Abstract
Long-lasting antibody responses rely upon the germinal center (GC), where B cells bearing high affinity antigen receptors are selected from a randomly mutated pool to populate the memory and plasma cell compartments. Signaling downstream of the B cell receptor (BCR) is dampened in GC B cells, raising the possibility that antigen presentation and competition for T cell help, rather than antigen-dependent signaling per se, drive these critical selection events. Here we use an in vivo reporter of BCR signaling, Nur77-eGFP, to demonstrate that although BCR signaling is reduced among GC B cells, a small population of cells exhibiting GC light zone phenotype (site of antigen and follicular helper T cell encounter) express much higher levels of GFP. We show that these cells exhibit somatic hypermutation, gene expression characteristic of signaling and selection, and undergo BCR signaling in vivo.
Introduction
Affinity maturation of B cell receptors (BCRs) is vital for optimizing antigen-specific protective antibodies. Upon antigen recognition in the context of cognate T cell help, a subset of activated B cells are recruited into the germinal center (GC)(1–5). GC B cells undergo random somatic hypermutation (SHM) to diversify their BCR repertoires and subsequently undergo selection and expansion that is regulated by resident follicular dendritic cells (FDCs) and recruited T follicular helper (Tfh) cells. GC B cells with highest affinity BCRs survive to populate the memory and long-lived plasma cell (LLPC) pools, thereby conferring potent and lasting humoral immunity. Antigen capture by the BCR and recruitment of Tfh help regulate GC B cell selection. However, it is not known whether BCR signaling contributes to affinity maturation. These two models are not mutually exclusive, and data in favor of both have been reported (2–4, 6, 7).
The GC has been histologically divided into light and dark zones (LZ and DZ), both populated by GC B cells. Importantly, Tfh and FDC are enriched in the LZ, suggesting that this may be the site of antigen-driven selection (1, 4). GC B cells shuttle rapidly (on the order of hours) between these two zones in a chemokine-dependent manner (8, 9). This is thought to be the dynamic spatiotemporal correlate of iterative rounds of SHM and selection in the GC reaction. LZ and DZ GC B cells can be identified reliably according to expression of the chemokine receptor CXCR4 and the costimulatory ligand CD86 (7, 10). Gene expression profiling of these subsets has revealed a signature of both BCR and CD40-dependent signaling in LZ GC B cells (7, 10). However, recent work by Shlomchik and colleagues has demonstrated that BCR signaling in GC B cells is markedly dampened due to phosphatase activity (6).
Since only a small fraction of GC B cells ultimately undergo selection, there must be hidden heterogeneity among GC B cells with respect to signaling and gene expression that is obscured by bulk analysis. Here we use an in vivo sensor of BCR signaling, Nur77-eGFP BAC transgenic line (‘Nur77-GFP’), to unmask such heterogeneity (11). In contrast to prior reports that BCR signaling is largely undetectable in GC B cells, our data demonstrate directly that BCR signaling, while markedly reduced relative to activated B cells, nevertheless occurs in vivo among a sub-population of LZ GC B cells.
Material and Methods
Mice
Nur77-GFP mice were previously described (11). C57/Bl6, BoyJ, and B1-8i mice obtained from Jackson labs (12). All mice were housed in a specific pathogen free facility at UCSF according to University and NIH guidelines.
Antibodies/reagents
Abs to B220, CD4, CD45.2, CD69, CD83, CD86, CXCR4, GL-7, Fas, IgD, IgM, IgG, and λ1 light chain conjugated to: biotin, PE, PECy7, PerCPCy5.5, APC, PB, and QDot605 (eBiosciences or BD Biosciences); NP conjugated to PE or KLH (Biosearch technologies); Gt anti-mouse IgM fab’2 (Jackson Immunoresearch); anti-CD3ε (2C11 clone; Harlan), anti-CD40 (hm40-3 clone, Pharmingen), ibrutinib (Jack Taunton, UCSF).
Immunization/infection
Mice were either immunized with 100ug NP-KLH (Biosearch) mixed 1:1 with alum injected IP or infected i.p. with 2×105 PFU of LCMV Armstrong.
Adoptive transfer
Splenocytes from CD45.2 B1-8i reporter mice were loaded with Cell Trace Violet (Invitrogen) per protocol. 2×106 cells were adoptively transferred into CD45.1 BoyJ hosts which were then immunized with NP-KLH as above. 3 days later splenocytes were surface stained, and analyzed by FACS.
Lymphocyte activation assay
Previously described (13).
Flow Cytometry and data analysis
Cells were collected on BD Fortessa and analyzed on FlowJo (v9.7.6; Treestar).. Graphs were generated with Prism v6 (GraphPad Software). Bulk cells were sorted on Moflo and single cells on Aria. BCR sequence data analyzed with IMGT/V-QUEST (imgt.org).
Single cell sorting and VH186.2 sequencing
10 days after NP-KLH immunization, B6 reporter splenocytes were stained with Fas, GL7, CXCR4, CD86, fixed, and single cell sorted (gating in Fig. S1B) into 96 well plates with catch media. Dump gating and pre-purification were not used. Plates were frozen and subjected to nested PCR and Sanger sequencing as described (14) except: secondary nested PCR run with Amplitaq DNA pol (Applied Biosystems) and 1x PCR buffer (Roche).
Sorting and qPCR
9 days after LCMV infection, B6 reporter splenocytes were negatively selected with Abs to IgD, CD4, CD8 to enrich for GC B cells. Cells were then stained for IgD, CXCR4, CD86, Gl7, Fas, CD19, and DAPI and sorted (gating in Fig. S2E) into Trizol (Invitrogen) and stored at −80°C. cDNA was prepared with Superscript III kit (Invitrogen). qPCR reactions were run on a QuantStudio 12K Flex thermal cycler (ABI) using either TaqMan Assays (Bcl2A1, Pax5, Bcl6, and GAPDH) with Taqman Universal PCR Master Mix (ABI) or 250nM (each) primer pairs (Aicda, Irf4 (15), Cxcr4, Ccnd2, Ccnb2, cMyc (7)) with FastStart Universal SYBR Green Master Mix (Roche).
Ibrutinib treatment
Mice were infected with LCMV as above, and on day 10 post-infection were injected i.p. 2x/day for 3 days with either vehicle or ibrutinib at 12.5 mg/kg/dose dissolved in Captex355.
Results and Discussion
Nur77-GFP reporter identifies B cells activated by antigen in vivo
The B cell response of C57BL/6 mice to immunization with the hapten NP is characterized by predominant use of heavy chain Vh186.2 (16). The B1-8i mouse strain was created by targeted insertion of an unmutated VH186.2 (DFL16.2)JH2 gene into the H chain locus (12). B1-8i transgenic B cells expressing endogenous λ1 light chain are capable of binding NP with a precursor frequency of 2–3% in the pre-immune B cell repertoire (17). To track antigen-specific B cells both before and after immunization, we generated B1-8i Nur77-GFP reporter mice. We adoptively transferred B1-8i reporter splenocytes loaded with CellTrace Violet (CTV) dilutional dye into congenically marked hosts, and immunized recipients with NP coupled to the protein antigen KLH. As expected, after 3 days we observed expanded cell number, GFP upregulation, and concurrent dye dilution in transferred NP-specific λ1+ B cells (Fig. 1A, 1B, Supplemental Fig. 1A).
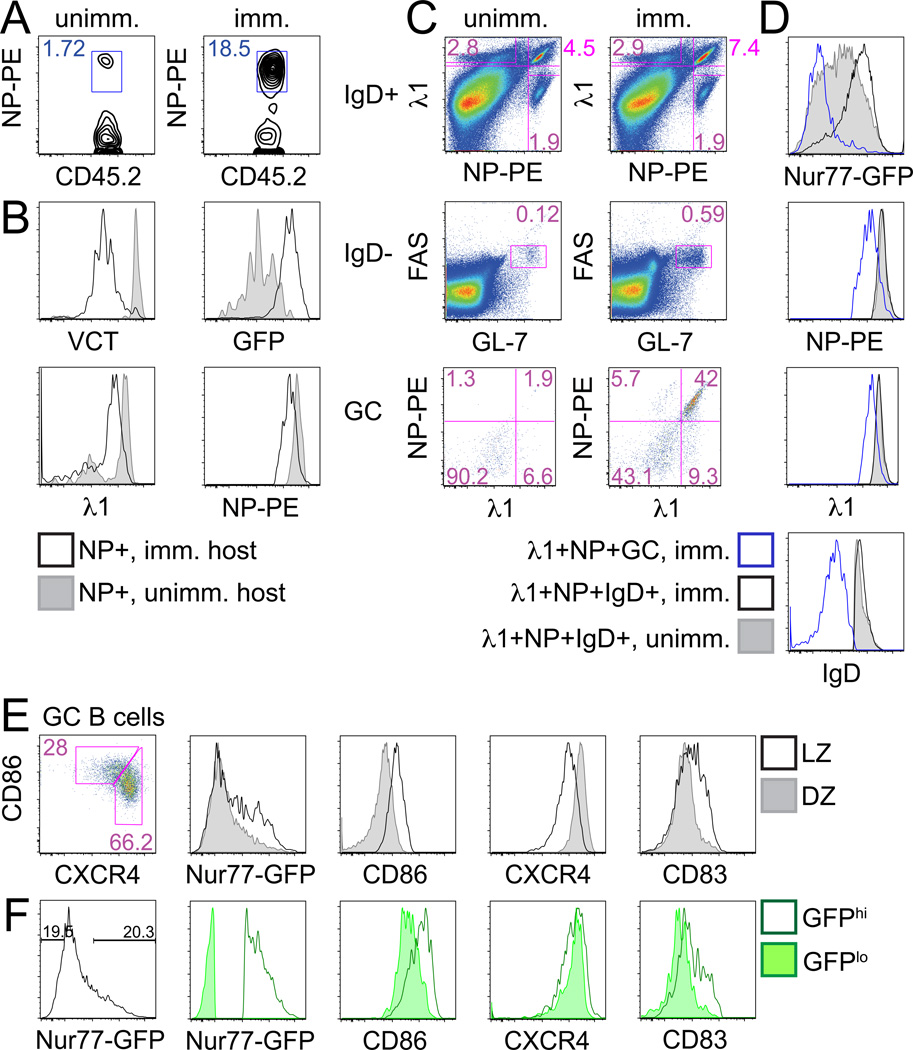
Figure 1. Nur77-GFP reporter identifies B cells activated by antigen in vivo.
(A, B) B1-8 reporter splenocytes loaded with CTV were adoptively transferred into CD45.1 BoyJ recipients. Recipient splenocytes were stained 3 days after NP-KLH immunization. (A) Plots represent CD45.2+ donor cells from unimmunized (left) and immunized (right) hosts stained with NP-PE. (B) Histograms represent VCT, GFP, surface NP, and λ1 expression in NP+ donor B cells from immunized (black line) or unimmunized (shaded histogram) recipients as gated in (A). Data are representative of 4 biological replicates.
(C–F) B1-8 reporter splenocytes were harvested and stained d9 after NP-KLH immunization. (C) Plots depict gating to identify: IgD+λ1+NP+ B cells (top panels), IgDnegFashiGL-7hi GC B cells (middle panels), and λ1+NP+GC B cells (bottom panels) from unimmunized (left) and immunized (right) mice. (D) histograms represent GFP, NP, λ1, and IgD expression in λ1+NP+ GC B cells (blue line), λ1+NP+IgD+ B cells from immunized mouse (black line) or λ1+NP+ IgD+ B cells from unimmunized mouse (shaded histogram) as gated in (C). (E) Histograms represent GFP, CD86, CXCR4, and CD83 expression in overlayed LZ (CD86hiCXCR4lo) and DZ (CD86loCXCR4hi) phenotype GC B cells from immunized B1-8 mice as gated in left-hand panel plot. (F) Right hand histograms depict same markers in GFP-low and GFP-high GC B cells gated as in left hand histogram. Data are representative of 4 biological replicates.
Nur77-GFP expression in GC B cells is low, but heterogeneous
To further characterize BCR signaling in GC B cells, we immunized unmanipulated B1-8i reporter mice with NP-KLH and assessed GFP expression in λ1+NP+ B cells 9 days later. Although GFP was upregulated in λ1+NP+IgD+ B cells, GFP expression in λ1+ NP+ GC B cells was markedly reduced (Fig. 1C, D). It is possible that GFP protein dilution upon cell division contributes to reduced GFP expression; however, GFP was markedly upregulated in dividing NP-specific adoptively transferred B cells despite such dilution (Fig. 1A, B). Although BCR expression was reduced among GC B cells by about 50%, reduction in GFP expression in GC B cells was disproportionate and did not correlate with surface BCR (Fig. 1D; data not shown). Therefore, we hypothesize that GC B cells ‘see’ less antigen and/or signaling is suppressed.
Although GFP expression is very low in GC B cells relative to naïve or activated B cells, we nevertheless observe a “shoulder” of high GFP-expressing cells, suggesting that a subset of GC B cells is undergoing active signaling (Fig. 1D). We further subdivided GC B cells into LZ and DZ phenotype cells on the basis of CD86 and CXCR4 expression (Fig. 1E)(7). High GFP expression is enriched among LZ phenotype GC B cells (CD86hiCXCR4lo), consistent with previously identified gene expression suggestive of active BCR and CD40 signaling in this compartment (7). Moreover, GFP expression was correlated with CD86 and CD83 surface expression but not CXCR4 (Fig. 1F). To reduce the high precursor frequency of NP-specific clones in the B1-8i model system, we immunized unrestricted repertoire B6 reporter mice with NP-KLH. We observed that GFP expression was again markedly reduced in GC B cells relative to naïve B cells (Fig. 2A), similarly enriched among LZ phenotype cells, and correlated with CD86 expression in GC B cells (Fig. 2B, C).
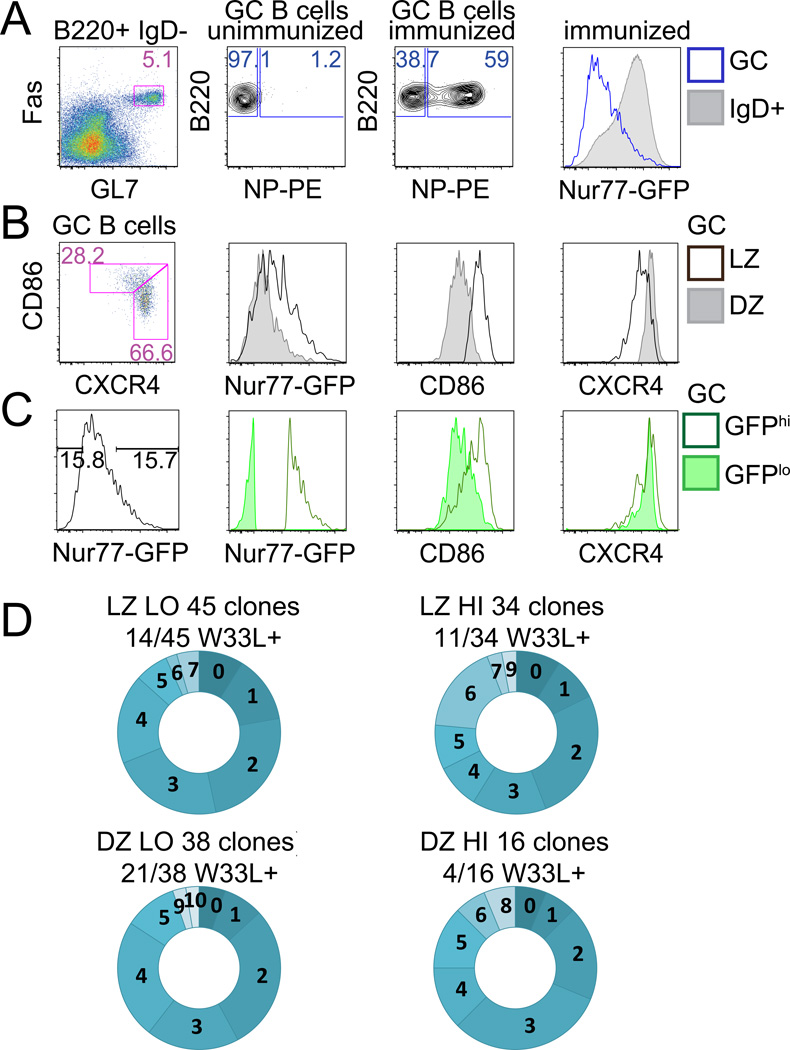
Figure 2. High Nur77-GFP-expressing GC B cells exhibit SHM.
(A–C) C57/Bl6 reporter splenocytes were stained d14 after NP-KLH immunization. (A) Plots represent gating to identify B220+IgDnegFashiGL7hi GC B cells (left hand panel), and NP-binding GC B cells (middle panels). Overlayed histograms (right hand panel) represent GFP expression from IgD+ (shaded histogram) and GC B cells (blue line). (B,C) Plots and histograms as described in Fig. 1E, F. Data are representative of 8 biological replicates.
(D) Vh186.2 heavy chains isolated from singly sorted GC B cells (see Fig. S1B for gating) from B6 reporter mice d10 after NP-KLH immunization were sequenced. Graphs represent number of mutations in individual clones (see Fig. S1C for mutation distribution). Numbers above graphs show proportion of clones with the W33L mutation.
High Nur77-GFP-expressing GC B cells exhibit SHM and are not recent arrivals in the GC
To determine whether heterogeneity of GFP expression in the GC was due to timing of GC entry such that ‘recent recruits’ might retain higher GFP expression, we assessed the extent of SHM among GFP-high and GFP-low GC subsets. In the first two weeks after immunization with NP coupled to a protein antigen, mutation of a single residue in the Vh186.2 heavy chain, 33WL, arises in immunized mice with very high frequency (90%) and increases antibody affinity for NP by a factor of 10 (16). Analysis of the ratio of coding to silent mutations (r/s ratio) in the CDR sequences of the Vh186.2 heavy chain has also been used as a proxy measure for affinity maturation (17). We assessed Vh186.2 sequences from GFP-high and GFP-low LZ and DZ phenotype GC B cells sorted 10 days after NP-KLH immunization of B6 reporter mice (see gating in Supplemental Fig. 1B). We observed comparable levels of missense mutations in Vh186.2 germline sequence among all sorted subsets, suggesting that neither high nor low GFP-expressing GC B cell compartments were preferentially enriched with unmutated recent arrivals (Fig. 2D, Supplemental Fig. 1C)(14).
Nur77-GFP expression correlates with LZ/DZ gene expression signature
We next infected reporter mice with lymphocytic choriomeningitis virus (LCMV) in order to determine whether GFP expression is similarly regulated in the context of a complex, physiologic stimulus. GC B cell number was markedly expanded at 8 and 15 days following infection (data not shown). At both of these time points, we observed a marked reduction in overall GFP expression among GC B cells relative to IgD+ B cells, as well as a unique “shoulder” of GFP-high GC B cells among LZ but not DZ-phenotype B cells (Fig. 3A,B, Supplemental Fig. 2A–C). We confirmed that these GC B cells are not only Fashi and GL-7hi, but also CD38lo and PNAhi (data not shown)(3, 4). Importantly, we found no correlation between either isotype or extent of surface BCR expression and GFP levels within LZ GC B cells (data not shown).
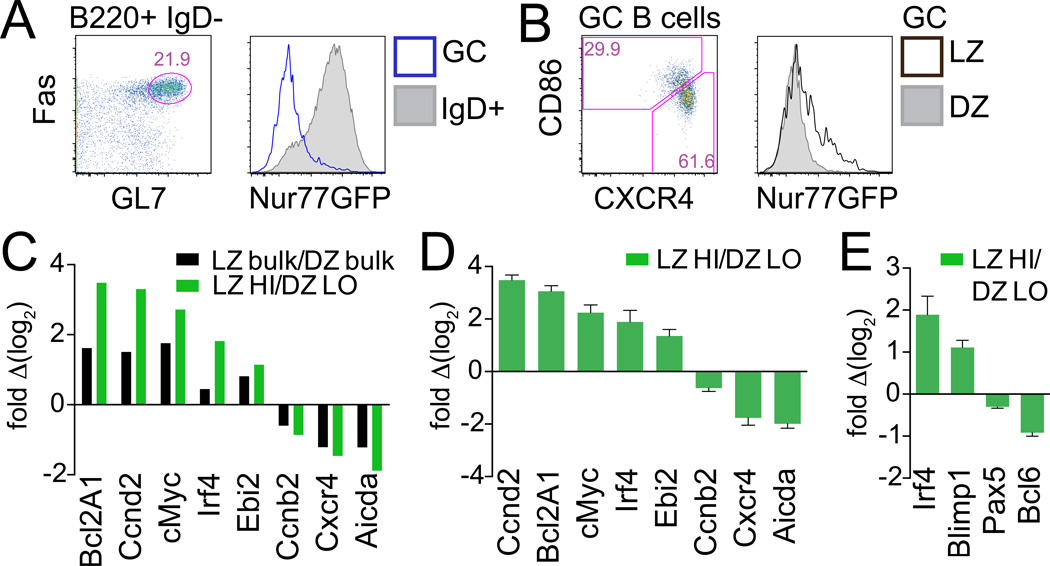
Figure 3. Nur77-GFP expression correlates with LZ gene expression signature.
(A–B) B6 reporter splenocytes were stained d8 after LCMV infection. (A) Plots represent gating to identify B220+IgDnegFashiGL7hi GC B cells (lefthand panel). Overlayed histograms (righthand panel) represent GFP expression from IgD+ (shaded histogram) and GC B cells (blue line). (B) Plot and histogram as described in Fig. 1E. Data are representative of at least 2 independent experiments with 2–3 mice per time point per experiment.
(C–E) GC B cells from B6 reporter mice infected with LCMV were sorted (see Fig. S2E for gating), and transcript abundance was assessed by qPCR. (C) shows enrichment of LZ-DZ gene expression signatures in GFP-sort relative to bulk sort; data from one sort shown here is representative of 3 independent sorts. (D and E) present qPCR data from 3 independent sorts +/− SEM to demonstrate reproducibility.
The Nur77-GFP reporter marks a subset of LZ GC B cells that may represent the actively signaling subpopulation of cells within the LZ compartment. To test this hypothesis we assessed expression of LZ and DZ “signature” genes in bulk LZ and DZ cells sorted with or without further sub-division on the basis of GFP expression from reporter mice infected with LCMV (see gating in Supplemental Fig. 2D). We observed predicted enrichment of relevant gene expression in bulk-sorted LZ and DZ populations (Fig. 3C black bars). Importantly, by enriching LZ GC B cells for active signaling by collecting high GFP-expressing cells, and, conversely, depleting the DZ of “contaminating” GFP-high cells, we see further enrichment of the putative LZ and DZ gene expression signatures relative to bulk sort (Fig. 3C, D green bars).
High GFP expression in GC B cells marks cells undergoing selection into the LLPC pool
Critical cell fate decisions in the GC control which clones become quiescent memory B cells, which populate the LLPC pool, and which continue to undergo further rounds of SHM (2–4). The transcriptional regulators Blimp1, Irf4 and Xbp-1 favor plasma cell fate and are counteracted by Bcl6 and Pax5, which maintain undifferentiated GC B cells (2, 4, 18, 19). LZ-phenotype genes Irf4 and Blimp1 were enriched in high GFP GC B cells while Pax5 and Bcl6 expression were reduced (Fig. 3E). Importantly, GFP-high GC B cells do not express CD138, and conversely fully differentiated plasma cells that do express CD138 as well as intra-cellular IgG do not express GFP (data not shown). Therefore, high-GFP-expressing GC B cells gate are not merely contaminating plasma cells or plasmablasts (18, 19). Rather, we propose that this high-GFP-expressing population may contain plasma cell precursors undergoing selection and differentiation in the germinal center.
Nur77-GFP expression in GC B cells is dependent upon BCR signaling
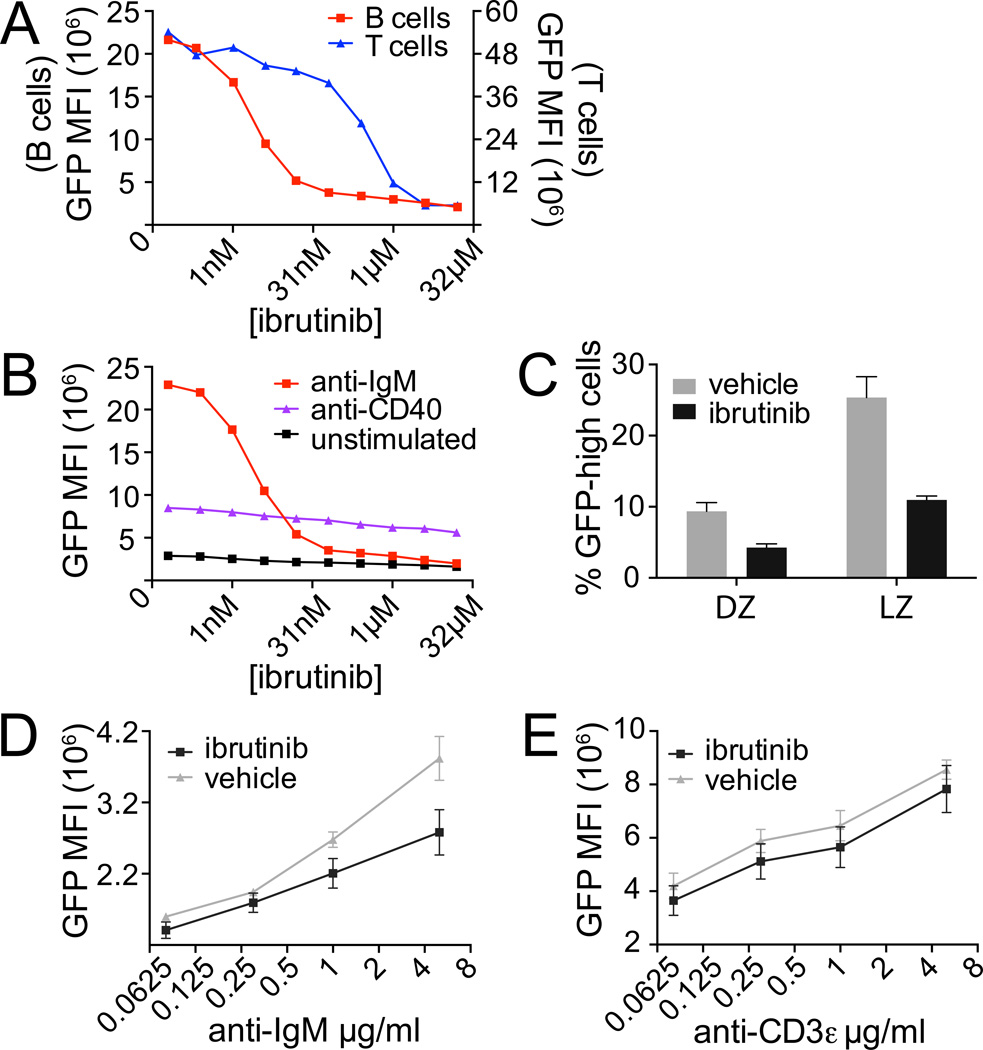
GFP expression in naïve Nur77-GFP reporter B cells is dependent upon antigen and BCR signaling (11). However, we have previously shown that signals other than BCR, including ligation of CD40, are capable of inducing GFP expression in vitro, albeit much less efficiently (11). We therefore wanted to determine whether GFP expression among GC B cells specifically requires BCR-dependent signaling, or may be driven by indirect consequences of antigen capture such as Tfh engagement and CD40-dependent signaling. To distinguish between these possibilities, we used the well-characterized irreversible Btk inhibitor ibrutinib (20). We found about a 60-fold selectivity of ibrutinib for inhibition of BCR signaling over TCR signaling in vitro (Fig. 4A, Supplemental Fig. 2E). CD40-induced GFP upregulation was insensitive to ibrutinib at all doses used (Fig. 4B). We next treated reporter mice that had been infected 10 days earlier with LCMV, a time point when germinal centers had already been well-established, for 3 additional days with either ibrutinib or vehicle. GC cell number was modestly reduced by about half among inhibitor-treated mice, but relative LZ and DZ composition remained unaffected (Supplemental Fig. 2F, G). Importantly, we found that LZ GFP expression was markedly reduced relative to vehicle-treated infected animals, implying that a Btk-dependent, BCR-specific signal was driving GFP expression in reporter GC B cells in vivo (Fig. 4C, Supplemental Fig. 2H, I, J). We restimulated lymphocytes taken from ibrutinib- or vehicle-treated mice immediately ex vivo (without further supplementation of inhibitor) through AgR for 4 hours. We observed persistent inhibition of AgR-mediated GFP and CD69 upregulation among B, but not T cells (Fig. 4D, E, Supplemental Fig. 2K, L). These data confirm in vivo specificity of ibrutinib treatment for Btk over Itk at doses used in our assay, and thereby suggest that GFP expression in reporter GC B cells is indeed specifically driven by BCR signaling in vivo.
Figure 4. Nur77-GFP expression in GC B cells is dependent upon BCR signaling.
(A) Reporter LN cells were treated with 5µg/ml of either anti-IgM or anti-CD3ε along with varying concentrations of ibrutinib and then stained after 16 hrs for B220, CD4, and CD69 expression. Graph depicts GFP MFI of CD4 T cells or B cells. Data are representative of 2 independent experiments.
(B) Reporter LN cells were treated with 4µg /ml of anti-IgM, 1 µg /ml of anti-CD40, or neither along with varying concentrations of ibrutinib and then stained after 16 hrs as above. Graph depicts GFP MFI of B cells. Data are representative of 2 independent experiments.
(C) LN cells harvested d13 from mice treated with vehicle or ibrutinib on days 10, 11, and 12 following LCMV infection were stained and gated as in Supplemental Figure 2I, J. Graph depicts % GFP-high LZ and DZ B cells.
(D, E) LN cells harvested from mice treated as in (C) were restimulated with varying doses of either anti-IgM (D) or anti-CD3ε (E) for 4 hrs without additional inhibitor, after which cells were stained as in (A). Graph depicts GFP MFI of B cells (D) or T cells (E) normalized to GFP MFI of individual PMA-treated samples (positive control). Data plotted in C–E are the mean +/− SEM of 3 biological replicates.
Although a variety of signals are integrated by GC B cells in vivo to make decisions about selection and differentiation, the contribution of active BCR signaling to this process has been controversial (2–4, 6, 7). The Nur77-eGFP reporter reveals a marked reduction in BCR signaling among GC B cells relative to activated B cells in vivo. This may render GC B cells especially dependent upon antigen and/or T cell help for survival and selection, enhancing competition and selection pressure within the GC. Furthermore, dampened yet active BCR signaling could resolve subtle affinity differences among competing clones.
BCR-dependent GFP expression is detected among a small subset of LZ-phenotype cells and correlates with gene expression characteristic of GC B cells undergoing selection as well as differentiation into the plasma cell compartment. These high GFP-expressing GC B cells exhibit high rates of SHM, confirming their status as bona fide GC resident B cells rather than recent immigrants or contaminating activated B cells. These cells may correspond partly to those GC B cells identified in recent work with a c-myc reporter (15, 21). Whether heterogeneity in BCR signaling as revealed by GFP expression among GC B cells is in turn controlled by limiting antigen, BCR affinity, and/or cell-intrinsic differences in signaling machinery remains to be determined as does the contribution of BCR signaling to affinity maturation. The Nur77-eGFP reporter marks cells undergoing BCR signaling and possibly selection in the germinal center and has great potential as a new tool to further refine our understanding of this complex and dynamic process.
Supplementary Material
Acknowledgements
We thank Chris Allen for critical reading of the manuscript and SHM protocol, Jack Taunton for providing ibrutinib, Wan-Lin Lo for help with Aria sorting, and Al Roque for help with animal husbandry.
Funding: K08 AR059723 (to JZ); Rheumatology Research Foundation R Bridge award (to JZ and MM).
Contributor Information
Mehrdad Matloubian, Email: julie.zikherman@ucsf.edu.
Julie Zikherman, Email: mehrdad.matloubian@ucsf.edu.
References
- 1.Allen CDC, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 3.Tarlinton DM. Evolution in miniature: selection, survival and distribution of antigen reactive cells in the germinal centre. Immunology and Cell Biology. 2008;86:133–138. doi: 10.1038/sj.icb.7100148. [DOI] [PubMed] [Google Scholar]
- 4.Victora GD, Nussenzweig MC. Germinal centers. Annual Review of Immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 5.Chan TD, Brink R. Affinity-based selection and the germinal center response. Immunol Rev. 2012;247:11–23. doi: 10.1111/j.1600-065X.2012.01118.x. [DOI] [PubMed] [Google Scholar]
- 6.Khalil AM, Cambier JC, Shlomchik MJ. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 2012;336:1178–1181. doi: 10.1126/science.1213368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal Center Dynamics Revealed by Multiphoton Microscopy with a Photoactivatable Fluorescent Reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen CDC, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 9.Allen CDC, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. 2004;5:943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 10.Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 13.Zikherman J, Hermiston M, Steiner D, Hasegawa K, Chan A, Weiss A. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. The Journal of Immunology. 2009;182:4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang A, Sullivan BM, Allen CDC. Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity. 2012;36:857–872. doi: 10.1016/j.immuni.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Calado DP, Sasaki Y, Godinho SA, Pellerin A, Köchert K, Sleckman BP, de Alborán IM, Janz M, Rodig S, Rajewsky K. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nature immunology. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen D, Simon T, Sablitzky F, Rajewsky K, Cumano A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response. The EMBO journal. 1988;7:1995–2001. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le T-vL, Kim TH, Chaplin DD. Intraclonal competition inhibits the formation of high-affinity antibody-secreting cells. J Immunol. 2008;181:6027–6037. doi: 10.4049/jimmunol.181.9.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin Immunol. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Oracki SA, Walker JA, Hibbs ML, Corcoran LM, Tarlinton DM. Plasma cell development and survival. Immunological reviews. 2010;237:140–159. doi: 10.1111/j.1600-065X.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 20.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, Buggy JJ. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, Dalla-Favera R. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nature immunology. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.