SUMMARY
The Mec1/Tel1 kinases (human ATR/ATM) play numerous roles in the DNA replication stress response. Despite the multi-functionality of these kinases, studies of their in vivo action have mostly relied on a few well-established substrates. Here we employed a combined genetic-phosphoproteomic approach to monitor Mec1/Tel1 signaling in a systematic, unbiased and quantitative manner. Unexpectedly, we find that Mec1 is highly active during normal DNA replication, at levels comparable or higher than Mec1’s activation state induced by replication stress. This “replication-correlated” mode of Mec1 action requires the 9-1-1 clamp and the Dna2 lagging-strand factor, and is distinguishable from Mec1’s action in activating the downstream kinase Rad53. We propose that Mec1/ATR performs key functions during ongoing DNA synthesis that are distinct from their canonical checkpoint role during replication stress.
INTRODUCTION
During DNA replication, cells are prone to accumulate genomic instabilities. Progression of the replication machinery is often impeded by barriers such as DNA adducts, DNA-RNA hybrids and protein-DNA complexes (Lambert and Carr, 2013). Replication forks often stall upon encountering these hard-to-replicate regions, leading to exposure of single-stranded DNA (ssDNA), which in turn, is a major signal for the activation of the evolutionarily conserved PI3K-like sensor kinase ATR (yeast Mec1) (MacDougall et al., 2007). Once activated, ATR and Mec1 initiate a signaling response that induces key effects such as cell cycle arrest, inhibition of origin firing and stabilization of stalled replication forks (Branzei and Foiani, 2010; Santocanale and Diffley, 1998). The importance of ATR is highlighted by the fact that deletion or mutations that affect its activity are associated with embryonic lethality, chromosomal fragmentation and increasing sensitivity to genotoxic drugs (Brown and Baltimore, 2000; Wright et al., 1998). In budding yeast, strains with mec1 mutations were shown to accumulate gross chromosomal rearrangements (GCRs) (Myung et al., 2001) and be exquisitely sensitive to genotoxic drugs that induce replication stress (Weinert et al., 1994). Like ATR, the PI3K-like sensor kinase ATM (yeast Tel1) is also important during DNA damage responses. Cells lacking ATM show sensitivity to DNA damaging agents and elevated levels of mitotic recombination (Meyn, 1993), but differently from ATR, which is a sensor for ssDNA accumulation, ATM responds mainly to DNA double strand breaks (DSBs) (Shiloh and Ziv, 2013). In yeast, tel1Δ null mutants are viable and show no significant sensitivity to DNA damaging agents. However, mec1Δtel1Δ double mutants are more sensitive to replication stress and display a more severe growth defect than the single deletion mutants, revealing functionally redundant roles for these kinases (Morrow et al., 1995).
Over the last decade, others and we have identified many candidate substrates of Mec1/Tel1 and ATR/ATM using large-scale mass spectrometry-based approaches (Chen et al., 2010; Matsuoka et al., 2007; Smolka et al., 2007). However, our understanding of how these kinases promote a systemic cellular response that safeguards genomic integrity and allows cells to better cope with the effects of replication stress is still limited. A major limitation towards a more comprehensive characterization of Mec1/Tel1 and ATR/ATM action is posed by the difficulty of reproducibly and quantitatively monitoring the many substrates identified by mass spectrometry (MS). Consequently, the use of antibody-based approaches to monitor well-established substrates remains the method of choice. Substrates commonly monitored using western blotting techniques include the histone variant H2AX (yeast H2A) and the downstream checkpoint kinases CHK1 and CHK2 (yeast Rad53). Despite the biological relevance of these substrates, the use of their phosphorylation as readouts for the checkpoint response has introduced a marked bias in studies aiming at characterizing Mec1/Tel1 action. To address this problem, here we employed a combined genetic-proteomic approach (which we refer as QMAPS: Quantitative Mass-Spectrometry Analysis of Phospho-Substrates) for identifying and monitoring multiple in vivo kinase substrates in a systematic, unbiased and quantitative manner. Using QMAPS we show that Mec1 is robustly activated during unperturbed DNA replication, in a manner that correlates with the extent of DNA replication and that is distinct from a canonical checkpoint. Collectively, our results demonstrate the importance of unbiased and quantitative analysis of kinase substrates to comprehensively characterize the in vivo action of multi-functional kinases.
RESULTS
Unbiased delineation of Mec1 and Tel1 action using a genetic-proteomic approach
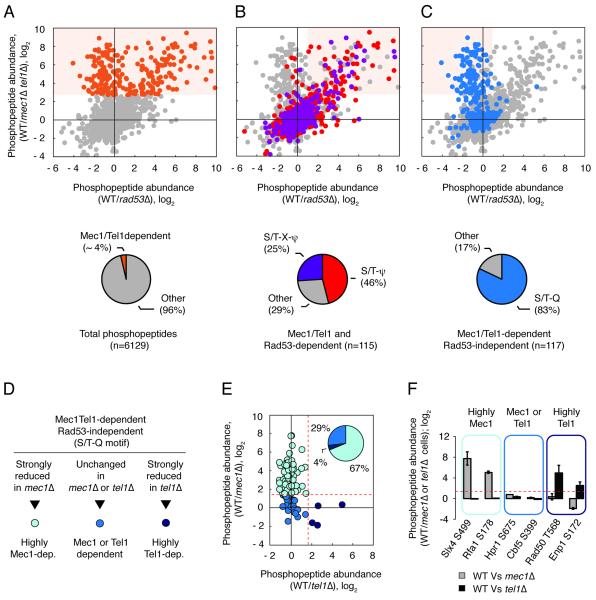
Our current understanding of Mec1 and Tel1 action is biased towards the use of a few established substrates as reporters of the in vivo activity of these kinases. In particular, the activation state of the major downstream kinase Rad53 has been extensively used as a key indicator of Mec1 and Tel1 activation status. To circumvent this bias and be able to comprehensively characterize the action of Mec1 and Tel1, we used quantitative MS analysis of kinase mutant strains to identify and monitor as many candidate substrates of these kinases as possible. First, we performed a proteomic screen to globally define the set of Mec1 and Tel1 candidate targets. Building on our previously published work (Smolka et al., 2007), we used quantitative MS to compare the phosphoproteome of wild-type (WT) and mec1Δtel1Δ cells treated with methyl methanesulfonate (MMS) or hydroxyurea (HU) to induce replication stress. To facilitate the classification of Mec1/Tel1-dependent phosphorylation sites into direct or indirect Mec1/Tel1 phosphorylation events, we also quantified the relative abundance of the phosphopeptides in cells lacking Rad53, the major kinase downstream of Mec1/Tel1. We were able to identify and quantify more than 6,000 phosphopetides over distinct biological replicates (Figure 1A, Table S1). Of interest, the abundance of 232 of the identified phosphopeptides was significantly reduced in cells lacking Mec1 and Tel1, and we refer to them as Mec1/Tel1-dependent events. Among the 232 Mec1/Tel1-dependent targets, 115 were found to be dependent on Rad53, and thus considered as indirect Mec1/Tel1-dependent events (Figure 1B). In our strategy, direct targets of Mec1/Tel1 should be present in the group of phosphopeptides carrying a Mec1/Tel1-dependent and Rad53-independent phospho-site. As shown in Figure 1C (Table S1), analysis of the amino acid in the +1 position of Mec1/Tel1-dependent and Rad53-independent phospho-sites revealed a strong enrichment of the S/T-Q motif, consistent with previous work indicating this preferential motif for Mec1 and Tel1 (Kim et al., 1999; Smolka et al., 2007). Of the 117 Mec1/Tel1-dependent and Rad53-independent phosphorylation events, 97 are in the preferred S/T-Q motif and we considered them as directly targeted by Mec1 or Tel1. On the other hand, Rad53 showed a bias towards the S/T-bulky amino acid (ψ) motif (Figure 1B and Table S1). For more than 60% of the proteins found to have a Mec1/Tel1-dependent phosphorylation, we were able to also detect at least one Mec1/Tel1-independent phosphorylation event, supporting that most of the observed changes are not due to changes in protein abundance (Figure S1 and Table S1).
Figure 1. Proteome-wide identification of Mec1/Tel1-dependent phosphorylation events using quantitative MS.
(A) Identification of Mec1/Tel1-dependent phosphopeptides (cells treated with 0.2M HU or 0.04% MMS). Orange dots correspond to 238 Mec1/Tel1-dependent phosphopeptides. See text for details. (B) Mec1/Tel1 and Rad53-dependent phosphorylation events (light orange shade) are biased towards the S/T-ψ (red) and S/T-X-ψ (purple) motifs. (C) Mec1/Tel1-dependent and Rad53-independent phosphorylation events (light orange shade) are biased towards the S/T-Q motif (blue). (D-E) The phosphoproteome of WT cells was compared to the phosphoproteome of mec1Δ or tel1Δ cells (all cells treated with 0.04% MMS) and phosphopeptides carrying phosphorylation in the S/T-Q motif were categorized according to the observed change in abundance. Dotted red lines represent the established cutoff of 3-fold increase in WT relative to mec1Δ or tel1Δ cells. (F) Examples of phosphopeptides of each of the indicated groups. Data are represented as fold change in phosphopeptide abundance; log2 +/− SEM (n≥2). See also Table S1.
To sort out the relative contribution of Mec1 or Tel1 in the response, we performed similar analyses as described above, but comparing WT cells to cells lacking either Mec1 or Tel1 (Figure 1D). Of the Mec1 and Tel1 direct phospho-events identified above, 67% were found to heavily depend mostly on Mec1 (Figure 1E and Table S1). Only four phospho-sites were found to heavily depend exclusively on Tel1, consistent with the fact that cells lacking Tel1 don’t exhibit significant sensitivity to replication stress-inducing agents (Morrow et al., 1995). Importantly, about 29% of Mec1/Tel1-dependent sites were found to remain robustly phosphorylated in cells lacking either Mec1 or Tel1, and represent a set of common candidate substrates of these kinases (Figures 1D-F and Table S1). These results establish a large set of Mec1 and Tel1 targets and define their relative level of dependency for each of these kinases. This defined set of phosphorylation sites targeted by Mec1 and/or Tel1 forms the basis of our unbiased strategy to characterize the action of these kinases in different growth conditions and genetic backgrounds. The output of this analysis of substrates is a quantitative map, herein named QMAPS, revealing the relative levels of phosphorylation of identified phosphopeptides in two different conditions being tested (see Figure 2A).
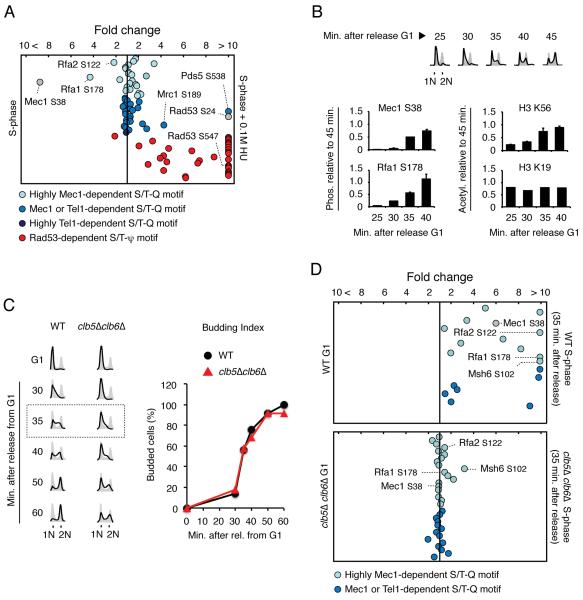
Figure 2. Quantitative analysis of Mec1/Tel1-dependent phosphorylation during normal DNA replication.
(A) QMAPS showing the relative abundance of phosphopeptides categorized according to results from Figure 1. Phosphopeptides carrying Mec1 autophosphorylation or Mec1-dependent Rad53 phosphorylation are indicated in grey. α-factor arrested cells were released from arrest in normal SILAC media or SILAC media containing 0.1M HU for 45 minutes. Abscissa indicates fold change in phosphopeptide abundance (linear scale) between S-phase cells treated with 0.1M HU and untreated. (B) Protein extracts were prepared from WT cells at indicated times after release from α-factor-arrest into fresh media. Mec1 (and Mec1-associated Rfa1) was pulled-down and phosphopeptides containing Mec1 autophosphorylation at S38 and Rfa1 phosphorylation at S178 were monitored by quantitative MS analysis. FACS analysis and H3K56 acetylation were used as positive controls for DNA replication progression while acetylation of H3K19 was used as a constitutive control. Data are represented as mean +/− SEM (n≥2). (C) FACS analysis and budding index of WT and clb5Δclb6Δ mutant cells following α-factor arrest and release in drug-free SILAC media. (D) QMAPS showing the relative abundance of phosphopeptides carrying Mec1/Tel1-dependent S/T-Q motifs. Indicated cells were released from α-factor arrest in drug-free SILAC media for 35 minutes. For all the QMAPS in Figure 2, each dot corresponds to a different phosphopeptide identified at least 3 times in 2 independent biological replicates. See also Table S2.
QMAPS reveals robust activation of Mec1 during normal DNA replication
It is currently accepted that activation of Mec1 is strongly induced by replication stress. This notion is mainly based on the fact that HU-induced replication fork stalling leads to a robust activation of Rad53 (Tercero et al., 2003). To test if our unbiased QMAPS approach could reveal new insights into the action of Mec1 or Tel1, we compared the phosphorylation level of Mec1/Tel1 candidate substrates in cells undergoing normal S-phase with cells treated with HU. In both cases, cells were arrested in G1 with α-factor and then released from the arrest in media containing HU or not for 45 minutes. As shown in Figure 2A and Table S2, nearly all phosphopeptides carrying a Rad53-dependent phosphorylation site were induced by HU. Unexpectedly, only a minor fraction of Mec1 and/or Tel1 candidate substrates was induced by HU treatment. This fraction included a phosphorylation site in Rad53 (serine 24) and a phosphorylation in the Mrc1 protein (serine 189), the adaptor known to transduce signals from Mec1 to Rad53 in response to HU. Most phosphorylation events in Mec1 and/or Tel1 targets were either only slightly induced by HU or did not change at all when comparing cells going through normal replication with cells experiencing HU-induced replication stress. Remarkably, a Mec1 autophosphorylation site (serine 38) and phosphorylation of Rfa1 and Rfa2 (serines 178 and 122, respectively), which are highly dependent on Mec1, were in fact inhibited by HU. Targeted analysis of purified Mec1 complexes further confirmed that the Mec1 autophosphorylation site and phosphorylation of Rfa1 are indeed induced during normal S-phase and accumulate as more DNA is replicated, following a similar trend observed for the acetylation of H3K56, which is a well-established replication mark (Figure 2B) (Masumoto et al., 2005). To test if Mec1 activation in normal S-phase is dependent on DNA replication, we used QMAPS to compare the phosphorylation levels of its targets in WT cells as well as in cells lacking the S-phase cyclins Clb5 and Clb6, which display delayed replication initiation due to delayed CDK activation, but undergo normal budding dynamics as they progress through S-phase (Figures 2C-D) (Donaldson et al., 1998). As shown in Figure 2D, several Mec1 candidate substrates are highly induced during S-phase in WT cells but are not induced in clb5Δclb6Δ cells at the 35 minute time point, when only limited DNA replication had occurred in the mutant (Figures 2C-D and Table S2). Taken together, these results show that Mec1 action in normal S-phase depends, at least partially, on DNA replication. While the MS analysis could detect many Mec1/Tel1-dependent phosphopeptides in G1 in clb5Δclb6Δ cells (Figure 2D), we attributed this basal phosphorylation level to the potential accumulation of these phospho-events in the extended and deregulated S-phase from the previous cell cycle. Very few Rad53-dependent phosphopeptides were detected in the absence of drug-induced replication stress (data not shown). Even in cells lacking three phosphatases known to act on Rad53, namely Ptc2, Ptc3 and Pph3 (Heideker et al., 2007), we were not able to detect robust Rad53 action during a normal S-phase as we still identified a very limited set of targets (Figure S2). Nonetheless, we were able to observe an increase in the level of phosphorylation of the detected Rad53 targets in ptc2Δptc3Δpph3Δ triple mutant cells compared to WT cells, suggesting that phosphatases play a role in counteracting Rad53 activation during normal DNA replication.
Collectively, the QMAPS results shown in Figure 2 reveal that Mec1 is robustly activated during normal DNA replication, and that this mode of Mec1 signaling is partially uncoupled from Rad53 activation. On the other hand, HU-induced replication stress leads to an increase in the phosphorylation of most Rad53 targets, but to minor changes in the phosphorylation of a large fraction of Mec1 targets, or even inhibition of some of them. We therefore propose that Mec1 can operate in two distinct modes of signaling during DNA replication, one correlated with ongoing DNA synthesis (“replication-correlated”) and another correlated with the extent of replication stress that involves strong Rad53 activation (canonical checkpoint response).
The 9-1-1 clamp and the lagging-strand factor Dna2 are important for “replication-correlated” Mec1 activation
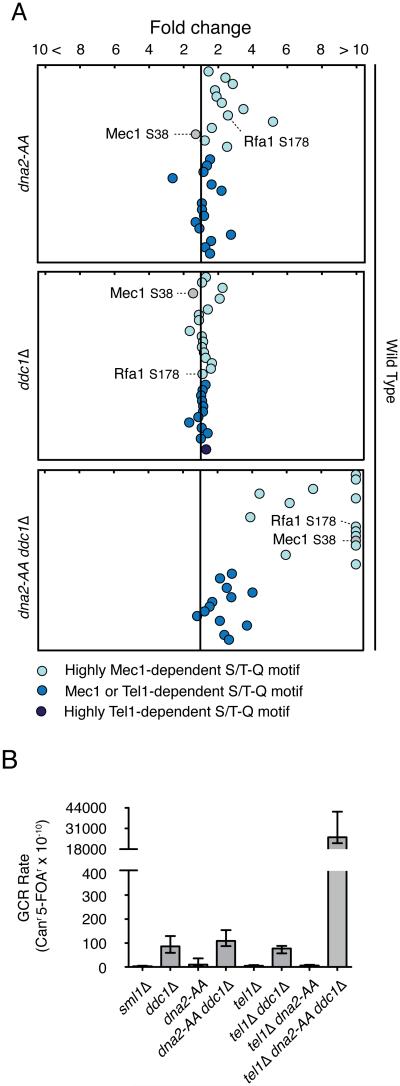
Recent work revealed that activation of the Mec1 kinase in response to replication stress or DNA damage requires the action of factors such as Ddc1, Dna2 and Dpb11, all of which possess an unstructured region that can tether Mec1 for activation (Kumar and Burgers, 2013; Navadgi-Patil and Burgers, 2009; Puddu et al., 2008). To test if the replication-correlated mode of Mec1 action also requires these factors for activation, we used QMAPS to compare phosphorylation of substrates in WT and mutants of Mec1-activating factors. As shown in Figure 3A and Table S3, mutation of two residues (W352A and Y544A) in Dna2 previously shown to be required for the ability of Dna2 to activate Mec1 has mild effects on the ability of Mec1 to target some of its specific targets, such as Rfa1, Spt7 and Dad1. Deletion of DDC1 had almost no effect in most targets (Figure 3A and Table S3) suggesting that Dna2 has a more prominent role in activating Mec1 during normal DNA replication. Importantly, deletion of DDC1 also prevents the recruitment of Dpb11 and its ability to activate Mec1 (Navadgi-Patil and Burgers, 2009). Finally, combination of the dna2 WY-AA mutation (herein referred as dna2-AA) with DDC1 deletion had a significant impact on the phosphorylation levels of targets that highly depend on Mec1, suggesting that Dna2 and Ddc1 function redundantly to activate Mec1 during normal DNA replication (Figure 3A and Table S3). This is consistent with the fact that these proteins are known to localize and function on the lagging strand of the replication fork. These results suggest that Mec1 may be activated mostly at the lagging strand of a moving replication fork during normal DNA replication.
Figure 3. Importance of Dna2 and Ddc1 for replication-correlated mode of Mec1 activation.
(A) QMAPS showing the relative abundance of phosphopeptides carrying Mec1/Tel1-dependent S/T Q motifs. WT, dna2-AA, ddc1Δ and dna2-AA ddc1Δ cells were released from α-factor arrest in SILAC media for 45 minutes. See also Table S3. (B) Effects of the dna2-AA mutation on accumulation of gross-chromosomal rearrangements in Ddc1 and Tel1 defective mutants. All strains are sml1Δ. Error bars indicate 95% confidence intervals (CI).
Tel1 phosphorylates a specific group of Mec1 targets to prevent GCR and support robust DNA replication in the absence of Mec1
Analysis of GCRs revealed that activation of Mec1 via Dna2 or Ddc1 during replication becomes particularly important in the absence of Tel1, as shown by the dramatic increase in GCR in tel1Δddc1Δdna2-AA cells (Figure 3B). This result highlights the key role of Tel1 in compensating for the loss of Mec1 during normal DNA replication. Consistent with this data, while ddc1Δdna2-AA cells exhibit a major loss of phosphorylation of most Mec1-specific phosphorylation, we could still observe robust phosphorylation of targets common to Mec1 and Tel1 during an unchallenged S-phase (Figure 3A). We interpret this result as Tel1 acting in the absence of Mec1 activation during a normal S-phase. Similar to Mec1’s “replication-correlated” mode, the action of Tel1 during normal S-phase (and in the absence of Mec1 activation) does not result in higher phosphorylation of Rad53 targets (Figure S3). Of importance, while ddc1Δdna2-AA cells can still replicate DNA and progress through S-phase at WT rates (data not shown), ddc1Δdna2-AA cells lacking TEL1 display severe replication defects (Kumar and Burgers, 2013). These results suggest that phosphorylation events in one, or several, common Mec1 and Tel1 targets play an important role in promoting robust DNA replication and preventing the accumulation of GCRs. As shown in Table S3 most of these targets are proteins involved in transcription, RNA processing and chromatin regulation, and several of them are either essential or required for efficient S-phase progression. These results reveal that Tel1 also plays a role during replication-correlated signaling, in a manner that is uncoupled from Rad53 activation. But differently from Mec1, Tel1 does not rely on Ddc1 and Dna2 for activation during replication, so it remains unclear how Tel1 engages and becomes active at sites of ongoing DNA replication. Taken together, this analysis uncovers a subset of Mec1 targets (common to Tel1) whose phosphorylations are correlated with the ability of cells to suppress GCRs and maintain robust DNA replication.
Dna2 and Ddc1 are not essential for activation of the canonical Mec1-Rad53 signaling response following replication stress
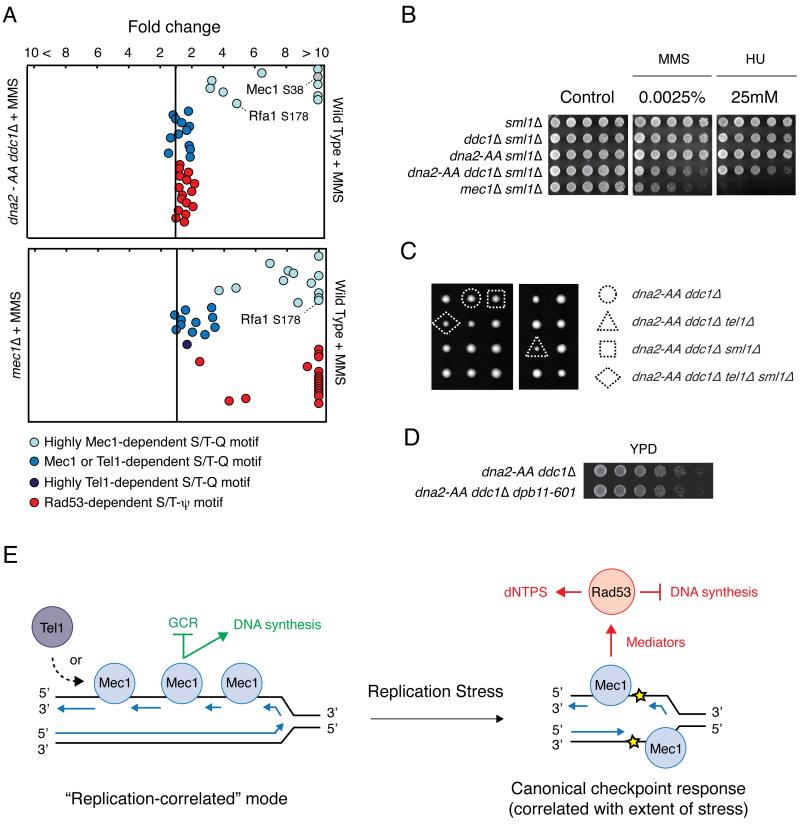
To determine the extent in which Dna2 and Ddc1 are necessary for activation of the canonical Mec1-Rad53 response during replication stress we performed QMAPS analysis comparing WT cells versus ddc1Δdna2-AA cells treated with MMS, which leads to robust Rad53 activation. As shown in Figure 4A (Table S4), ddc1Δdna2-AA cells exhibit strong defects in Mec1 activation during MMS treatment, but unexpectedly, activation of Rad53 under this condition does not seem to be greatly affected. On the other hand, similar QMAPS analysis comparing WT and mec1Δ cells revealed a strong impact in the phosphorylation of Rad53 targets in the absence of Mec1. These results show that ddc1Δdna2-AA cells do not phenocopy mec1Δ cells regarding Rad53 activation and suggest the existence of additional factors that may activate Mec1 to specifically activate Rad53, consistent with a recent paper (Bandhu et al., 2014). In support of the idea of additional Mec1 activator(s), ddc1Δdna2-AA cells are not as sensitive to MMS or HU as mec1Δ cells (Figure 4B). Also, while mec1Δ and rad53Δ cells are well known to require deletion of the ribonucleotide reductase inhibitor SML1 for viability (Zhao et al., 1998), we found that ddc1Δdna2-AA cells do not require SML1 deletion for viability (Figure 4C). Of note, even tel1Δddc1Δdna2-AA cells do not require SML1 deletion for viability despite these cells showing the dramatic increase in GCR rates that is characteristic of mec1Δtel1Δ cells. We could exclude the possibility of a Ddc1-independent role for Dpb11 in the activation of Mec1 under MMS in ddc1Δdna2-AA cells as removal of the C-terminal region of Dpb11, which is required for its ability to activate Mec1 (Navadgi-Patil and Burgers, 2008), did not cause loss of viability or major growth defect (Figure 4D). As shown in the working model in Figure 4E, we propose two distinct modes of Mec1 action during DNA replication, one correlated with DNA replication and another correlated with the extent of replication stress as part of a canonical checkpoint signaling. In our model, the replication-correlated mode of Mec1 action functions redundantly with Tel1 to ensure robust DNA replication and prevent GCR. On the other hand, the canonical checkpoint mode leads to the well-established effects of inhibition of DNA replication and increased production of dNTPs.
Figure 4. Dna2 and Ddc1 are not essential for Mec1 activation during replication stress.
A) QMAPS analysis comparing WT and indicated mutant cells. Cells were arrested with α-factor and released from arrest in SILAC media containing 0.04% MMS for 45 minutes. See also Table S4. (B) 5-fold serial dilutions of indicated cells with sml1Δ background were plated on YPD plates containing indicated drugs and incubated at 30°C for 48h. (C) Meiotic tetrads from a DNA2/dna-AA DDC1/ddc1Δ TEL1/tel1Δ SML1/sml1Δ diploid strain were dissected on YPD plates and incubated at 30°C for 72h. (D) 4-fold serial dilutions of indicated cells were plated on YPD plates and incubated at 30°C for 36h. (E) Model depicting distinct modes of Mec1 action during DNA replication. See text in the discussion. Blue arrows correspond to newly synthesized DNA strands.
DISCUSSION
The ATR and ATM kinases, and their yeast orthologs, regulate hundreds of substrates, but our ability to fully capture their multi-functional action in vivo has been hampered by the common use of one or a few classical substrates as readouts of their activity. Here we used a quantitative MS approach to monitor in vivo Mec1/Tel1 kinase action in a systematic, unbiased and quantitative manner. Our analysis revealed surprising insights into how Mec1 functions during DNA replication and provided evidence of a non-canonical mode of Mec1 action, which we propose is distinct from Mec1’s established role in the checkpoint response (see model in Figure 4E).
By quantitatively monitoring the phospho-status of dozens of Mec1 candidate substrates, we found that Mec1 is highly active during normal DNA replication. In fact, genetic data support the idea that Mec1 functions during normal DNA replication. For example, cells lacking MEC1 and TEL1 exhibit high rates of GCR in an assay performed in the absence of any exogenously-induced DNA damage (Myung et al., 2001). But the prevailing hypothesis has been that the ability of Mec1 to suppress spontaneous GCR accumulation is attributed to a residual action of Mec1 in response to spontaneous DNA damage generated during DNA replication. Distinct from the notion of residual Mec1 activation during normal replication, our work supports a model in which Mec1 is highly engaged onto sites of ongoing DNA synthesis to become activated in a “replication-correlated” manner. Also, distinct from the established role of Mec1 in checkpoint signaling, our results reveal that the action of Mec1 during normal DNA replication is partially uncoupled from the action of the downstream kinase Rad53. Our results are consistent with the idea that Mec1 is either continuously activated during ongoing DNA synthesis or is activated at many sites in the genome that pose moderate level of difficulty for replication forks to pass. At these sites, forks would only dynamically pause, allowing sufficient ssDNA exposure for Mec1 recruitment and activation but not for robust Rad53 activation, which requires further recruitment and/or phosphorylation of mediator proteins to mount a full checkpoint response. Nonetheless, it is important to mention that Rad53 also needs to be activated during normal DNA replication. Cells lacking Rad53 are not viable, unless the RNR inhibitor SML1 is also deleted (Zhao et al., 1998). But contrary to Mec1’s action, our quantitative analysis reveals that the activity of Rad53 in normal DNA replication is significantly lower than drug-induced Rad53 activity (Figure 2A). We speculate that during normal DNA replication Rad53 becomes preferentially activated at specific genomic sites that pose major challenges for replication, such as hard-to-replicate transcriptional barriers. Interestingly, our results suggest that phosphatases such as Pph3, Ptc2 and Ptc3 may also function during normal S-phase to prevent excess Rad53 activation, consistent with a recent report showing a constitutive Mec1-Pph3 interaction (Hustedt et al., 2014).
The identification of a replication-correlated mode of Mec1 action leads to a paradox, as Rad53 has established roles in inhibiting DNA synthesis as part of a canonical checkpoint response to replication stress (Santocanale and Diffley, 1998). We hypothesize that Mec1 positively regulates DNA replication when functioning uncoupled from Rad53 activation in the replication-correlated mode (Figure 4E). Consistent with this hypothesis, the Bell lab has shown that Mec1 phosphorylates the MCM complex to prime it for activation (Randell et al., 2010). We further speculate that the replication-correlated mode of Mec1 signaling plays a major role in facilitating the movement of replication forks by preemptively opening chromatin and/or removing RNA and transcriptional machineries from DNA. Consistent with this notion, we found that Mec1 targets several proteins involved in transcription, RNA processing and chromatin remodeling during unchallenged DNA replication. Also, we showed that during normal DNA replication Tel1 partially compensates for the lack of Mec1 by targeting substrates involved in transcription and chromatin regulation. The fact that cells lacking both Mec1 and Tel1 are extremely slow growing, further strengthens the idea that the set of Mec1 substrates that can also be phosphorylated by Tel1 comprise a critical set of proteins involved in promoting robust DNA replication. Previous reports have functionally connected Mec1 to chromatin and transcription regulation (Rodriguez and Tsukiyama, 2013; Seeber et al., 2013). Our work suggests that regulation of these processes by Mec1 is actually part of the normal replication program that positively controls ongoing DNA synthesis. The delineation of which substrates are common to Mec1 and Tel1 should provide the framework of targets that will help better understand the mechanisms by which Mec1 and Tel1 positively impact DNA synthesis. Finally, the observation that replication-correlated mode of Mec1 and Tel1 action does not efficiently relay signaling to Rad53 activation is consistent with these ideas, as it is well known that Rad53 activation leads to outputs that would antagonize the potential role of Mec1 as a positive regulator of DNA replication.
EXPERIMENTAL PROCEDURES
Cell Culture
Yeast strains used in this study are listed in Table S5. For stable isotope labeling of amino acids in cell culture (SILAC) auxotrophic yeast strains for lysine and arginine were grown in -Arg -Lys synthetic dropout media supplemented with either normal l-arginine and l-lysine (light culture) or l-lysine 13C6, 15N2 and l-arginine 13C6, 15N4 (heavy culture) as described in (Ohouo et al., 2013).
Phosphopeptide Enrichment
Phosphopeptide enrichment was performed as described in (Ohouo et al., 2013). See supplemental experimental procedures for further details.
Mass Spectrometry Analysis
Phosphopeptides were subjected to LC-MS/MS analysis using a Q-Exactive Orbitrap or an Orbitrap XL mass spectrometer. See supplemental experimental procedures for further details.
Supplementary Material
ACKNOWLEDGMENTS
We thank Beatriz S. Almeida for technical support. This work is supported by M.B.S. grants from the National Institute of Health (R01-GM097272), American Cancer Society (RSG-11-146-01-DMC), K.H.S. grant from National Institute of Health (R01-GM081425), H.Y. grants from the National Institute of Health (R01-GM104424 and R01-GM097358) and Cornell Fleming Research Fellowship to F.M.B.d.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The mass spectrometry proteomics data have been deposited to the Peptide Atlas database (http://www.peptideatlas.org/) with the dataset identifier PASS00651 and PASS00652.
SUPPLEMENTAL INFORMATION
Supplemental information contains Supplemental Experimental Procedures, 3 figures and 5 tables and can be found within this article online at
REFERENCES
- Bandhu A, Kang J, Fukunaga K, Goto G, Sugimoto K. Ddc2 mediates Mec1 activation through a Ddc1- or Dpb11-independent mechanism. PLoS genetics. 2014;10 doi: 10.1371/journal.pgen.1004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Heideker J, Lis ET, Romesberg FE. Phosphatases, DNA damage checkpoints and checkpoint deactivation. Cell Cycle. 2007;6:3058–3064. doi: 10.4161/cc.6.24.5100. [DOI] [PubMed] [Google Scholar]
- Hustedt N, Seeber A, Sack R, Tsai-Pflugfelder M, Bhullar B, Vlaming H, van Leeuwen F, Guenole A, van Attikum H, Srivas R, et al. Yeast PP4 Interacts with ATR Homolog Ddc2-Mec1 and Regulates Checkpoint Signaling. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Kumar S, Burgers PM. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–321. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Carr AM. Replication stress and genome rearrangements: lessons from yeast models. Curr Opin Genet Dev. 2013;23:132–139. doi: 10.1016/j.gde.2012.11.009. [DOI] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Meyn MS. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Nair KR. Table of confidence intervals for the median in samples from any continuous population. Sankhya. 1940;4:551–558. [Google Scholar]
- Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008 doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohouo PY, Bastos de Oliveira FM, Liu Y, Ma CJ, Smolka MB. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature. 2013;493:120–124. doi: 10.1038/nature11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Tsukiyama T. ATR-like kinase Mec1 facilitates both chromatin accessibility at DNA replication forks and replication fork progression during replication stress. Genes Dev. 2013;27:74–86. doi: 10.1101/gad.202978.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Seeber A, Dion V, Gasser SM. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev. 2013;27:1999–2008. doi: 10.1101/gad.222992.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci U S A. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.