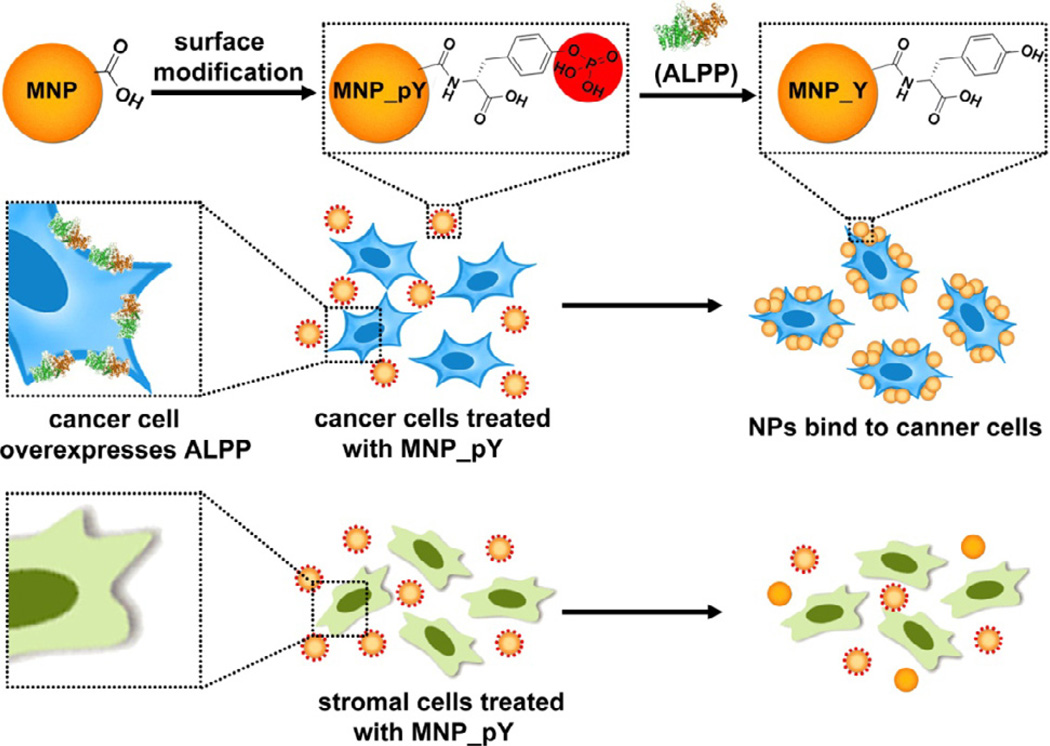
Abstract
Enzymatic switch, such as phosphorylation and dephosphorylation of proteins, is the most important mechanism for cellular signal transductions. Inspired by Nature and encouraged by our recent unexpected observation of the dephosphorylation of D-tyrosine phosphate-contain small peptides, we modify the surface of magnetic nanoparticles (MNP) with D-tyrosine phosphate that is a substrate of alkaline phosphatase (ALP). Our studies find that ALP is able to remove the phosphate groups from the magnetic nanoparticles. Most importantly, placental alkaline phosphatase (ALPP), an ectoenzyme that locates on cell surface with catalytic domains outside the plasma membrane and is overexpressed on many cancer cells, dephosphorylate the D-tyrosine phosphates on the surface of the magnetic nanoparticle and enable the magnetic nanoparticles to adhere selectively to the cancer cells, such as HeLa cells. Unlikely commonly used antibodies, the selectivity of the magnetic nanoparticles to cancer cells originates from the enzymatic reaction catalyzed by ALPP. The use of enzymatic reaction to modulate the surface of various nanostructures may lead to a general method to broadly target cancer cells without relying on specific ligand-receptor interactions (e.g., antibodies). This work, thus, illustrates a fundamentally new concept to allow cells to actively engineer the surface of colloids materials, such as magnetic nanoparticles, for various applications.
Keywords: enzyme, surface engineering, magnetic, nanoparticles, cancer cells
Introduction
The intersection of nanotechnology and molecular cell biology, over the last decade, has developed into an emerging research area: nanobiotechnology. As a versatile functional colloid material, magnetic nanoparticles offer controlled size, ability to be manipulated externally, and enhancement of contrast in magnetic resonance imaging (MRI).[1–13] Consequently, magnetic nanoparticles have already been explored for many applications in biology and medicine,[14–24] including protein purification,[25, 26] bacteria capture and inhibition,[27, 28] drug delivery,[29, 30] band gap materials,[31, 32] and medical imaging.[33–35] In order to make magnetic nanoparticles to be biofunctional, one of most common strategy is to attach antibodies to magnetic nanoparticles to target cell specifically.[36, 37] Despite its effectiveness, this approach relies on the tight ligand-receptor binding, which becomes ineffective when the receptors on the cells mutate and no longer bind to the ligand on the nanoparticles. This generic drawback of ligand-receptor interactions highlights the need of a new approach to use nanoparticles to target the cell without relying on tight and specific ligand-receptor binding. Recently, we unexpectedly observed that alkaline phosphatase (ALP) catalytically removes the phosphate group from the D-tyrosine phosphate residue of a D-peptide at (almost) the same rate as that from the corresponding L-peptide.[38] The insensitivity of ALP to stereochemistry of its substrates lead to another surprise that placental alkaline phosphatase (ALPP), an ectoenzyme that locates on cell surface with catalytic domains outside the plasma membrane, overexpressed on HeLa cells results in the formation a hydrogel/nanonet on the cancer cell surface, thus selectively inhibit the cancer cells.[39, 40] These results indicate that ALPP on cancer cells may dephosphorylate D-peptides on magnetic nanoparticles, thus allowing the nanoparticles to interact with cancer cells selectively.
Based on the above rationale, we simply decorate magnetic nanoparticles with D-tyrosine phosphates to be a new type of substrate (MNP_pY) of phosphatases. We find that ALP is able to dephosphorylate the D-tyrosine phosphates on the magnetic nanoparticles. Moreover, ALPP overexpressed on the surface of cancer cells[41, 42] catalytically dephosphorylate the phosphate-bearing magnetic nanoparticles to form tyrosine coated magnetic nanoparticles (MNP_Y) that adhere to the cancer cells. On the contrary, HS-5 cells, a stromal cell that has low level expression of ALPP,[43] fail to trigger the adherence of the magnetic nanoparticles to HS-5 cells. Moreover, MNP_Y adheres to neither HeLa cells nor HS-5 cells, suggesting that the enzymatic switch of the surface of the magnetic nanoparticles is responsible for the selective binding of the magnetic nanoparticles to the cancer cells. This result represents a fundamentally new approach for selectively targeting cancer cells because it relies on enzymatic reaction near the cell surface rather than on a specific ligand-receptor interaction. The use of the ectoenzyme on cells to actively engineer the surface of nanoparticles, thus, provides a new opportunity to design the applications of nanoparticles, based on the spatiotemporal distribution of a specific enzyme, for disease diagnosis and treatment.
Experimental Methods
Materials and Instruments
Iron oxide nanoparticles with amphiphilic polymer coating and a monolayer of oleic acid were purchased from the Ocean NanoTech company. Transmission electron microscope images were collected by using Morgagni 268 microscope. Confocal images were taken with the Leica TCS SP2 Spectral Confocal Microscope. The cells were counted by a hemacytometer.
Phosphate assay
We quantify the amount of phosphate on MNP_pY with the purchased phosphate assay kit (colorimetric) (ab65622, abcam). First, we made the 0, 1, 2, 3, 4, 5 nmol of phosphate standard solutions, mixed 200 μl these standard solutions and 30 μl phosphate reagent for 30 min, and read the absorbance at 620 nm using a plate reader. Similarly, when quantifying the amount of phosphate on MNP_pY, we followed the same protocol. After incubating 40 μg MNP_pY with 100 μl dH2O or 100 μl dH2O containing 30 U alkaline phosphatase (ALP) for 24 hrs, we centrifugalized the nanoparticles and the solutions. We took 20 μl treated solution out and adjusted the volume to 200 μl with dH2O. With the treatment with 30 μl phosphate reagent for 30 min at room temperature, we read the absorbance by using a plate reader.
Cell culture
The HeLa-GFP and HS-5 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Both of the cells were incubated in a fully humidified incubator containing 5% CO2 at 37°C with the growth medium, Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen Life Technologies, 10829-018) supplemented with 10% fetal bovine serum (FBS, Invitrogen Life Technologies, 10082-147), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Invitrogen Life Technologies, 15070-063).
Confocal microscopy
Initially, 1.0×106 cells were seeded in 6 cm cell culture dish with 5 mL growth medium. The cells were allowed for 12 h to adhere on culture dish. New medium containing MNP_pY, MNP_Y, or MNP at the concentration of 40 μg/mL was added into each dish after the old medium was removed. With 4 h of incubation, cells were washed with growth medium for 3 times and detached with 0.25% (w/v) Trypsin- 0.53 mM EDTA solution. As described in the main text, after harvesting all the cells, we separated extraction from supernatant by using one small magnet, rinsed the two cells for three times with growth medium, and then seeded them back onto the confocal dishes. 4 h later, the cells were rinsed for three times in PBS, and then kept in the PBS buffer for imaging.
Results and discuss
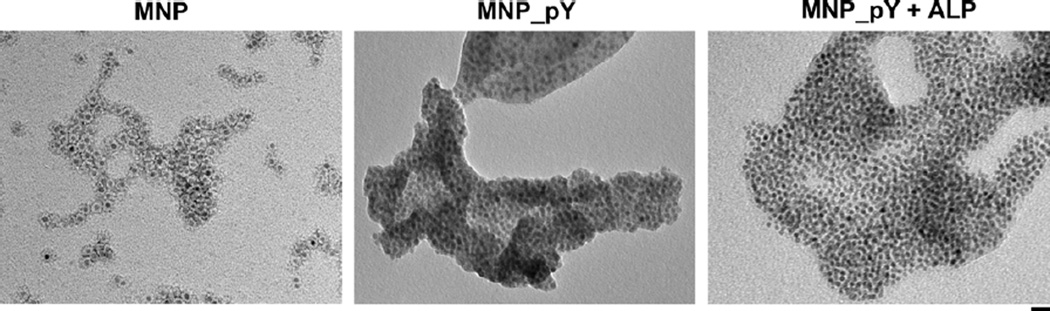
The synthesis of MNP_pY is fast and straightforward. Starting from the well-established iron oxide nanoparticles (MNP) that are surface-functionalized with carboxylic acid groups (and are commercially available), we use N-hydroxysuccinimide (NHS) to activate the carboxylic acid groups on the nanoparticles for directly coupling them with D-tyrosine phosphate. Following three times rinsing by methanol and water, respectively, centrifugation separates the final MNP_pY dispersed in water for use. Transmission electron microscopy (Figure 1) confirms that, except an increased clustering on the TEM grids, there is little morphological change of the iron oxide nanoparticles before and after functionalization by D-tyrosine phosphates (Figure 1). The nanoparticles MNP, coated with a layer of amphiphilic polymer and a layer of oleic acid, already cluster to form small aggregates that consist of the particles with a diameter of around 10 nm. Similarly, both MNP_pY and MNP_Y (which results from the treatment of MNP_pY with ALP), also form aggregates of nanoparticles that still have well-defined iron oxide cores and amphiphilic coatings.
Figure 1.
Transmission electron microscope (TEM) images of iron oxide nanoparticles MNP (Left), MNP_pY (Middle), and MNP_pY + ALP (Right). The nanoparticles are dissolved in water at the concentration of 2,000 μg/mL (pH = 7.4). The scale bar is 20 nm.
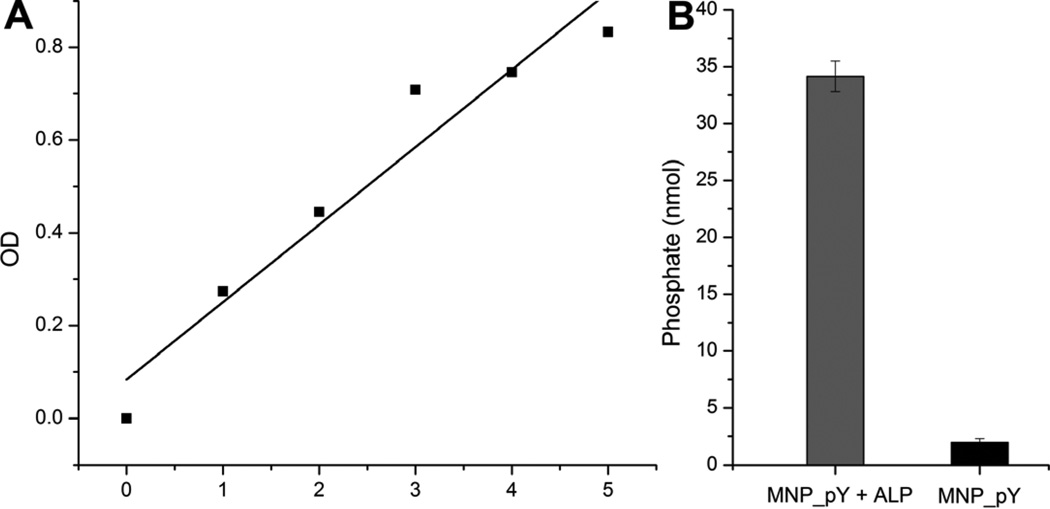
To determine the number of D-tyrosine phosphate molecules decorated on the surface of the MNP, we utilized the phosphate assay to quantify the amount of phosphate on MNP_pY (Figure 2). According to the phosphate standard curve (Figure 2A), we can conclude that there are at least 34 nmol D-tyrosine phosphate molecules on the surface of 40 μg MNP_pY (Figure 2B), considering that the treatment of MNP_pY with ALP for 24 hours unlikely remove all the phosphates from the nanoparticles. According to the manufacture data of the commercial iron oxide nanoparticles, there are 6.9 nmol particles of 1 mg iron oxide, which means 40 μg MNP_pY contains 0.28 nmol nanoparticles. Thus, this quantification of phosphate on MNP_pY indicates that, on average, there are at least 124 D-tyrosine phosphate molecules on each MNP_pY nanoparticles.
Figure 2.
(A) Phosphate standard curve performed according to the phosphate assay (ab65622, abcam). (B) Average amount of the phosphates on 40 μg MNP_pY. Gray bar indicates amount of phosphates in MNP_pY treated with ALP for 24 hrs; and black bar indicates amount of phosphates from the MNP_pY treated with di H2O for 24 hrs.
After determine the numbers of the D-tyrosine phosphates on the surface of the MNP, we incubated the nanoparticles and the relevant controls to prove the selective interaction between the nanoparticles and cancer cells. The procedure for testing the selective binding of these nanoparticles to cells is exceptionally simple. After seeding about 1.0×106 HeLa-GFP or HS-5 cells per culture dish (6 cm) overnight,[44] we add MNP_pY, MNP_Y, or MNP (40 μg/mL) to incubate the cells for 4 hours. After removing the growth medium containing nanoparticles and rinsing the cells for three times, we use trypsin solution (0.25% (w/v) in 0.53 mM EDTA) to detach the cells. Following aspirating the cells to obtain the cell suspension by gently pipetting, we place a small magnet outside the Eppendorf tube for 1 min to divide the cell suspension into two portions: supernatant and extraction. After the centrifugation and rinse of the supernatant or extraction, the pellets of cells are reseeded onto confocal petri dishes for imaging which acts as a way to verify the results of the interactions between nanoparticles and cells.
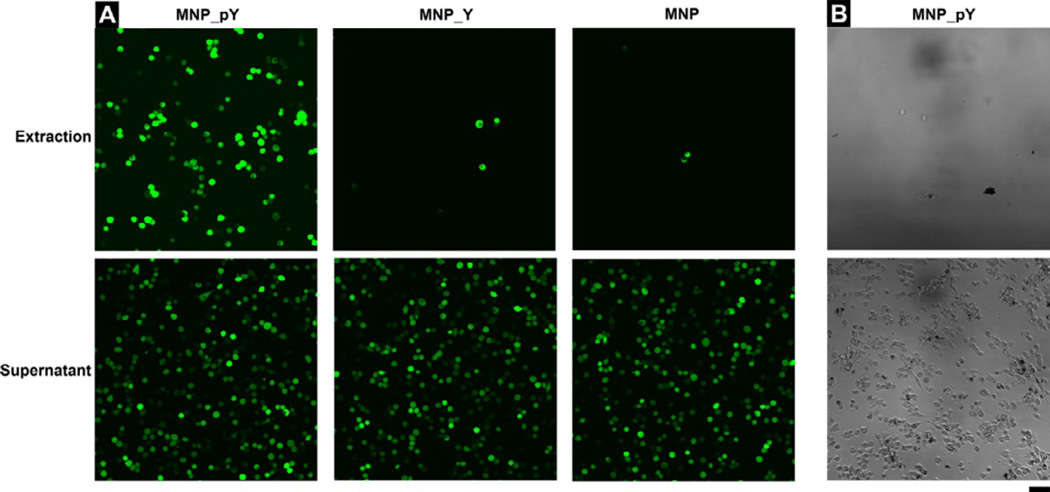
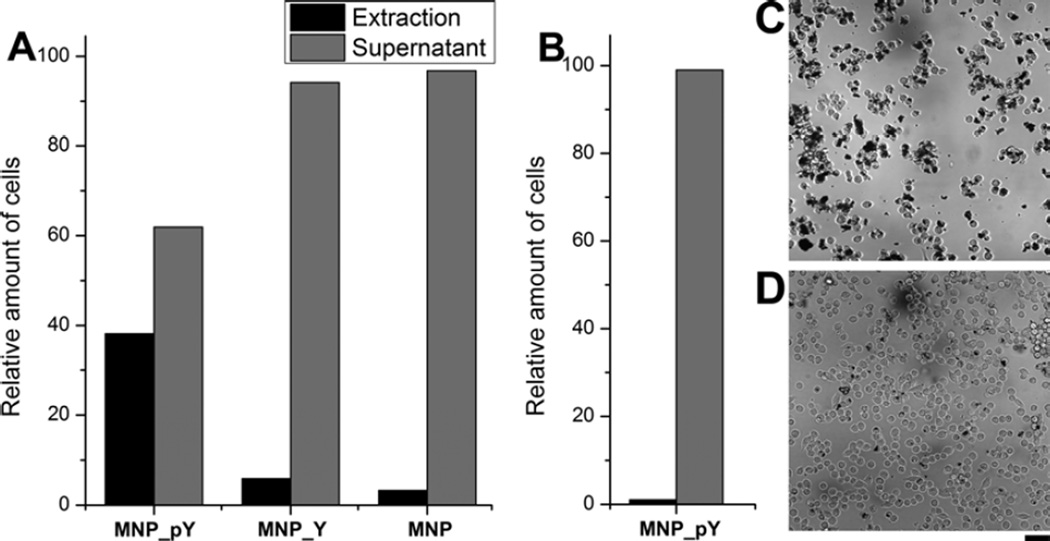
Figure 3 and Figure 4 show the results of the binding of HeLa-GFP cells (Figure 3A and 4A) and HS-5 cells (Figure 3B and 4B) treated with MNP-pY, MNP_Y, or MNP. As shown in Figure 3A, after the treatment by MNP_pY and the magnetic separation, around 40% HeLa-GFP cells (Figure 4A) are extracted from all the cells with bright green fluorescence. Meanwhile, the bright field images also confirm that many magnetic nanoparticles (MNP_Y) adhere on the surface of the cancer cells extracted by the magnet (Figure 4C), which likely results from the dephosphorylation of D-tyrosine phosphates on the iron oxide nanoparticles by the overexpressed ALPP on the surface of cancer cells. To confirm that enzymatic dephosphrylation is responsible for the adhesion of the magnetic nanoparticles to the HeLa cells, we use MNP_Y as a control and repeat the same experiment as that of MNP_pY. After the treatment by MNP_Y and magnetic separation, almost no cell (less than 4%, Figure 4A) is observed from the extraction portion after reseeding, but the corresponding supernatant (i.e., from the sample treated by MNP_Y) contains (almost) all the fluorescent (HeLa-GFP) cells (Figure 3A). Agreeing with this observation, after the incubation of the cells with the control nanoparticles (MNP_Y), the bright field images reveal that the nanoparticles hardly adhere on the surface of cancer cells (Figure 4D). Another control iron oxide nanoparticle (MNP) shows the similar results to those of MNP_Y when being treated with HeLa-GFP cells (Figure 3A). Moreover, the incubation of MNP_pY with HS-5 cells hardly results in HS-5 cells in the extraction portion, and there are almost no nanoparticles on the HS-5 cells in the supernatant (Figure 3B). These results, together, confirm that MNP_pYs, being catalytic dephosphorylated by the ectophosphatases overexpressed on the HeLa cells, selectively adher to HeLa cells.
Figure 3.
(A) The fluorescent images (×20 dry objective lens) of the extraction and supernatant portions of cells after adding MNP_pY (Left), MNP_Y (Middle), and MNP (Right) to HeLa-GFP cells. (B) The bright field images (×20 dry objective lens) of the extraction and supernatant portions of cells after adding MNP_pY to HS-5 cells. Cells were incubated with the growth medium, Dulbecco’s Modified Eagle Medium (DMEM), containing 40 μg/mL nanoparticles for 4 hrs (top: the cells extracted by magnet; bottom: the cells remained in supernatant). The initial number of cells is 1.0×106 per 6 cm culture dish. The scale bar is 100 μm.
Figure 4.
(A) Relative amount of cells (%) in the extraction or supernatant of all the HeLa-GFP cells collected after the treatment by 40 μg/mL MNP_pY, MNP_Y, or MNP. (B) Relative amount of cells (%) in the extraction or supernatant of all the HS-5 cells collected after the treatment by 40 μg/mL MNP_pY. The bright field microscope images (×20 dry objective lens) of HeLa-GFP cells after treated with (C) MNP_pY and (D) MNP_Y show the interaction between cells and nanoparticles. Cells were incubated with the growth medium, Dulbecco’s Modified Eagle Medium, containing 40 μg/mL nanoparticles for 4 hrs. The initial number of cells is about 1.0×106 per 6 cm culture dish. The scale bar is 100 μm.
Conclusion
In conclusion, this work, for the first time, demonstrates the use of enzymatic reaction from ectoenzymes (i.e., ALPP) to actively modulate the surface composition of magnetic nanoparticles for selectively binding to cancer cells without involving specific ligand-receptor interactions or the use of antibodies. The application of isoenzymes, such as ALPP, for selective enzymatic transformation and self-assembly,[39, 45] promises a fundamentally new way to control cell-materials interactions. Although the cells usually internalize nanoparticles, the decorating of the MNP by D-tyrosine phosphates or D-tyrosine promote the nanoparticles to reside on the surface of the cells, similar to the case of the D-peptide nanofibrils.[39] While the detailed mechanism of this intriguing observation warrants further investigation, this strategy may find application for sorting or inhibiting cancer cells in a mixture of cells or co-culture, which is the on-going study of this research.
Scheme 1.
Ectoenzyme transforms magnetic nanoparticles for selectively binding with cancer cells.
Acknowledgement
This work was supported by National Institutes of Health (NIH R01CA142746). JZ is a HHMI international research fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Gu HW, Ho PL, Tsang KWT, Wang L, Xu B. J. Am. Chem. Soc. 2003;125:15702. doi: 10.1021/ja0359310. [DOI] [PubMed] [Google Scholar]
- 2.Zhai Y, Dou Y, Liu X, Park SS, Ha C-S, Zhao D. Carbon. 2011;49:545. [Google Scholar]
- 3.Zhang F, Braun GB, Pallaoro A, Zhang Y, Shi Y, Cui D, Moskovits M, Zhao D, Stucky GD. Nano Lett. 2012;12:61. doi: 10.1021/nl202949y. [DOI] [PubMed] [Google Scholar]
- 4.Kircher MF, Mahmood U, King RS, Weissleder R, Josephson L. Cancer Res. 2003;63:8122. [PubMed] [Google Scholar]
- 5.Rabin O, Perez JM, Grimm J, Wojtkiewicz G, Weissleder R. Nat. Mater. 2006;5:118. doi: 10.1038/nmat1571. [DOI] [PubMed] [Google Scholar]
- 6.Pan Y, Du X, Zhao F, Xu B. Chem. Soc. Rev. 2012;41:2912. doi: 10.1039/c2cs15315g. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, Liang G, Cheung JS, Pan Y, Kuang Y, Zhao F, Zhang B, Zhang X, Wu EX, Xu B. J. Am. Chem. Soc. 2008;130:11828. doi: 10.1021/ja803920b. [DOI] [PubMed] [Google Scholar]
- 8.Long MJC, Pan Y, Lin H-C, Hedstrom L, Xu B. J. Am. Chem. Soc. 2011;133:10006. doi: 10.1021/ja202767g. [DOI] [PubMed] [Google Scholar]
- 9.Gao JH, Zhang B, Zhang XX, Xu B. Angew. Chem. Int. Edit. 2006;45:1220. doi: 10.1002/anie.200503486. [DOI] [PubMed] [Google Scholar]
- 10.Chi H, Liu B, Guan G, Zhang Z, Han M-Y. Analyst. 2010;135:1070. doi: 10.1039/c000285b. [DOI] [PubMed] [Google Scholar]
- 11.Zhang ZP, Han MY. J. Mater. Chem. 2003;13:641. [Google Scholar]
- 12.Ge J, Hu Y, Biasini M, Beyermann WP, Yin Y. Angew. Chem. Int. Edit. 2007;46:4342. doi: 10.1002/anie.200700197. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Delikanli S, Zeng H, Ferkey DM, Pralle A. Nat. Nanotechnol. 2010;5:602. doi: 10.1038/nnano.2010.125. [DOI] [PubMed] [Google Scholar]
- 14.Hajipour MJ, Fromm KM, Ashkarran AA, Jimenez de Aberasturi D, Ruiz de Larramendi I, Rojo T, Serpooshan V, Parak WJ, Mahmoudi M. Trends Biotechnol. 2012;30:499. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Pellegrino T, Kudera S, Liedl T, Javier AM, Manna L, Parak WJ. Small. 2005;1:48. doi: 10.1002/smll.200400071. [DOI] [PubMed] [Google Scholar]
- 16.Hoppe CE, Rivadulla F, Arturo Lopez-Quintela M, Carmen Bujan M, Rivas J, Serantes D, Baldomir D. J. Phys. Chem. C. 2008;112:13099. [Google Scholar]
- 17.Vazquez-Vazquez C, Lopez-Quintela MA, Bujan-Nunez MC, Rivas J. J. Nanopart. Res. 2011;13:1663. [Google Scholar]
- 18.Farrell D, Cheng Y, McCallum RW, Sachan M, Majetich SA. J. Phys. Chem. B. 2005;109:13409. doi: 10.1021/jp050161v. [DOI] [PubMed] [Google Scholar]
- 19.Majetich SA, Sachan M. J. Phys. D. Appl. Phys. 2006;39:R407. [Google Scholar]
- 20.Murray CB, Kagan CR, Bawendi MG. Annu. Rev. Mater. Sci. 2000;30:545. [Google Scholar]
- 21.Wong C, Stylianopoulos T, Cui J, Martin J, Chauhan VP, Jiang W, Popovic Z, Jain RK, Bawendi MG, Fukumura D. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2426. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun SH, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX, Li GX. J. Am. Chem. Soc. 2004;126:273. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 23.Gao J, Gu H, Xu B. Acc. Chem. Res. 2009;42:1097. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]
- 24.Zheng RK, Gu HW, Xu B, Zhang XX. Phys. Rev. B. 2005;72 [Google Scholar]
- 25.Pan Y, Long MJC, Lin H-C, Hedstroma L, Xu B. Chem. Sci. 2012;3:3495. [Google Scholar]
- 26.Pan Y, Long MJC, Li X, Shi J, Hedstrom L, Xu B. Chem. Sci. 2011;2:945. [Google Scholar]
- 27.Hergt R, Dutz S, Mueller R, Zeisberger M. J. Phys-condens. Mat. 2006;18:S2919. [Google Scholar]
- 28.Shevchenko EV, Talapin DV, Kotov NA, O'Brien S, Murray CB. Nature. 2006;439:55. doi: 10.1038/nature04414. [DOI] [PubMed] [Google Scholar]
- 29.Chen D, Jiang M, Li N, Gu H, Xu Q, Ge J, Xia X, Lu J. J. Mater. Chem. 2010;20:6422. [Google Scholar]
- 30.Lee J-H, Kim J-w, Cheon J. Mol. Cells. 2013;35:274. doi: 10.1007/s10059-013-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu GL, Yin Y, Kunchakarra S, Mukherjee B, Gerion D, Jett SD, Bear DG, Gray JW, Alivisatos AP, Lee LP, Chen FF. Nat. Nanotechnol. 2006;1:47. doi: 10.1038/nnano.2006.51. [DOI] [PubMed] [Google Scholar]
- 32.Lu Z, Yin Y. Chem. Soc. Rev. 2012;41:6874. doi: 10.1039/c2cs35197h. [DOI] [PubMed] [Google Scholar]
- 33.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanomedicine. 2011;6:715. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao ZH, Zhou ZJ, Bao JF, Wang ZY, Hu J, Chi XQ, Ni KY, Wang RF, Chen XY, Chen Z, Gao JH. Nat. Commun. 2013;4:7. doi: 10.1038/ncomms3266. [DOI] [PubMed] [Google Scholar]
- 35.Ling D, Hackett MJ, Hyeon T. Nano Today. 2014;9:457. [Google Scholar]
- 36.Gupta AK, Naregalkar RR, Vaidya VD, Gupta M. Nanomedicine. 2007;2:23. doi: 10.2217/17435889.2.1.23. [DOI] [PubMed] [Google Scholar]
- 37.Ito A, Shinkai M, Honda H, Kobayashi T. J. Biosci. Bioeng. 2005;100:1. doi: 10.1263/jbb.100.1. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Kuang Y, Gao Y, Du X, Shi J, Xu B. J. Am. Chem. Soc. 2013;135:542. doi: 10.1021/ja310019x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuang Y, Shi J, Li J, Yuan D, Alberti KA, Xu Q, Xu B. Angew. Chem. Int. Edit. 2014;53:8104. doi: 10.1002/anie.201402216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Du X, Yuan D, Zhou J, Zhou N, Huang Y, Xu B. Biomacromolecules. 2014;15:3559. doi: 10.1021/bm5010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pospisil P, Iyer LK, Adelstein SJ, Kassis AI. Bmc Bioinformatics. 2006;7:11. doi: 10.1186/1471-2105-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang PJ, Goldblum JR, Pawel BR, Fisher C, Pasha TL, Barr FG. Modern. Pathol. 2003;16:229. doi: 10.1097/01.MP.0000056630.76035.F3. [DOI] [PubMed] [Google Scholar]
- 43.Roecklein BA, Torokstorb B. Blood. 1995;85:997. [PubMed] [Google Scholar]
- 44.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, Mitsiades N, Schlossman RL, Munshi NC, Kung AL, Griffin JD, Richardson PG, Anderson KC, Mitsiades CS. Nat. Med. 2010;16:483. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Liang G, Xu B. Acc. Chem. Res. 2008;41:315. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]