Abstract
The chemokine receptor CCR9 controls the immigration of multipotent hematopoietic progenitor cells into the thymus to sustain T cell development. Post-immigration, thymocytes down regulate CCR9 and migrate toward the subcapsular zone where they recombine their T cell receptor β-chain and γ-chain gene loci. CCR9 is subsequently upregulated and participates in the localization of thymocytes during their selection for self-tolerant receptor specificities. While the dynamic regulation of CCR9 is essential for early T cell development, the mechanisms controlling CCR9 expression have not been determined. Here, we show that key regulators of T cell development, Notch1 and the E protein transcription factors E2A and HEB, coordinately control the expression of Ccr9. E2A and HEB bind at two putative enhancers upstream of Ccr9 and positively regulate CCR9 expression at multiple stages of T cell development. In contrast, the canonical Notch signaling pathway prevents the recruitment of p300 to the putative Ccr9 enhancers, resulting in decreased acetylation of histone H3 and a failure to recruit RNA polymerase II to the Ccr9 promoter. While Notch signaling modestly modulates the binding of E proteins to one of the two Ccr9 enhancers, we found that Notch signaling represses Ccr9 in T cell lymphoma lines in which Ccr9 transcription is independent of E protein function. Our data support the hypothesis that activation of Notch1 has a dominant negative effect on Ccr9 transcription and that Notch1 and E proteins control the dynamic expression of Ccr9 during T cell development.
The development of functional T lymphocytes occurs in the thymus and is maintained by the periodic immigration of multipotent progenitor cells (MPPs) from either the embryonic liver or the adult bone marrow (1). In adult animals, MPPs enter the thymus through venules at the cortical medullary junction (CMJ), rapidly loose B cell differentiation potential, and give rise to early thymic progenitors (ETPs) (2). Differentiation from ETPs is associated with migration of progenitors through the cortex away from the CMJ where DN2 (CD4−CD8− double negative, DN) cells undergo a final stage of lineage restriction to become T lymphocyte lineage committed DN3 thymocytes that reside in the subcapsular zone (SCZ) of the cortex (1, 3). Upon rearrangement of a functional T cell receptor (TCR) β chain, DN3 cells undergo pre-TCR-dependent selection (β-selection) and migrate back toward the CMJ. Major histocompatibility antigen class (MHC) I and class II reactive TCRαβ+ cells are positively selected on cortical thymic epithelial cells (cTEC), migrate into the medulla where they are negatively selected on medullary (m) TEC, and mature into CD8+ and CD4+ T cells (3). The basis for the developmental migration of thymocytes is not fully understood but clearly involves multiple essential receptors that dictate thymocyte adhesion and chemotaxis.
At least three chemokine receptors have been implicated in the immigration of MPPs into the thymus. Deficiency in either one or a combination of chemokine (C-C motif) receptor 7 (CCR7), CCR9, and chemokine (C-X-C motif) receptor 4 (CXCR4) reduces the number of ETPs in the thymus and severely limits T cell production in competitive reconstitution assays (4-10). CCR7 and CCR9 are dynamically expressed on thymocytes and both proteins are required for the migration of CD4-CD8- (double negative/DN) thymocytes toward the SCZ (4, 11). Neonatal thymocytes that lack CCR9 fail to migrate away from the CMJ toward the SCZ (12) and the forced expression of CCR9 on thymocytes arrests T cell development at the DN3 stage when the cells are migrating toward the SCZ (13). Despite the important role that the appropriate control of CCR9 expression plays in thymic immigration and intrathymic migration, the mechanisms controlling Ccr9 transcription, surface expression and function are not well characterized.
The early stages of T cell development are critically dependent on the activation of the transmembrane receptor Notch1 by its ligand Delta-like 4 (DL4) (14, 15). The interaction of Notch1 with its ligands results in a series of proteolytic cleavage events that culminate in the release of the intracellular domain of Notch1 (ICN1) from the plasma membrane by γ-secretase (16). ICN1 translocates to the nucleus and converts the DNA bound transcription factor CSL/RBPJk into a transcriptional activator by recruiting the MAML co-activator and its associated proteins (17). Numerous targets of the ICN/CSL/MAML complex have been identified in T cell progenitors and many of these have critical functions that contribute to T cell differentiation and transformation (18). Among these targets are Ptcra, which encodes the pre-T cell receptor alpha chain (19), and Hes1, which encodes a transcriptional repressor that limits myeloid potential in ETPs and promotes the survival of DN3 cells (20, 21). The ICN/CSL/MAML complex also activates the transcription of Tcf7, which encodes the T cell specification transcription factor TCF1 (22, 23). Genomic mutations that affect the Notch signaling pathway play a major role in the development of both human and mouse T cell leukemia (24-30). These mutations can be divided into at least two classes: those that lead to the ligand independent activation of Notch1 and those that stabilize the active form of Notch1. The second category includes mutations in the PEST domain of Notch1 and mutations in the E3 ligase Fbw7, both of which inhibit the rapid degradation of ICN1 by the ubiquitin-proteasome pathway (31). While many targets of the Notch signaling pathway have been identified, a complete understanding of how this pathway mediates its many biological functions in the thymus has not been achieved.
Notch signaling was proposed to antagonize either the expression or DNA binding activity of the E protein transcription factors (32-34), which play a critical role in B lymphocyte specification (35). However, the interactions between Notch1 and E proteins are complex and the E proteins play multiple important functions during stages of T cell development when Notch signaling is active (36, 37), and Notch1 itself is a transcriptional target of the E proteins (38, 39). The E proteins can also synergize with ICN1 to induce Hes1 expression in T cell progenitors (39). Therefore, the interaction E proteins and Notch 1 in immature DN thymocytes cannot be explained by a simple model in which Notch signaling inhibits either the expression or DNA binding of E proteins. The E proteins encoded by the E2A/Tcf3 gene, E12 and E47, promote the development of the lympho-myeloid and common lymphoid progenitors that seed the thymus and are required for the upregulation of Ccr9 in these cells (38, 40, 41). Another E protein, HEB, is upregulated as ETPs commit to the T cell lineage and HEB expression peaks at the DP stage (42), after Notch signaling subsides. In DP thymocytes, E2A and HEB form dimers that control survival, induce Tcra rearrangement and enforce positive selection (43).
Here, we have investigated a role for the Notch1 signaling pathway in the regulation of Ccr9 in primary T cell progenitors and in Notch1-dependent T cell lymphomas. We show that Notch signaling represses Ccr9 transcription through the canonical pathway involving the MAML co-activator protein. Notch signaling prevents the recruitment of the histone acetyltransferase p300 and the acetylation of histones at two putative enhancers upstream of Ccr9 and prevents the recruitment of RNA polymerase (Pol) II to the Ccr9 promoter. We show that the E protein transcription factors promote CCR9 expression at multiple stages of T cell development and that E2A and HEB bind to the putative enhancers upstream of Ccr9 in primary cells and lymphoma cell lines. The opposing effects of Notch1 and the E protein transcription factors at the Ccr9 gene are not mediated by a global effect of Notch1 on either E protein expression or DNA binding since E proteins bind to one of the two Notch regulated enhancers even in the presence of Notch signals. Our data reveal a negative regulatory function for the Notch1 signaling pathway at the Ccr9 gene and indicate that the integration of Notch signaling and E protein function cooperatively to guide T cell progenitor immigration and migration.
Materials and Methods
Mice
Mice were housed at the University of Chicago and all procedures were approved by the University of Chicago Institutional Animal Care and Use Committee. Fetal liver (FL) MPPs (Ter119−Gr1−CD117+CD27+ cells) were isolated from E13.5-14.5 embryos derived from timed breedings of C57BL/6J mice. E47−/− mice have been described previously and were on an FvB/NJ background (44). Cd4Cre;Hebf/f mice were on a C57BL/6 background and have been described previously (45).
Isolation and treatment of primary cells and cell lines
The 531026 T cell lymphoma cell line was described previously (46). The E13.5-E14.5 fetal liver MPPs used to initiate the in vitro cultures were isolated by magnetic bead depletion (MACS, Miltenyi Biotech, San Diego CA) of Gr1+ and Ter119+ cells followed by flow-assisted cell sorting for CD117+CD27+ cells. The primary cells were cultured for 7 days on OP9-DL1 stromal cells (plated at 1.5 × 105 cells per well of a 6 well plate, 24hrs before use) in OPTI-MEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 mg/ml streptomycin, 29.2 mg/ml glutamine and 80 mM of 2-mercaptoethanol, Flt3L (5 ng/ml, Peprotech), IL-7 (1:100 dilution of culture supernatant from the J558-IL7 cell line) and Kit Ligand (KL) (5 ng/ml, Peprotech) prior to analysis. The cells were cultured at 37°C in a humidified incubator with 5% CO2. DMSO (vehicle control) or γ-secretase inhibitor (10 uM) (DAPT, Sigma Aldrich) were included for the indicated time. The cell lines were cultured in OPTI-MEM supplemented as above but lacking IL-7, Flt3 and KL.
Retroviral transduction
The MigR1, MigR1-ICN, MigR1-DNMAML, and MigR1-HES1 retroviral vectors were described previously (33, 47). Retroviral supernatants were produced in Plat-E cells using the Fugene 6 transfection reagent per the manufacturer’s instructions (Promega). The cells were transduced by spin inoculation as previously described (26).
Flow Cytometry
The flow cytometry was performed on a Fortessa flow cytometer (BD Biosciences) using FACS Diva software and analyzed using FlowJo (Tree Star, Inc.). The cells were sorted on a FACS Aria using the FACS Diva software.
The bone marrow cells, thymocytes, FL MPPs and 531026 cells were isolated and stained with antibodies from BD Biosciences or eBiosciences. Non-specific antibody staining was blocked by incubating the cells with a CD16/CD32 antibody for 10 minutes. The cells were then stained with biotin-conjugated antibodies and fluorescently labeled antibodies for 30 minutes on ice. The cells were washed with FACS buffer (1XPBS, 2.5% FBS, 0.02% Sodium azide) and stained with PerCPCy5.5 conjugated Streptavidin for 15 minutes. The cells were washed with FACS buffer and re-suspended in 500ul of FACS buffer containing propidium iodide to allow dead cells to be excluded from the analysis. The antibodies used included: CD3ε (145-2C11), CD4 (GK1.5), CD8α (53-6.7), TCRβ (H57-597), TCRγδ (UC7-13D5), NK1.1 (PK136), CD11c (HL3), Ter-119 (Ter-119), CD11b (M1/70), Gr1 (RB6-8C5), B220 (RA3-6B2), CD19 (1D3), CD25 (PC61.5), CD117 (2B8), Sca-1 (D7), Flt3 (A2F1D), IL7R-a (A7R34) and CCR9 (CW1.2). The lineage cocktail included CD8, CD3ε, TCRβ, TCRγ, NK1.1, CD11c, Ter119, CD11b, Gr1, B220 and CD19.
Quantitative Real Time PCR (qPCR)
The RNA from 5,000-20,000 FACS sorted primary cells was extracted using the RNeasy micro kit (Qiagen) per the manufacturer’s protocol. The RNA from 500,000 – 5 × 10^6 cells T lymphoma cells was extracted using the Trizol reagent per the manufacturer’s protocol (Invitrogen). The qPCR reactions contained 1ul of either cDNA or ChIP DNA, gene specific primers and SYBR green master mix (Biorad) in a total volume of 25 μl. The amplification was performed in a MyiQ iCycler (BioRad). The data were analyzed in Microsoft Excel using the ΔΔCT method with Hprt as a reference gene for normalization. The primers used were: a. Ccr9 For: 5' CAA TCT GGG ATG AGC CTA AAC AAC 3', Ccr9 Rev: 5' ACC AAA AAC CAA CTG CTG CG 3'; b. Hprt For: 5’ ACC TCT CGA AGT GTT GGA TA 3’, Hprt Rev: 5’ CAA CAA CAA ACT TGT CTG GA 3’; c. Hes1 For: 5’ TCC TGA CGG CCA ATT TGC 3’, Hes1 Rev: 5’ GGA AGG TGA CAC TGC GTT AGG 3’; d. Deltex1 Exon For: 5' CTG AGG ATG TGG TTC GGA GGT 3', Deltex1 Exon Rev: 5' CCT CAT AGC CAG ATG CTG TGA CC 3'. The data for the ChIP samples are displayed relative to the signal for input DNA. The primers used were: a. −88.5Kb For: 5' ATG CCA TAC TGA CCC CAG AAC G 3', −88.5Kb Rev: 5' CGA CCG CAA GTT GAA ACA CCA G 3'; b. −13 kb For: 5’ AGA TGG CTT CAG TGG ACC AG 3’, −13 kb Rev: 5’ ACC CCA AAC CAC ACA GTT TC 3’; c. −10 kb For: 5’ AGG GTG AAC CAC ACT CAA CC 3’, −10 kb Rev: 5’ GGCCCTCAGCACTATGCTAC 3’; d. P1 For: 5’ ACC CTG GTG GTC CTT AGC TT 3’, P1 Rev: 5’ TGT GGG TTC TGA GCA GAC AG 3’; e. P2 For: 5’ TAC TCC GCC AGT GAC AAC AC 3’, P2 Rev: 5’ GCA AAG AGC ATG GTA CAC ATG 3’; f. EX1 For: 5’ GTG TTA CTA GAA TCT GCA GC 3’, EX1 Rev: 5’ TGA GCA GAC AGC TAT CCG C 3’; g. EX2 For: 5’ CTG ATA TGC TGC TAC AGT CCG 3’, EX2 Rev: 5’ GAG AGA GTA AGT GTT CTG AGG 3’; h. EX3 For: 5’ CTC CAC TGC TTC CAC AGA TGA CTA C 3’, EX3 Rev: 5’ TTG CCC AAG GTG CCC ACA ATG AAC 3’; i. Deltex1 P1 For: 5’ GCA TGG ATT GTA GGT CGA TG 3’, Deltex1 P1 Rev: 5’ GTG TGG GAG TGG CTC AAT G 3’; j. β-globin For: 5’ GCC ATC GTT AAA GGC AGT TAT CA 3’, β-globin Rev: 5’ TGC TAT CAT GGG TAA TGC CAA A 3’; and Ebf1 For: 5’ TGA AGG TGT CAC TTG AGC AGT CC 3', Ebf1 Rev: 5’ ACT TTC CCA AAC CCC TAT GGC 3'.
Chromatin Immunoprecipitation
Ten million cells were fixed with 1% formaldehyde in 1X PBS at room temperature for 15 minutes. The reaction was quenched by the addition of 2M glycine to reach a final concentration of 0.125M and the samples were incubated for an additional 5 minutes at room temperature. The cells were washed 3 times with 1X PBS containing 1 mM PMSF and 0.1% protease inhibitor cocktail and then sonicated to shear the DNA into 400-800 bp fragments. For the histone modification ChIPs, the sonication was performed in 1X SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH8.1). For the E protein, p300 and RNA Pol II ChIPs, the sonication was performed in 0.8 M RIPA buffer (10 mM Tris pH 7.4, 1 mM EDTA, 1% Triton X-100, 0.1% Sodium De-oxycholate, 0.8 M NaCl, 0.1% SDS). The protein-DNA complexes were detected using 5 μg of antibody against either RNA Pol II (N-20)(Santa Cruz, sc-899x), p300 (C-20)(Santa Cruz, sc-585x), HEB (A-20, Santa Cruz, sc-357) or E2A (N-649, Santa Cruz, sc-763x) in one ml of 0.8 M RIPA and incubated overnight at 4°C under rotating conditions. The complexes were precipitated by an overnight incubation at 4°C on a rotator with 20 μl of Protein A (Millipore) conjugated magnetic beads. The protein-DNA complexes were also precipitated with antibodies against acetyl Histone H3 (Millipore 06-599), trimethyl Histone H3 Lysine 4 (Millipore 07-473) and trimethyl Histone 3 Lysine 36 (Millipore ABE305) in 1 ml ChIP dilution buffer containing protein A magnetic beads and incubated overnight at 4°C with rotation. The protein-DNA complexes were washed multiple times, reverse cross-linked and the DNA was purified and eluted using a PCR purification kit (Qiagen) per the manufacturer’s protocol. The specific DNA enrichment was quantified by real time PCR. The ChIP sequencing for HEB was performed with thymocytes from 4-week-old wild-type mice and the enrichment peaks were generated using the MACS algorithm (model-based analysis of ChIP-Seq data) through the use of the appropriate input controls, as described previously (48).
Results
The chemokine receptor CCR9 is repressed upon thymic immigration coincident with Notch signaling
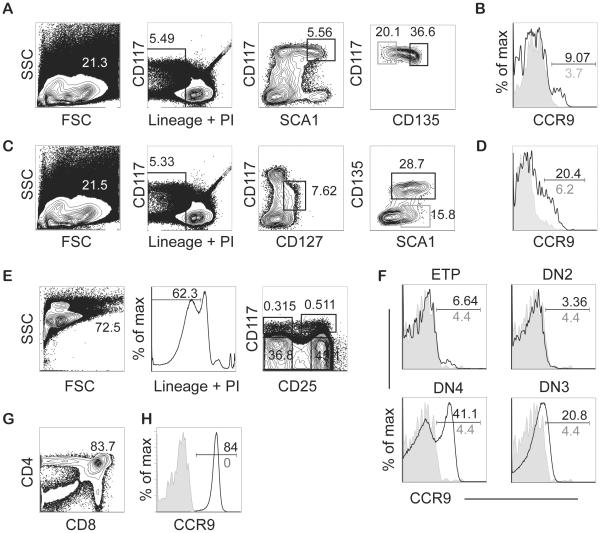
We examined CCR9 expression on the surface of MPPs and their thymic progeny using flow cytometry to gain insight into the regulation of CCR9. We found that CCR9 was expressed on a very small fraction of LMPPs and on a larger fraction of CLPs (Fig. 1A-D). However, the ETPs and DN2 cells, which had recently immigrated to the thymus, expressed almost no CCR9 on their surface (Fig. 1E and F). Approximately 25% of the thymocytes re-expressed CCR9 at the DN3 stage, a number that coincides with the frequency of cells that have passed through β-selection. The frequency of CCR9 expressing cells increased after the DN4 stage and essentially all the DP cells were CCR9 positive (Fig. 1G and H). These data demonstrate that CCR9 expression is transiently decreased when Notch signaling is most active in T cell progenitors and lead us to hypothesize that Notch signaling controls the expression of CCR9.
Figure 1. Dynamic expression of the chemokine receptor CCR9 during T lymphopoiesis.
Flow cytometric analysis of CCR9 on bone marrow derived LMPPs and CLPs or thymus-derived ETP, DN2, DN3, DN4 and DP thymocytes. (A) Gating strategy for the identification of LMPPs (Lin/PI−CD117+SCA1+FLT3+) and HSCs (Lin/PI−CD117+SCA1+Flt3−), which served as a negative control, among bone marrow cells. Gates used for selection are indicated as are the frequency of cells in each gate. (B) CCR9 expression on LMPPs (black) and HSCs (grey shaded), as indicated in the last plots showing gating in (A). (C) Gating strategy for the identification of CLPs (Lin/PI−CD117intCD127+SCA1+FLT3+) and pre-pro-NK cells (Lin/PI−CD117intCD127+SCA1+FLT3−), which served as a negative control, among bone marrow cells. (D) CCR9 expression on CLPs (black) and pre-pro-NK cells (grey shaded), as indicated in the final gating in (C). (E) Gating strategy for identification of CD4−CD8− (DN) thymocyte subsets. Thymocytes were depleted of Lineage positive cells by MACS and then gated on ETPs (Lin/PI−CD117+CD25−), DN2 (Lin/PI−CD117+CD25+), DN3 (Lin/PI−CD117−CD25+) and DN4 (Lin/PI−CD117−CD25−). (F) CCR9 expression (black) on ETP, DN2, DN3 and DN4 thymocytes as indicated. Grey shading is HSC from BM isolated and stained in the same experiment. (G) CD4 and CD8 expression on total thymocytes with the gate used for identification of DP thymocytes. (H) CCR9 expression on DP thymocytes (black) with HSCs as a negative control (grey). In (B), (D), (F), (H) the frequency of CCR9+ cells is indicated in the experimental (black) and control (grey) population. One of at least 3 experiments with similar outcomes is shown for all populations.
Notch signaling represses Ccr9/CCR9 expression in primary MPPs
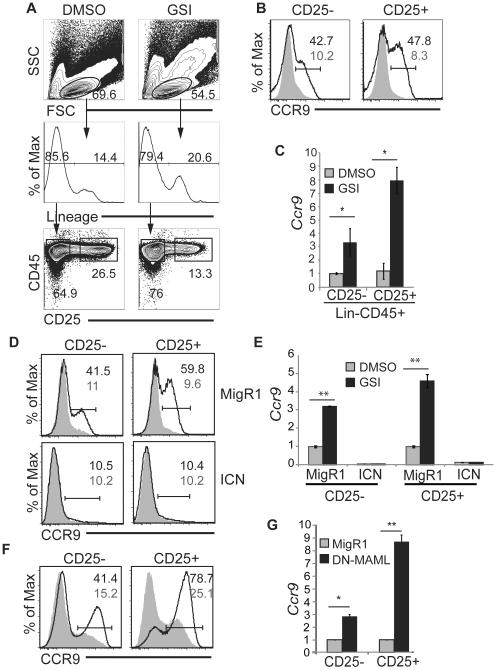
We cultured wild-type (WT) FL-derived MPPs on the stromal cell line OP9-DL1, which expresses the Delta-like 1 (DL1) Notch1 ligand, to determine whether Notch signaling could inhibit the expression of CCR9 on T cell progenitors (49). After 7 days in culture, the CD25+ T lymphocyte lineage specified and CD25− MPPs were isolated from by flow cytometry and examined for their expression of CCR9 (Fig. 2A). Regardless of their expression of CD25, less than 11% of the cells expressed CCR9 on their surface (Fig. 2B). In contrast, CCR9 expression was substantially increased when these cells were cultured with a γ-secretase inhibitor (GSI) for the final 48 hours of culture (Fig. 2B). In the presence of GSI, 42.7% of CD25− and 47.8% of the CD25+ cells expressed CCR9. Ccr9 mRNA levels also increased in both the CD25− and CD25+ populations after treatment with GSI compared to the cells treated with DMSO (Fig. 2C).
Figure 2. Notch signaling represses Ccr9 mRNA and CCR9 protein expression in primary T cell progenitors.
(A) Gating strategy for identification of Lin−CD45+CD25− and Lin−CD45+CD25+ progeny of FL MPPs after 7 days of culture in which DMSO or GSI (10 μM) were added for the final 48 hours. The frequency of cells in each of the indicated gates is shown. (B) Expression of CCR9 on Lin−CD45+CD25− (CD25−) and Lin−CD45+CD25+ (CD25+) cells cultured on OP9-DL1 for 7 days including a 48 hour treatment with DMSO (shaded histogram) or GSI (DAPT, 10 uM) (open histogram). The frequency of CCR9+ cells in GSI (black) and DMSO (grey) treated samples is indicated. (C) QPCR analysis for Ccr9 in Lin−CD45+CD25− and Lin−CD45+CD25+ FL MPPs cultured in vitro for 7 days including a 48 hour treatment with DMSO (grey) or GSI (black) as in (B). Hprt mRNA was used for normalization. Error bars = standard deviation. (D) FL MPPs were transduced with MigR1 or MigR1-ICN on day 4 of culture and treated with DMSO (shaded histogram) or GSI (open histogram) for 48 hours starting on day 5 of culture. CCR9 on Lin−CD45+GFP+CD25− (CD25−) and Lin−CD45+GFP+CD25+ (CD25+) cells is shown. The frequency of cells expressing CCR9 in GSI (black) and DMSO (grey) treated cultures is indicated. (E) QPCR analysis for Ccr9 mRNA in the Lin−CD45+GFP+CD25− and Lin−CD45+GFP+CD25+ in the progeny of FL MPPs treated with DMSO (grey) or GSI (black) as described in (D) after infection with MigR1 or MigR1-ICN. (F) CCR9 expression on the Lin−CD45+GFP+CD25− and Lin−CD45+GFP+CD25+ progeny of FL MPPs that were transduced with MigR1 (shaded histogram) or MigR1-DN-MAML (open histogram) on day 4 of culture (D) and analyzed on day 7. (G) QPCR analysis for Ccr9 mRNA in the Lin−CD45+GFP+CD25− and Lin−CD45+GFP+CD25+ progeny of FL MPPs as described in (F) after infection with MigR1 (grey) or MigR1-DNMAML (black). All experiments are representative of > 3. For QPCR analysis, * = p<0.05, ** = p<0.01.
GSIs inhibit the cleavage event that liberates the intracellular domain of the Notch1 protein (ICN1) (16). However, γ-secretase can also cleave other targets that might impact Ccr9/CCR9 expression. We transfected primary MPPs with a retrovirus encoding a constitutively activated form of ICN1, which is impervious to GSI to demonstrate that the GSI-induced increase in Ccr9 mRNA and CCR9 protein was a consequence of reduced Notch signaling. Surface CCR9 expression was strongly induced by GSI treatment in the CD25− and CD25+ cells that were transduced with a control virus (MigR1) (Fig. 2D). In contrast, CCR9 was not induced by GSI treatment in the CD25− and CD25+ cells that were transduced with the ICN producing retrovirus (Fig. 2D). At day 7 of culture, Ccr9 transcription was barely detectable in either the CD25− or the CD25+ cells that were constitutively expressing ICN1 regardless of the presence GSI (Fig. 2E). The repression of basal Ccr9 transcription in DMSO treated cells by ectopic ICN1 suggests that the background level of CCR9 detected on these T cell progenitors may be a consequence of insufficient Notch signaling.
While our data demonstrate that Notch signals can repress Ccr9, they do not demonstrate that Notch signaling is essential for the repression of Ccr9. We transduced the day 5 progeny of WT FL MPPs with a retrovirus encoding a dominant negative version of MAML (DN-MAML), which is the co-activator for the ICN/CSL complex, or a control virus to determine whether Notch signaling was essential to repress Ccr9 (50-52). In comparison to the cells that were transduced with the control retrovirus, both the CD25− and CD25+ cells that were transduced with the retrovirus encoding DN-MAML up-regulated Ccr9 mRNA and CCR9 protein, even in the absence of GSI (Fig. 2F). Both the frequency and the intensity of CCR9 expression increased in the CD25− and CD25+ cells that were transduced with the DN-MAML encoding retrovirus (Fig. 2F). Ccr9 transcripts were also increased in CD25− and CD25+ cells that had been transduced with the DN-MAML encoding retrovirus (Fig. 2G). These data demonstrate that Notch signaling represses Ccr9 mRNA and CCR9 protein expression in the progeny of FL MPPs. Our findings are consistent with the hypothesis that the decreased expression of CCR9 on ETPs following thymic immigration is a consequence of Notch signals.
CCR9 expression on primary MPPs and thymocytes requires E protein transcription factor activity
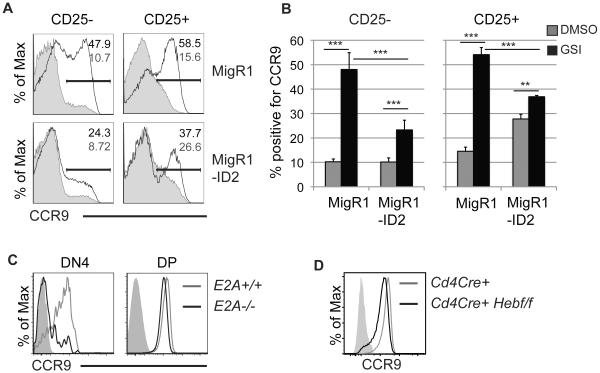
We previously demonstrated that CCR9 expression is E protein dependent in LMPPs (38). We transduced FL MPPs with a retrovirus encoding ID2, an inhibitor of E protein DNA binding, to determine whether the induction of CCR9 on the progeny of primary FL MPPs after the inhibition of Notch signaling also requires E protein activity (35). The transduction of MPPs cultured on OP9-DL1 for 5 days with ID2-producing virus did not influence the frequency of CD25− cells expressing CCR9 after 48 hours, in comparison with control virus transduced cells (Fig. 3A and B). There was a slight increase in the frequency of ID2-producing virus transduced CD25+ cells that expressed CCR9 (Fig. 3A and B). Control virus transduced cells and untransduced cells both strongly up-regulated CCR9 expression upon treatment with GSI. In contrast, the induction of CCR9 expression upon GSI treatment was blunted in ID2-expressing cells (Fig. 3A and B). The frequency of CCR9 expressing cells was increased approximately 5-fold in control virus transduced CD25− and CD25+ cells by 48 hours after GSI treatment. In contrast, the frequency of CCR9 expressing ID2 virus transduced CD25− cells was increased approximately 2-fold. There was almost no increase in the frequency of CCR9 expressing CD25+ ID2 virus transduced cells (Fig. 3A and B). These data indicate that E protein transcription factors contribute to the expression of CCR9 on FL MPPs following the withdrawal of Notch signaling.
Figure 3. The E protein transcription factors contribute to CCR9 expression on primary T cell progenitors.
(A) FACS analysis for CCR9 on CD25− and CD25+ Lin−CD45+GFP+ FL MPPs culture in vitro for 3 days prior to infection with MigR1 or MigR1-ID2 retrovirus and treated with DMSO (shaded histogram) or GSI (open histogram) on day 5 for 48hrs. The frequency of CCR9+ cells on GSI (black) or DMSO (grey) treated cells is indicated. One of four representative experiments is shown. (B) Average + standard deviation of the percent of CD25− and CD25+ Lin−CD45+GFP+ cells expressing CCR9 after treatment as in (A). n=4, ** = p<0.01 and *** = p<0.001. (C) FACS analysis for CCR9 protein on Lin−CD44−CD117−CD25− DN4 thymocytes and CD4+CD8+ DP thymocytes isolated from an E2A+/+ or E2A−/− mouse. One of three experiments is shown. (D) FACS analysis for CCR9 protein on DP thymocytes from a Cd4Cre/- or Cd4Cre+/-Hebf/f mouse.
We next tested whether E proteins were required for CCR9 expression on thymocytes. We found that CCR9 was substantially reduced on DN4 thymocytes in the absence of E2A (Fig. 3C). There was a small decrease in the relative MFI of CCR9 expression on E2A−/− DP thymocytes compared to the WT controls (Fig. 3C). Because the E protein HEB is highly expressed in DP thymocytes, we tested whether HEB was required for CCR9 expression in DP thymocytes. We found a mild decrease in CCR9 expression on DP thymocytes in Cd4CreHebf/f mice, similar to the decrease observed in E2A−/− DP thymocytes (Fig. 3D). These data indicate that the E2A proteins plays an essential role in regulating CCR9 expression on DN thymocytes and that E2A and HEB have either non-essential or possibly redundant functions in regulating CCR9 expression on DP thymocytes.
E proteins bind putative enhancers near Ccr9
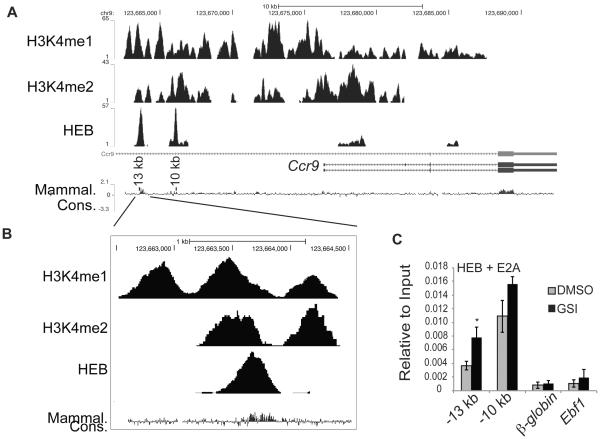
To determine whether E proteins could bind to the Ccr9 gene we examined HEB ChIP-seq data from DP thymocytes. We identified two peaks of HEB binding approximately 13 kb and 10 kb upstream of the Ccr9 transcription start site in regions that are highly conserved between multiple species (Fig. 4A, B). In addition, these conserved regions overlapped with regions containing the histone modifications H3K4me1 and H3K4me2, both of which are enriched at enhancers (Fig 4A and B). Additional experiments revealed that p300 binding, a definitive marker of enhancer activity, was also enriched in these regions. Based on these findings, we identified these two HEB binding regions as putative enhancers for Ccr9 in DP cells.
Figure 4. Identification of E protein binding sites near the Ccr9 gene.
(A) Occupancy by the HEB transcription factor as well as H3K4me1 and H3K4me2 at the Ccr9 locus, as determined by ChIP-seq and visualized by the UCSC browser. Schematic of the Ccr9 gene is shown below the ChIP-seq histograms. The UCSC track of sequence conservation in mammals is also shown and the −13 kb and −10 kb conserved regions are indicated. The chromosome locations are indicated above the tracks. (B) Enlarged view of the −13 kb conserved region showing the overlap between HEB binding, H3K4me1, and H3K4me2. (C) QPCR analysis of chromatin immunoprecipitated by antibodies directed against the E proteins HEB and E2A from the day 7 Lin−CD45+ progeny of FL-MPPs treated for 48hrs treated with DMSO (grey) or GSI (black). DNA was amplified using primers within the −13 kb and −10 kb conserved regions or using primers to β-globin or Ebf1, which served as negative controls. Data is expressed as enrichment normalized to input and is averaged from three independent experiments. * = p<0.05, ** = p<0.01.
To determine whether these putative enhancers were bound by E proteins in T cell progenitors we performed ChIP on the progeny of FL MPPs that were expanded on OP9-DL1 for 5 days and then treated with either DMSO or GSI for 48 hours. E protein binding was detected above background levels (i.e. binding at the β-globin or the Ebf1 loci) at both the −13 kb and the −10 kb regions when the cells were treated with DMSO (Fig. 4C). The relevant E protein in these cells is likely E47 since that is the major E protein expressed in MPPs. The addition of GSI to block Notch signaling resulted in a 2.2 fold increase in E protein binding at the −13 kb region and a mild, but not significant, increase at the −10 kb region (Fig. 4C). The overall higher enrichment for E proteins at the −10 kb region compared to the −13 kb region may reflect the number of E-box sites at these putative enhancers. These data indicate that E proteins can bind to both the −13 kb and the −10 kb regions upstream of Ccr9 and that E protein binding at the −13kb region is only modestly influenced by Notch signals.
Lymphoma lines also show Notch-dependent repression of Ccr9
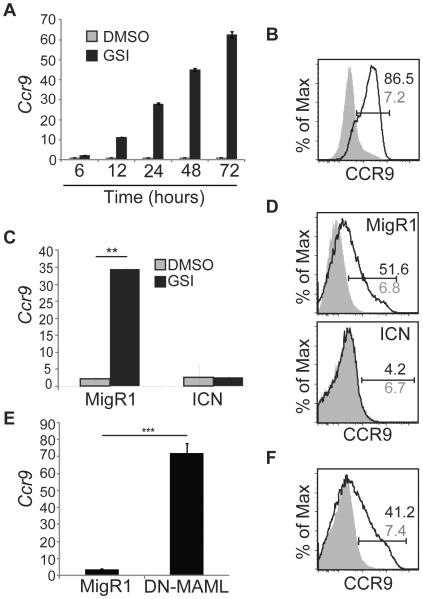
We examined Notch dependent T cell lymphoma cell lines to gain further insight into how Notch signaling regulates Ccr9 transcription. We tested 3 lymphoma cell lines with differing genotypes (E2A−/−, E2A−/−Rag1−/−, and p53−/−). All of these cell lines expressed very low levels of Ccr9 mRNA and CCR9 protein, but CCR9 and Ccr9 mRNA expression was increased by treatment with GSI (Fig. 5A, B and data not shown). We focused our subsequent analysis on the E2A−/−Rag1−/− lymphoma line 531026 because it underwent growth arrest when treated with GSI but remained viable for longer than 96 hours (data not shown). In comparison to treatment with DMSO, treatment with GSI led to a rapid increase in Ccr9 mRNA that could be detected as early as 6 hours after treatment and continued out to 72 hours (Fig. 5A). More than 80% of the cells were positive for CCR9 on their cell surface by 48 hours following GSI treatment (Fig. 5B). Importantly, Ccr9 mRNA was not induced by GSI treatment when the lymphoma cells were transduced with a retrovirus encoding ICN1 (Fig. 5 C, D). Transduction of the lymphoma cells with the retroviruses encoding DN-MAML resulted in expression of CCR9 and Ccr9 mRNA without the need for GSI (Fig. 5E, F). These data demonstrate that Ccr9 mRNA is repressed by the Notch signaling pathway in both primary MPPs and T cell lymphomas.
Figure 5. Notch signaling represses Ccr9 mRNA and CCR9 protein expression in immature T cell lymphomas.
(A) The 531026 T cell lymphoma line was treated with DMSO (grey) or GSI (black) for the indicated time and analyzed for Ccr9 mRNA by QPCR. Hprt mRNA was used for normalization. (B) FACS analysis for CCR9 protein on 531026 cells 48 hours after treatment with DMSO (shaded histogram) or GSI (open histogram). The frequency of CCR9+ cells is indicated for GSI (black) and DMSO (grey) treated cells. (C) Relative expression of Ccr9 mRNA in GFP+ 531026 cells transduced with MigR1 or MigR1-ICN and treated with DMSO (grey) or GSI (black) for 48 hours. Hprt mRNA was used for normalization. (D) FACS analysis for CCR9 on GFP+ cells isolated 48 hours after treatment of MigR1 or MigR1-ICN infected cells with DMSO (shaded histogram) or GSI (open histogram). The frequency of CCR9+ cells in GSI (black) and DMSO (grey) treated cells is indicated by the gated region. (E) Relative expression of Ccr9 mRNA in GFP+ 531026 cells 40 hours after transduced with MigR1 or MigR1-DN-MAML. Hprt mRNA was used for normalization. (F) FACS analysis for CCR9 on GFP+ 531026 cells 40 hours after being transduced with MigR1 (shaded histogram) or DN-MAML (open histogram). The frequency of CCR9+ cells on MigR1 (grey) or MigR1-DN-MAML (black) infected cells is indicated in the gated region. For QPCR experiment, * = p<0.05, ** = p<0.01, *** = p<0.003. All experiments are representative of at least three.
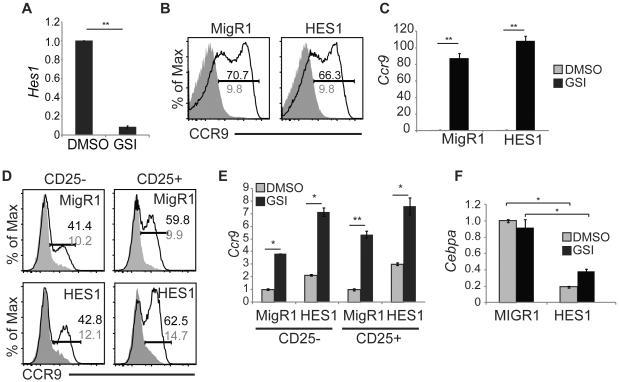
Notch1 can repress gene expression by inducing the transcription factor HES1 and Hes1 mRNA levels decline in lymphoma lines after treatment with GSI (Fig. 6A) (21, 53). Therefore, we ectopically expressed HES1 in 531026 cells and examined CCR9 expression after treatment with either DMSO or GSI to determine whether HES1 was sufficient to repress Ccr9. If HES1 was sufficient to repress Ccr9 then GSI treatment would not induce CCR9 upregulation in HES1 expressing lymphomas. However, CCR9 protein and Ccr9 mRNA was increased in the HES1 expressing lymphoma line after treatment with GSI (Fig. 6B, C). HES1 also failed to repress Ccr9 when ectopically expressed in primary MPPs cultured on OP9-DL1 in the presence of GSI (Fig. 6D, E) Importantly, the retrovirally encoded HES1 was functional because it repressed Cebpa mRNA expression in primary MPPs (Fig. 6F) (21). Therefore, while HES1 is a transcriptional repressor induced by activated Notch1, its expression is not sufficient to repress Ccr9.
Figure 6. HES1 is not sufficient to repress Ccr9 mRNA.
(A) QPCR analysis for Hes1 mRNA in 531026 cells treated with DMSO or GSI for 48 hours. Hprt mRNA was used for normalization. (B) FACS analysis for CCR9 on 531026 cells treated with DMSO (shaded histogram) or GSI (open histogram) for 48 hours following infection with MigR1 or MigR1-HES1. The frequency of CCR9+ cells on GSI (black) and DMSO (grey) treated cells is indicated by the gated region. (C) QPCR for Ccr9 mRNA in 531026 cells treated with DMSO (grey) or GSI (black) for 48 hours following infection with MigR1 or MigR1-HES1. Hprt mRNA was used for normalization. (D). FACS analysis for CCR9 protein on CD25− or CD25+ Lin−CD45+GFP+ FL MPPs treated with DMSO (shaded histogram) or GSI (open histogram) for 48hrs following infection with MigR1 or MigR1-HES1. The frequency of CCR9+ cells on GSI (black) and DMSO (grey) treated cells is indicated by the gated region. (E) QPCR analysis for Ccr9 mRNA in the same DMSO (grey) and GSI (black) populations as in (D). Hprt mRNA was used for normalization. (F) QPCR analysis for Cebpα mRNA in Lin−CD45+ FL-MPPs treated with DMSO (grey) or GSI (black) for 48 hours. * = p<0.05, ** = p<0.01. All experiments were performed at least 3 times.
Notch signaling impacts the transcriptional status of the Ccr9 gene
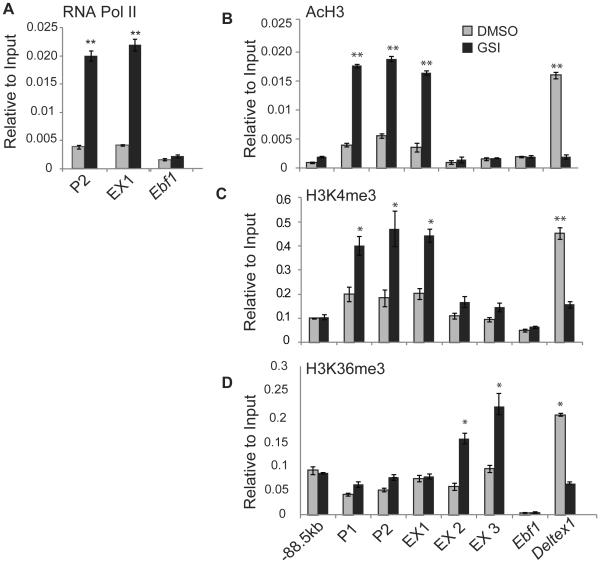
Our data indicate that Notch signaling influences the expression of Ccr9 mRNA and protein in primary MPPs and T cell lymphomas. We examined the recruitment of RNA polymerase II (PolII) to the Ccr9 promoter and histone modifications associated with active transcription across the Ccr9 gene to determine whether Notch signals impact Ccr9 transcription. In the presence of Notch signaling, RNA PolII was not highly enriched at the Ccr9 promoter (P2) or at exon 1 (EX1) when compared to the negative control Ebf1 locus (Fig. 7A). However, in the presence of GSI, RNA PolII binding near the Ccr9 promoter and exon 1 increased substantially (Fig. 7A). Our EX1 primers lie immediately downstream of the Ccr9 transcription start site and therefore likely detect events occurring at the promoter. We also examined the effect of Notch signaling on H3K4 trimethylation (H3K4me3) and H3 acetylation (AcH3), which are both positively correlated with active promoters (54). The inhibition of Notch signaling was associated with an increase in H3K4me3 and H3Ac at the promoter (P1 and P2) and at exon 1. These histone modifications were not as prevalent at exons 2 and 3 of Ccr9, consistent with the enrichment of these modifications at promoters but not at more distal exons (Fig. 7B, C). H3K36me3, a marker of histones in actively transcribed exons, was increased at exon 2 and exon 3 after the inhibition of Notch signaling (Fig 7D). The H3K36me3 modification is generally enriched toward the 3’ end of the gene rather than at the promoter (54, 55). Taken together, these data indicate that Notch signaling prevents the recruitment of RNA PolII to the Ccr9 promoter and inhibits the transcription of Ccr9.
Figure 7. Notch signaling represses Ccr9 transcription.
Chromatin immunoprecipitation was performed using antibodies against (A) RNA Polymerase II (PolII), (B) AcH3, (C) H3K4me3, and (D) H3K36me3 on extracts from 531026 cells isolated 48 hours after treatment with DMSO (grey) or GSI (black). The precipitated DNA was amplified by QPCR using primers located near the Ccr9 promoter (primers P1 and P2) or within the Ccr9 exons 1 (EX1), 2 (EX2) and 3 (EX3). Primers in a region −88.5 kb upstream of Ccr9 and at the Ebf1 or Deltex1 genes served as controls. Primers at the Deltex1 promoter were used for H3K4me3 and AcH3 and to Deltex1 exon 7 were used for H3K36me3. The data are expressed as enrichment normalized to input and are averaged from three independent experiments. * = p<0.05, ** = p<0.01.
Notch signaling regulates recruitment of the histone acetyltransferase p300 to the −13kb and −10kb regions of the Ccr9 gene
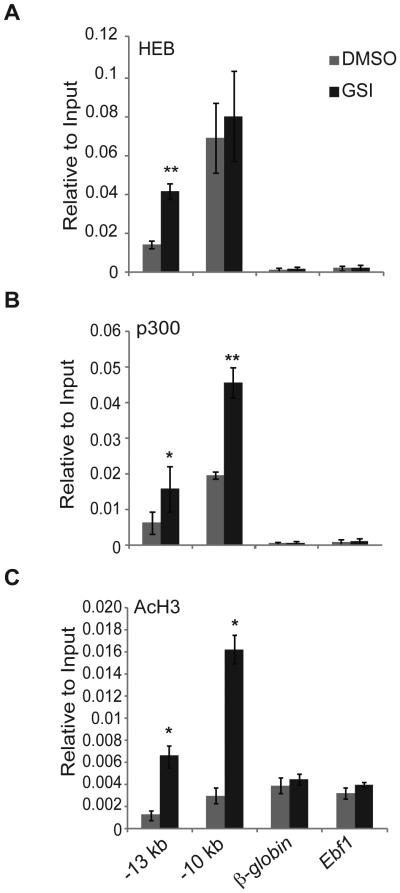
Our lymphoma lines are E2A-deficient, thus Ccr9 transcription in these cells is either independent of E proteins or dependent on HEB, unlike in primary MPPs. By ChIP, we found a substantial enrichment of HEB binding at the −10 kb region in this lymphoma even in the presence of Notch signaling (Fig. 8A). HEB also bound the −13 kb region in the presence of Notch signals although, as in primary MPPs (Fig. 4C), the binding was significantly increased by GSI treatment (Fig. 8A). However, we conclude that Ccr9 transcription is E protein independent in these lymphomas because ectopic retrovirus-driven expression of Id2 did not affect lymphoma cell surface expression of CCR9 after treatment with GSI (data not shown). These findings are consistent with our previous conclusion that Notch represses Ccr9 expression independent of E protein function.
Figure 8. Notch signaling prevents recruitment of p300 and E proteins to a distal enhancer of Ccr9.
Chromatin immuno-precipitation using (A) HEB, (B) p300, and (C) AcH3 antibodies was performed on 531026 cells 48 hours after treatment with DMSO (grey) or GSI (black). The purified DNA was amplified by QPCR using primers within the −13 kb and −10 kb regions of the Ccr9 locus. Primers to β-globin and Ebf1 served as negative controls. Data is expressed as enrichment normalized to input and is averaged from three independent experiments. * = p<0.05, ** = p<0.01.
To gain further insight into how Notch signaling might impact Ccr9 transcription we determined whether p300, a co-activator that modulates chromatin accessibility by histone acetylation at enhancers, was recruited to the putative Ccr9 enhancers. P300-mediated histone acetylation can increase transcription factor occupancy at enhancers such as we observed with the E proteins (56, 57). In the 531026 lymphoma, we found that Ccr9 transcription after Notch inhibition was associated with strong binding of p300 to the −13 kb and −10 kb regions upstream of Ccr9 (Fig. 8B). When compared to the negative control loci, p300 also bound these regions in the presence of Notch signals, but to a significantly lesser extent (Fig. 8B). Importantly, AcH3 was only substantially enriched at the −13 kb and −10 kb regions over the β-globin and Ebf1 control genes when Notch signaling was inhibited (Fig. 8C). Our data indicate that Notch signaling limits p300 recruitment and AcH3 at two regions upstream of the Ccr9 gene. Given that p300 recruitment is a major function of transcriptional enhancers, we propose that the −13 kb and −10 kb regions are enhancers of Ccr9 that are compromised, either directly or indirectly, by Notch signaling.
Discussion
The dynamic up and down regulation of CCR9 controls the migration of lymphocytes into and through the thymus (58, 59). We have shown that the E protein transcription factors promote Ccr9/CCR9 expression in MPPs and at the DN stages of T cell development. In contrast, Notch signaling is antagonistic to Ccr9/CCR9 expression in T cell progenitors and is sufficient to account for the immediate down-regulation of CCR9 after MPPs enter the Notch ligand-rich thymic environment. We showed that Notch signaling influences Ccr9 at the level of transcription. We identify two regions upstream of Ccr9 that show dynamic histone modifications consistent with their designation as enhancers and demonstrate that Notch signals prevent the recruitment of the co-activator protein p300 to these regions. In addition to its known roles in promoting thymocyte differentiation, our data reveal that Notch1 is an important regulator of DN thymocyte migration via the repression of Ccr9.
In both T cell lymphomas and primary T cell progenitors, Ccr9 repression is mediated by the canonical Notch signaling pathway but this repression is not executed directly by the Notch regulated transcriptional repressor HES1. HES1 is activated by Notch signaling in T cell progenitors and plays a role in restricting the development of myeloid cells through the regulation of Cebpa (21). Although ectopic HES1 represses Cebpa in MPPs, it does not prevent the induction of Ccr9 after treatment with GSI. Notch signaling induces multiple proteins that could repress Ccr9, including GATA3 and TCF1 (22, 23). However, neither of these transcription factors remains dependent on Notch signaling after their initial induction and therefore they are unlikely to be the target of Notch1 that transiently regulates Ccr9. In genome wide binding studies in mouse leukemia cells, ICN was shown to potentially associate with the transcriptional repressor ZNF143 at a subset of genes that lack evidence of activating histone modifications (60, 61), indicating that ICN does not activate but could repress these genes. Nonetheless, we favor a model in which the ICN1/CSL/MAML complex either activates or interacts with a factor that binds to the Ccr9 enhancers and either directly or indirectly prevents the recruitment of p300. Interestingly, in human T lineage acute lymphoblastic leukemia cell lines inhibition of Notch signaling with GSI resulted in a loss of Ccr9 expression (62). Genome-wide binding studies revealed that ICN can have divergent functions in human and mouse leukemic cells and that the supraphysiologic levels of ICN found in leukemic cells can result in divergent functions for Notch signaling in leukemic as compared to normal cells (63). Therefore, it is important to note that our studies show repression of Ccr9 by Notch signaling in both primary and transformed mouse cell lines. Nonetheless, further studies are needed to determine how ICN impacts the recruitment of p300 to the putative enhancers of Ccr9 and whether ICN function to regulate Ccr9 in both mouse and human T cell development.
It has been proposed that Notch signaling inhibits the function of the E protein transcription factors either directly, by limiting their expression, or indirectly, by inducing the expression of ID proteins that inhibit the ability of E proteins to bind DNA (33, 34, 64). Our data indicate that E proteins bind to the −10 kb region of Ccr9 even in the presence of Notch signals. Therefore, Notch signaling does not globally prevent E protein binding in these cells. However, there was a reduction of E protein binding at the −13 kb region when Notch signaling was active and this decrease could contribute to the reduced expression of Ccr9 in primary T cell progenitors after the activation of Notch1. Indeed, the E protein E47 is necessary for optimal CCR9 expression on FL MPPs and on DN4 thymocytes. While the E protein HEB is bound to the putative Ccr9 enhancers in DP thymocytes, neither E47 nor HEB is absolutely required for CCR9 expression, although the absence of either protein caused a mild decrease in CCR9. Therefore, E47 and HEB may play redundant roles in Ccr9 regulation in DP thymocytes, although this possibility remains to be tested. E proteins can bind p300 through their activation domains (65, 66) and they could, therefore, contribute to the stability of p300 binding to the Ccr9 locus in MPPs and T cell progenitors when Notch signaling is inhibited. However, E protein binding may not be sufficient to maintain p300. Alternatively, the decrease in E protein binding to the −13 kb region may be sufficient to destabilize p300 binding. If an interaction between the two Ccr9 enhancers stabilizes p300, such as via the formation of an “enhancer bridge”, then the loss of E protein binding to one of the putative enhancers may be sufficient to reduce enhancer function. However, further studies are needed to determine whether the decrease in E protein binding at the −13 kb region contributes to the loss of Ccr9 transcription in the presence of Notch signaling. Moreover, how Notch signals impact E protein binding at this region remains to be determined. Notch signaling could induce a factor that binds to this enhancer and occludes E protein binding to some of the E-boxes in this region. Alternatively, factors that cooperate with E proteins to allow their recruitment to this region when it is in a “closed” chromatin conformation (i.e. lacking AcH3) may be lacking when Notch signaling is active.
We found that neither E2A nor HEB were essential for Ccr9 expression in multiple T cell lymphomas after the inhibition of Notch signaling. This observation leads us to suggest that the ability of Notch signals to inhibit the recruitment of p300 to the putative Ccr9 enhancers is not a consequence of a direct effect on E protein function. Rather, Notch signaling likely regulates Ccr9 transcription either directly or through distinct factors that inhibit Ccr9 enhancer function. The liberation of Ccr9 transcription from E protein function in T cell lymphomas is of interest and suggests that while E proteins can regulate Ccr9, other factors are able to take over this function. The identity of these factors and their role in normal and malignant T cell migration could be of interest. Our data reveal opposing roles for E protein and Notch signaling in the dynamic expression of Ccr9 on thymocytes and their progenitors and indicate that both proteins function to control intrathymic T cell progenitor migration.
Acknowledgements
We would like to thank Dr. W.S. Pear for the MigR1-ICN, MigR1-DN-MAML, and MigR1-HES1 retroviruses, Dr. J.C. Zuniga-Pflucker for the OP9-DL1 cells, and Dr. J. Aster for discussions and for sharing unpublished data. We also thank members of the Kee lab for helpful discussions.
This work was supported by grants from the National Institutes of Health (R01 AI089213 and R01 CA099978/AI106352). B. L. K. was a Scholar of the Leukemia and Lymphoma Society.
References
- 1.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 3.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 4.Uehara S, Grinberg A, Farber JM, Love PE. A role for CCR9 in T lymphocyte development and migration. J Immunol. 2002;168:2811–2819. doi: 10.4049/jimmunol.168.6.2811. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 6.Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2009;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Forster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2009;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Saito F, Liu Z, Lei Y, Uehara S, Love P, Lipp M, Kondo S, Manley N, Takahama Y. Coordination between CCR7- and CCR9-mediated chemokine signals in prevascular fetal thymus colonization. Blood. 2006;108:2531–2539. doi: 10.1182/blood-2006-05-024190. [DOI] [PubMed] [Google Scholar]
- 9.Robertson P, Means TK, Luster AD, Scadden DT. CXCR4 and CCR5 mediate homing of primitive bone marrow-derived hematopoietic cells to the postnatal thymus. Exp Hematol. 2006;34:308–319. doi: 10.1016/j.exphem.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Calderon L, Boehm T. Three chemokine receptors cooperatively regulate homing of hematopoietic progenitors to the embryonic mouse thymus. Proc Natl Acad Sci U S A. 2011;108:7517–7522. doi: 10.1073/pnas.1016428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misslitz A, Pabst O, Hintzen G, Ohl L, Kremmer E, Petrie HT, Forster R. Thymic T cell development and progenitor localization depend on CCR7. J Exp Med. 2004;200:481–491. doi: 10.1084/jem.20040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benz C, Heinzel K, Bleul CC. Homing of immature thymocytes to the subcapsular microenvironment within the thymus is not an absolute requirement for T cell development. Eur J Immunol. 2004;34:3652–3663. doi: 10.1002/eji.200425248. [DOI] [PubMed] [Google Scholar]
- 13.Uehara S, Hayes SM, Li L, El-Khoury D, Canelles M, Fowlkes BJ, Love PE. Premature expression of chemokine receptor CCR9 impairs T cell development. J Immunol. 2006;176:75–84. doi: 10.4049/jimmunol.176.1.75. [DOI] [PubMed] [Google Scholar]
- 14.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 15.Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, Manley NR, Duarte A, R. Macdonald H, Radtke F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 17.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 18.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annu Rev Cell Dev Biol. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 19.Gounari F, Aifantis I, Martin C, Fehling HJ, Hoeflinger S, Leder P, von Boehmer H, Reizis B. Tracing lymphopoiesis with the aid of a pTalpha-controlled reporter gene. Nat Immunol. 2002;3:489–496. doi: 10.1038/ni778. [DOI] [PubMed] [Google Scholar]
- 20.Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Obaldia ME, Bell JJ, Wang X, Harly C, Yashiro-Ohtani Y, DeLong JH, Zlotoff DA, Sultana DA, Pear WS, Bhandoola A. T cell development requires constraint of the myeloid regulator C/EBP-alpha by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, Gounari F. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108:20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng AP, Ferrando AA, Lee W, Morris J. P. t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 25.O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, Alt FW, Kelliher M, Look AT. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reschly EJ, Spaulding C, Vilimas T, Graham WV, Brumbaugh RL, Aifantis I, Pear WS, Kee BL. Notch1 promotes survival of E2A-deficient T cell lymphomas through pre-T cell receptor-dependent and -independent mechanisms. Blood. 2006;107:4115–4121. doi: 10.1182/blood-2005-09-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, Kee B, Vooijs M, Radtke F, Redondo JM, Georgopoulos K. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity. 2010;33:685–698. doi: 10.1016/j.immuni.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin YW, Nichols RA, Letterio JJ, Aplan PD. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talora C, Campese AF, Bellavia D, Pascucci M, Checquolo S, Groppioni M, Frati L, von Boehmer H, Gulino A, Screpanti I. Pre-TCR-triggered ERK signalling-dependent downregulation of E2A activity in Notch3-induced T-cell lymphoma. EMBO Rep. 2003;4:1067–1072. doi: 10.1038/sj.embor.7400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 34.Nie L, Xu M, Vladimirova A, Sun XH. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 2003;22:5780–5792. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 36.Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nat Rev Immunol. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- 37.Miyazaki K, Miyazaki M, Murre C. The establishment of B versus T cell identity. Trends Immunol. 2014;35:205–210. doi: 10.1016/j.it.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci U S A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Anderson MK. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- 43.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spaulding C, Reschly EJ, Zagort DE, Yashiro-Ohtani Y, Beverly LJ, Capobianco A, Pear WS, Kee BL. Notch1 co-opts lymphoid enhancer factor 1 for survival of murine T-cell lymphomas. Blood. 2007;110:2650–2658. doi: 10.1182/blood-2007-04-084202. [DOI] [PubMed] [Google Scholar]
- 47.Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, Petrie HT, Georgopoulos K. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2011;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 50.Jeffries S, Robbins DJ, Capobianco AJ. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol Cell Biol. 2002;22:3927–3941. doi: 10.1128/MCB.22.11.3927-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 53.Kim HK, Siu G. The notch pathway intermediate HES-1 silences CD4 gene expression. Mol Cell Biol. 1998;18:7166–7175. doi: 10.1128/mcb.18.12.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 56.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uehara S, Song K, Farber JM, Love PE. Characterization of CCR9 expression and CCL25/thymus-expressed chemokine responsiveness during T cell development: CD3(high)CD69+ thymocytes and gammadeltaTCR+ thymocytes preferentially respond to CCL25. J Immunol. 2002;168:134–142. doi: 10.4049/jimmunol.168.1.134. [DOI] [PubMed] [Google Scholar]
- 59.Svensson M, Marsal J, Uronen-Hansson H, Cheng M, Jenkinson W, Cilio C, Jacobsen SE, Sitnicka E, Anderson G, Agace WW. Involvement of CCR9 at multiple stages of adult T lymphopoiesis. J Leukoc Biol. 2008;83:156–164. doi: 10.1189/jlb.0607423. [DOI] [PubMed] [Google Scholar]
- 60.Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, Blacklow SC, Liu XS, Aster JC. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci U S A. 2014;111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, Bernstein BE, Kieff E, Aster JC. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirandola L, Chiriva-Internati M, Montagna D, Locatelli F, Zecca M, Ranzani M, Basile A, Locati M, Cobos E, Kast WM, Asselta R, Paraboschi EM, Comi P, Chiaramonte R. Notch1 regulates chemotaxis and proliferation by controlling the CC-chemokine receptors 5 and 9 in T cell acute lymphoblastic leukaemia. J Pathol. 2011;226:713–722. doi: 10.1002/path.3015. [DOI] [PubMed] [Google Scholar]
- 63.Chiang MY, Shestova O, Xu L, Aster JC, Pear WS. Divergent effects of supraphysiologic Notch signals on leukemia stem cells and hematopoietic stem cells. Blood. 2012;121:905–917. doi: 10.1182/blood-2012-03-416503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, Zhuang Y, Pear WS. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, LeBrun DP. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J Biol Chem. 2004;279:55362–55371. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- 66.Hyndman BD, Thompson P, Bayly R, Cote GP, LeBrun DP. E2A proteins enhance the histone acetyltransferase activity of the transcriptional co-activators CBP and p300. Biochim Biophys Acta. 2012;1819:446–453. doi: 10.1016/j.bbagrm.2012.02.009. [DOI] [PubMed] [Google Scholar]