF2. Moreover, we found a large number of eQTL-associated genes whose expression levels were also associated with a variety of CVD phenotypes, suggesting the existence of 3-way relationships between genetic variants, gene expression, and phenotypes (Figure 5).
The underlying mechanism of downstream effects of disease-associated SNPs (trans-eQTL) has not yet been fully characterized. It has been suggested that expressed cis-eQTL genes can act as master trans regulators.48 Among 31 eQTLs with both cis and trans associations, we found that only SNPs in the FADS1 region (rs174546, rs174547, and rs174548) lost significance for association with LDLR and SREBF2 after conditioning on expression of the corresponding cis genes (for expression of FADS1 and FADS2, see Table XI in the online-only Data Supplement). Both the cis and trans associations were replicated in the Blood eQTL Browser, suggesting that the trans effects on LDLR and SREBF2 were mediated by FADS1 and FADS2 expression. Moreover, both the SNPs in GWAS and gene expression in our samples were associated with multiple lipids traits (HDL-C, LDL-C, and triglycerides), providing evidence of trans effects and implicating this cis-trans eQTL module (Figure 4C) in the link between FADS gene variation and CVD risk.
Common variants from GWAS explained only a small fraction of interindividual trait variance, yet they may provide important biological or therapeutic insights. For example, common variants in the introns of HMGCR and NPC1L1 confer small effects on plasma LDL-C (3 and 2 mg/dL, respectively), but they have dramatic effects on LDL-C when targeted by statins or ezetimibe, respectively12 Using mediation testing, we found that the genetic effects of variants (rs12916, rs3846663, and rs 12654264; pairwise R2 =0.94–1.0) in HMGCR on LDL-C may be mediated through HMGCR expression (P=0.034, P=0.036, and P=0.044, respectively). This analysis also revealed several known as well as potentially novel therapeutic targets. For example, we found that the expression of PCSK7 was not only cis associated with rs964184, a pleiotropic SNP in ZNF259 that is associated in GWAS with HDL-C, LDL-C, triglycerides, and CVD risk,4,12 but PCSK7 expression also was associated with LDL-C and triglyceride levels in FHS participants. Thus, part of the genetic effect of rs964184 on LDL-C (8%) and triglycerides (5%) was mediated through expression of PCSK7, providing orthogonal support for this gene as a potential therapeutic target. Of note, a rare coding variant in PCSK7 was recently found to be associated with HDL-C by analysis of exonic variants in individuals of African ancestry.49 Expression of another gene, FDFT1, revealed cis associations with SNPs associated in GWAS with HDL-C, LDL-C, triglycerides, and coronary disease (Figure 4E; light red represents long-range cis associations). The expression of FDFT1 was significantly associated with HDL-C and triglyceride levels (P=1.5×10−10 and 1.5×10−7, respectively) in FHS participants. Moreover, the mediation test for FDFT1 was significant (P <0.001 for average causal mediation effects) on HDL-C. A recent study found that expression of FDFT1 was significantly higher in atherosclerosis-resistant Japanese quail than in atherosclerosis-susceptible strains,50 suggesting that FDFT1 may represent another potential therapeutic target for the treatment of lipids and atherosclerotic CVD.
There are several limitations to this study. First, from this observational study, we can only infer the mediation effects of genetic variants. Causal relationships may be validated through randomized experiments or biological validation studies. Second, our gene expression data were derived from whole blood; some eQTLs may be highly tissue dependent. Therefore, the CVD modules and mediation effects may not be reflective of other tissues. Third, because each SNP only contributes a small effect on phenotypic variation, the combination of SNPs and their interactions may reveal a more complete picture of disease mechanisms.
In summary, integrating published GWAS with genetic variants, gene expression, and phenotype data from >5000 FHS participants allowed us to decipher the genetic architecture that underlies CVD and its risk factors at the population level. The integration of 3 levels of data not only afforded plausible functional explanations for disease but also revealed promising therapeutic targets.
Supplementary Material
CLINICAL PERSPECTIVE.
Cardiovascular diseases (CVDs) reflect a highly coordinated complex of traits. Although thousands of single nucleotide polymorphisms have been found to be associated with CVD traits, a key question that remains unanswered is as follows: How does DNA sequence variation cause disease? Answers to this question can be translated into new drug targets to improve patient care. In this study, we built a CVD network using single nucleotide polymorphism-CVD phenotype associations. The shared single nucleotide polymorphisms between CVD risk factors provide evidence of a genetic explanation for the clustering of metabolic risk factors in the same individuals. We incorporated transcriptomic data into genetic and phenotype network analysis using data from 5257 Framingham Heart Study participants to dissect the relationships between genetic variants, gene expression, and CVD phenotypes. We identified several putatively causal genetic variants that appear to exert their function by altering expression of associated genes that in turn appear to promote interindividual variation in CVD phenotypes. These variants and pathways identified by this approach point toward novel therapeutic targets for the treatment and prevention of CVD.
Acknowledgments
We thank all of the study participants who helped to create this valuable resource and supported this work. We thank the data management group of FHS for organizing and providing these data. We thank the National Institutes of Health Fellows Editorial Board members for their valuable edits and comments. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Sources of Funding
The FHS is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, and the Center for Information Technology, National Institutes of Health, Bethesda, MD, and a National Institutes of Health grant (U01 HG007019). Dr Tastan acknowledges support from Bilim Akademisi, The Science Academy, Turkey, under the BAGEP program and support from L’Oreal-UNESCO under the UNESCO-L’Oreal National Fellowships Program for Young Women in Life Sciences. B.O. is supported by The Scientific and Technological Research Council of Turkey (TÜBİTAK, 2211-C PhD Scholarship). J.B. Meigs is supported by K24 DK080140.
Footnotes
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/lookup/suppl/doi:10.1161/CIRCULATIONAHA.114.010696/-/DC1.
Disclosures
None.
References
- 1.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaziano TA, Bitton A, Anand S, Abrahams-Gessel S, Murphy A. Growing epidemic of coronary heart disease in low- and middle-income countries. Curr Probl Cardiol. 2010;35:72–115. doi: 10.1016/j.cpcardiol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schunkert H, König JR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Home BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Mühleisen TW, Muhlestein JB, Miinzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nöthen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Mörz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O’Donnell CJ, McPherson R, Erdmann J, Samani NJ CARDIoGRAM Consortium. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Völker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP, Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. BIRAC Consortium; YEAR Consortium. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C, Stevens H, Jackson L, Simmonds MJ, Bingley PJ, Gough SC, Todd JA. Type 1 Diabetes Genetics Consortium Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Döring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L ENGAGE Consortium. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ndiaye NC, Azimi Nehzad M, El Shamieh S, Stathopoulou MG, Visvikis-Siest S. Cardiovascular diseases and genome-wide association studies. Clin Chim Acta. 2011;412:1697–1701. doi: 10.1016/j.cca.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 10.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, ‘t Hoen PA, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Völker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JB, Franke L. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König ER, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lusis AJ, Weiss JN. Cardiovascular networks: systems-based approaches to cardiovascular disease. Circulation. 2010;121:157–170. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oppenheimer GM. Becoming the Framingham Study 1947–1950. Am J Public Health. 2005;95:602–610. doi: 10.2105/AJPH.2003.026419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joehanes R, Ying S, Huan T, Johnson AD, Raghavachari N, Wang R, Liu P, Woodhouse KA, Sen SK, Tanriverdi K, Courchesne P, Freedman JE, O’Donnell CJ, Levy D, Munson PJ. Gene expression signatures of coronary heart disease. Arterioscler Thromb Vase Biol. 2013;33:1418–1426. doi: 10.1161/ATVBAHA.112.301169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M, Feolo M. NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. (database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamin AM, Suchindran S, Pearce K, Rowell J, Lien LF, Guyton JR, McCarthy JJ. Gene by sex interaction for measures of obesity in the Framingham Heart Study. J Obes. 2011;2011:329038. doi: 10.1155/2011/329038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez AI, Bates DM, Rosa GJ, Gianola D, Weigel KA. Technical note: an R package for fitting generalized linear mixed models in animal breeding. J Anim Sci. 2010;88:497–504. doi: 10.2527/jas.2009-1952. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B Met. 1995;57:289–300. [Google Scholar]
- 23.Imai K, Keele L, Tingley D. A general approach to causal mediation analysis. Psychol Methods. 2010;15(4):309–334. doi: 10.1037/a0020761. [DOI] [PubMed] [Google Scholar]
- 24.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC. Defining functional DNA elements in the human genome. Proc Natl AcadSci USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. (Web server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Gierman HJ, Levy D, Plump A, Dobrin R, Goring HH, Curran JE, Johnson MP, Blangero J, Kim SK, O’Donnell CJ, Emilsson V, Johnson AD. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spinelli L, Gambette P, Chappie CE, Robisson B, Baudot A, Garreta H, Tichit L, Guènoche A, Brun C. Clust&See: a Cytoscape plugin for the identification, visualization and manipulation of network clusters. Biosystems. 2013;113:91–95. doi: 10.1016/j.biosystems.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Gambettea P, Guenoche A. Bootstrap clustering for graph partitioning. RAIRO Op Res. 2012;45:339–352. [Google Scholar]
- 30.Rutter MK, Meigs JB, Sullivan LM, D’Agostino RB, Sr, Wilson PW. Insulin resistance, the metabolic syndrome, and incident cardiovascular events in the Framingham Offspring Study. Diabetes. 2005;54:3252–3257. doi: 10.2337/diabetes.54.11.3252. [DOI] [PubMed] [Google Scholar]
- 31.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, Pellikka N, Wallaschofski H, Kettunen J, Henneman P, Baumert J, Strachan DP, Fuchsberger C, Vitart V, Wilson JF, Parè G, Naitza S, Rudock ME, Surakka I, de Geus EJ, Alizadeh BZ, Guralnik J, Shuldiner A, Tanaka T, Zee RY, Schnabel RB, Nambi V, Kavousi M, Ripatti S, Nauck M, Smith NL, Smith AV, Sundvall J, Scheet P, Liu Y, Ruokonen A, Rose LM, Larson MG, Hoogeveen RC, Freimer NB, Teumer A, Tracy RP, Launer LJ, Buring JE, Yamamoto JF, Folsom AR, Sijbrands EJ, Pankow J, Elliott P, Keaney JF, Sun W, Sarin AP, Fontes JD, Badola S, Astor BC, Hofman A, Pouta A, Werdan K, Greiser KH, Kuss O, Meyer zu Schwabedissen HE, Thiery J, Jamshidi Y, Nolte IM, Soranzo N, Spector TD, Völzke H, Parker AN, Aspelund T, Bates D, Young L, Tsui K, Siscovick DS, Guo X, Rotter JI, Uda M, Schlessinger D, Rudan I, Hicks AA, Penninx BW, Thorand B, Gieger C, Coresh J, Willemsen G, Harris TB, Uitterlinden AG, Järvelin MR, Rice K, Radke D, Salomaa V, Willems van Dijk K, Boerwinkle E, Vasan RS, Ferrucci L, Gibson QD, Bandinelli S, Snieder H, Boomsma DI, Xiao X, Campbell H, Hayward C, Pramstaller PP, van Duijn CM, Peltonen L, Psaty BM, Gudnason V, Ridker PM, Homuth G, Koenig W, Ballantyne CM, Witteman JC, Benjamin EJ, Perola M, Chasman DI. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segrè AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpeläinen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proença C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grössler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jørgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, König IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaløy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimäki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Parè G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstråle M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tönjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kähönen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Grönberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ MAGIC; Procardis Consortium. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a Web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han D, Cai X, Wen J, Matheson D, Skyler JS, Kenyon NS, Chen Z. Innate and adaptive immune gene expression profiles as biomarkers in human type 1 diabetes. Clin Exp Immunol. 2012;170:131–138. doi: 10.1111/j.1365-2249.2012.04650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikäinen LP, Kangas AJ, Soininen P, Würtz P, Silander K, Dick DM, Rose RJ, Savolainen MJ, Viikari J, Kähænen M, Lehtimäki T, Pietilainen KH, Inouye M, McCarthy MI, Jula A, Eriksson J, Raitakari OT, Salomaa V, Kaprio J, Järvelin MR, Peltonen L, Perola M, Freimer NB, Ala-Korpela M, Palotie A, Ripatti S. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Mägi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tönjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Döring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Järvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K ENGAGE Consortium. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, Albrecht E, Jackson CM, Evans DM, Cadby G, Fornage M, Manichaikul A, Lopez LM, Johnson T, Aldrich MC, Aspelund T, Barroso I, Campbell H, Cassano PA, Couper DJ, Eiriksdottir G, Franceschini N, Garcia M, Gieger C, Gislason GK, Grkovic I, Hammond CJ, Hancock DB, Harris TB, Ramasamy A, Heckbert SR, Heliövaara M, Homuth G, Hysi PG, James AL, Jankovic S, Joubert BR, Karrasch S, Klopp N, Koch B, Kritchevsky SB, Launer LJ, Liu Y, Loehr LR, Lohman K, Loos RJ, Lumley T, Al Balushi KA, Ang WQ, Barr RG, Beilby J, Blakey JD, Boban M, Boraska V, Brisman J, Britton JR, Brusselle GG, Cooper C, Curjuric I, Dahgam S, Deary IJ, Ebrahim S, Eijgelsheim M, Francks C, Gaysina D, Granell R, Gu X, Hankinson JL, Hardy R, Harris SE, Henderson J, Henry A, Hingorani AD, Hofman A, Holt PG, Hui J, Hunter ML, Imboden M, Jameson KA, Kerr SM, Kolcic I, Kronenberg F, Liu JZ, Marchini J, McKeever T, Morris AD, Olin AC, Porteous DJ, Postma DS, Rich SS, Ring SM, Rivadeneira F, Rochat T, Sayer AA, Sayers I, Sly PD, Smith GD, Sood A, Starr JM, Uitterlinden AG, Vonk JM, Wannamethee SG, Whincup PH, Wijmenga C, Williams OD, Wong A, Mangino M, Marciante KD, McArdle WL, Meibohm B, Morrison AC, North KE, Omenaas E, Palmer LJ, Pietilàinen KH, Pin I, Pola Sbreve EkO, Pouta A, Psaty BM, Hartikainen AL, Rantanen T, Ripatti S, Rotter JI, Rudan I, Rudnicka AR, Schulz H, Shin SY, Spector TD, Surakka I, Vitart V, Volzke H, Wareham NJ, Warrington NM, Wichmann HE, Wild SH, Wilk JB, Wjst M, Wright AF, Zgaga L, Zemunik T, Pennell CE, Nyberg F, Kuh D, Holloway JW, Boezen HM, Lawlor DA, Morris RW, Probst-Hensch N, Kaprio J, Wilson JF, Hayward C, Kahonen M, Heinrich J, Musk AW, Jarvis DL, Gläser S, Järvelin MR, Ch Strieker BH, Elliott P, O’Connor GT, Strachan DP, London SJ, Hall IP, Gudnason V, Tobin MD International Lung Cancer Consortium; GIANT Consortium. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, Ruppert A, Lodrup Carlsen KC, Roses A, Anderson W, Rennard SI, Lomas DA, Silverman EK, Goldstein DB ICGN Investigators. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ware JJ, van den Bree M, Munafὸ MR. From men to mice: CHRNA5/CHRNA3, smoking behavior and disease. Nicotine Tob Res. 2012;14:1291–1299. doi: 10.1093/ntr/nts106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Floyel T, Brorsson C, Nielsen LB, Miani M, Bang-Berthelsen CH, Friedrichsen M, Overgaard AJ, Berchtold LA, Wiberg A, Poulsen P, Hansen L, Rosinger S, Boehm BO, Ram R, Nguyen Q, Mehta M, Morahan G, Concannon P, Bergholdt R, Nielsen JH, Reinheckel T, von Herrath M, Vaag A, Eizirik DL, Mortensen HB, Storling J, Pociot F. Ctsh regulates beta-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc Natl Acad Sci USA. 2014;111:10305–10310. doi: 10.1073/pnas.1402571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell JE, Henders AK, McRae AF, Wright MJ, Martin NG, Dermitzakis ET, Montgomery GW, Visscher PM. Genetic control of gene expression in whole blood and lymphoblastoid cell lines is largely independent. Genome Res. 2012;22:456–166. doi: 10.1101/gr.126540.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, Sugiyama T, Jeon JP, Liu JJ, Takayanagi R, Kim SS, Aung T, Sung YJ, Zhang X, Wong TY, Han BG, Kobayashi S, Ogihara T, Zhu D, Iwai N, Wu JY, Teo YY, Tai ES, Cho YS, He J. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 45.Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Luan J, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu YJ, Lee SH, Liang L, Lin DY, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga JJ, Huffman JE, Jarick I, Johansson Å, Johnson T, Kanoni S, Kleber ME, König IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Müller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson WO, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, Ferrières J, Franke L, Frau F, Gejman PV, Grallert H, Grönberg H, Gudnason V, Hall AS, Hall P, Hartikainen AL, Hayward C, Heard-Costa NL, Heath AC, Hebebrand J, Homuth G, Hu FB, Hunt SE, Hyppönen E, Iribarren C, Jacobs KB, Jansson JO, Jula A, Kähönen M, Kathiresan S, Kee F, Khaw KT, Kivimaki M, Koenig W, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen JH, Lakka TA, Langenberg C, Launer LJ, Lind L, Lindström J, Liu J, Liuzzi A, Lokki ML, Lorentzon M, Madden PA, Magnusson PK, Manunta P, Marek D, März W, Mateo Leach I, McKnight B, Medland SE, Mihailov E, Milani L, Montgomery GW, Mooser V, Mtihleisen TW, Munroe PB, Musk AW, Narisu N, Navis G, Nicholson G, Nohr EA, Ong KK, Oostra BA, Palmer CN, Palotie A, Peden JF, Pedersen N, Peters A, Polasek O, Pouta A, Pramstaller PP, Prokopenko I, Putter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo TE, Sambrook JG, Sanders AR, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin SY, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stὸncakὸva A, Stark K, Stephens JC, Stirrups K, Stolk RP, Stumvoll M, Swift AJ, Theodoraki EV, Thorand B, Tregouet DA, Tremoli E, Van der Klauw MM, van Meurs JB, Vermeulen SH, Viikari J, Virtamo J, Vitart V, Waeber G, Wang Z, Widén E, Wild SH, Willemsen G, Winkelmann BR, Witteman JC, Wolffenbuttel BH, Wong A, Wright AF, Zillikens MC, Amouyel P, Boehm BO, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Cupples LA, Cusi D, Dedoussis GV, Erdmann J, Eriksson JG, Franks PW, Froguel P, Gieger C, Gyllensten U, Hamsten A, Harris TB, Hengstenberg C, Hicks AA, Hingorani A, Hinney A, Hofman A, Hovingh KG, Hveem K, Illig T, Jarvelin MR, Jöckel KH, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kuh D, Laakso M, Lehtimäki T, Levinson DF, Martin NG, Metspalu A, Morris AD, Nieminen MS, Njølstad I, Ohlsson C, Oldehinkel AJ, Ouwehand WH, Palmer LJ, Penninx B, Power C, Province MA, Psaty BM, Qi L, Rauramaa R, Ridker PM, Ripatti S, Salomaa V, Samani NJ, Snieder H, Sørensen TI, Spector TD, Stefansson K, Tönjes A, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Vollenweider P, Wallaschofski H, Wareham NJ, Watkins H, Wichmann HE, Wilson JF, Abecasis GR, Assimes TL, Barroso I, Boehnke M, Borecki IB, Deloukas P, Fox CS, Frayling T, Groop LC, Haritunian T, Heid IM, Hunter D, Kaplan RC, Karpe F, Moffatt MF, Mohlke KL, O’Connell JR, Pawitan Y, Schadt EE, Schlessinger D, Steinthorsdottir V, Strachan DP, Thorsteinsdottir U, van Duijn CM, Visscher PM, Di Blasio AM, Hirschhorn JN, Lindgren CM, Morris AP, Meyre D, Scherag A, McCarthy MI, Speliotes EK, North KE, Loos RJ, Ingelsson E. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmüller G, Spector TD, Soranzo N Multiple Tissue Human Expression Resource (MuTHER) Consortium. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, Thorsteindottir U, Shin SY, Richards HB, Soranzo N, Ahmadi KR, Lindgren CM, Stefansson K, Dermitzakis ET, Deloukas P, Spector TD, McCarthy MI GIANT Consortium; MAGIC Investigators; DIAGRAM Consortium; MuTHER Consortium. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peloso GM Group CLW. Novel rare coding variants underlie blood lipid levels in the population: an exome array association study in 55,000 whites and blacks; Paper presented at: Annual Meeting of the American Society of Human Genetics; October 2013; Boston, MA. [Google Scholar]
- 50.Li X, Schulte P, Godin DV, Cheng KM. Differential mRNA expression of seven genes involved in cholesterol metabolism and transport in the liver of atherosclerosis-susceptible and -resistant Japanese quail strains. Genet Sel Evol. 2012;44:20. doi: 10.1186/1297-9686-44-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.