SUMMARY
DNA methylation is a conserved epigenetic mark that plays important roles in plant and vertebrate development, genome stability, and gene regulation. Canonical Methyl-CpG-Binding Domain (MBD) proteins are important interpreters of DNA methylation that recognize methylated CG sites and recruit chromatin remodelers, histone deacetylases and histone methyltransferases to repress transcription. Here, we show that Arabidopsis MBD7 and Increased DNA Methylation 3 (IDM3) are anti-silencing factors that prevent gene repression and DNA hypermethylation. MBD7 preferentially binds to highly methylated, CG-dense regions and physically associates with other anti-silencing factors, including the histone acetyltransferase IDM1 and the alpha-crystallin domain proteins IDM2 and IDM3. IDM1 and IDM2 were previously shown to facilitate active DNA demethylation by the 5-methylcytosine DNA glycosylase/lyase ROS1. Thus, MBD7 tethers the IDM proteins to methylated DNA, which enables the function of DNA demethylases that in turn limit DNA methylation and prevent transcriptional gene silencing.
Keywords: MBD, IDM1, anti-silencing, DNA methylation, transposable elements
INTRODUCTION
DNA methylation, a conserved epigenetic mark in plants and vertebrates, plays important roles in development, genome stability, and in several well-known epigenetic phenomena such as genomic imprinting, paramutation and X chromosome inactivation (He et al., 2011; Law and Jacobsen, 2010; Tariq and Paszkowski, 2004). In mammals, 5-methylcytosine (5mC) occurs mainly at CG dinucleotides, whereas in plants, 5mC is found in three sequence contexts, CG, CHG, and CHH (where H is A, C, or T) (He et al., 2011; Law and Jacobsen, 2010). In plants, DNA methylation can be established through the RNA-directed DNA methylation (RdDM) pathway (Matzke and Mosher, 2014; Haag and Pikaard, 2011; Zhang and Zhu, 2011). Maintenance of CG and CHG methylation requires the Dnmt1 ortholog MET1, and the plant specific enzyme CMT3, respectively (He et al., 2011; Law and Jacobsen, 2010). CHH methylation is carried out by DRM2 through the RdDM pathway (Matzke and Mosher, 2014) and by CMT2, which requires the chromatin remodeling protein DDM1 (Zemach et al., 2013).
DNA methylation is associated with transcriptional silencing (He et al., 2011; Law and Jacobsen, 2010; Tariq and Paszkowski, 2004). Methyl-CpG-Binding Domain (MBD) proteins are important interpreters of DNA methylation (Hendrich and Bird, 1998). Generally, MBD proteins recognize methylated CG sites and recruit chromatin remodelers, histone deacetylases and histone methyltransferases to repress transcription (Ng et al., 1999). Recent analysis of the genome-wide binding sites of MBD proteins in mammalian cells confirmed that MBD proteins are typically enriched at methylated genomic regions, and methylation-dependent MBD protein binding at promoters coincides with gene silencing (Baubec et al., 2013). The Arabidopsis genome is predicted to encode 13 canonical MBD proteins, although only three of them, MBD5, MBD6 and MBD7, bind specifically to methylated CG sites in vitro (Zemach and Grafi, 2007). The functions of these plant MBD proteins are poorly understood, but the available information is consistent with a conserved role as effectors of DNA methylation that cause transcriptional repression (Zemach and Grafi, 2007).
DNA methylation is dynamically controlled by both methylation and demethylation reactions (Zhu, 2009). In vertebrates, active DNA demethylation is initiated by oxidation or deamination of 5-methylcytosine (5mC) by TET enzymes or AID/APOBECs, respectively followed by the DNA glycosylase TDG or MBD4 (Rai et al., 2008; Kohli and Zhang, 2013), whereas in plants, active DNA demethylation is initiated by the ROS1/DME family of 5mC DNA glycosylases (Zhu, 2009). In Arabidopsis, ROS1 is necessary for preventing DNA hypermethylation at thousands of genomic regions (Qian et al., 2012). How the DNA demethylation enzymes are targeted to specific genomic loci is poorly understood. At a subset of these genomic regions, ROS1 function requires the histone acetyltransferase Increased DNA Methylation 1 (IDM1) (Qian et al., 2012) and its partner protein IDM2 (Qian et al., 2014).
In this study, we employed a previously described forward genetic screen to uncover MBD7 and IDM3 as anti-silencing factors in Arabidopsis. Our screen has identified several anti-silencing factors, such as Anti-silencing 1 (ASI1), Enhanced Downy Mildew 2 (EDM2) and IDM2, in addition to new alleles of known anti-silencing factors, such as IDM1 and ROS1 (Lei et al., 2014; Wang et al., 2013). We found that both MBD7 and IDM3 limit DNA methylation, particularly at transposable elements (TEs). MBD7 is enriched at highly methylated, CG dense sites throughout the genome, and this enrichment coincides with its function in preventing DNA methylation spread or hypermethylation. Protein interaction analyses revealed that MBD7 associates with IDM2 and IDM3 (an IDM2-like protein) in vitro and in vivo, and also associates with IDM1 in vivo. These results suggest a model in which MBD7 recognizes methylated DNA and recruits regulators of active DNA demethylation to prevent DNA methylation spread and transcriptional silencing.
RESULTS
ASI2 is an anti-silencing factor that prevents spread of DNA methylation
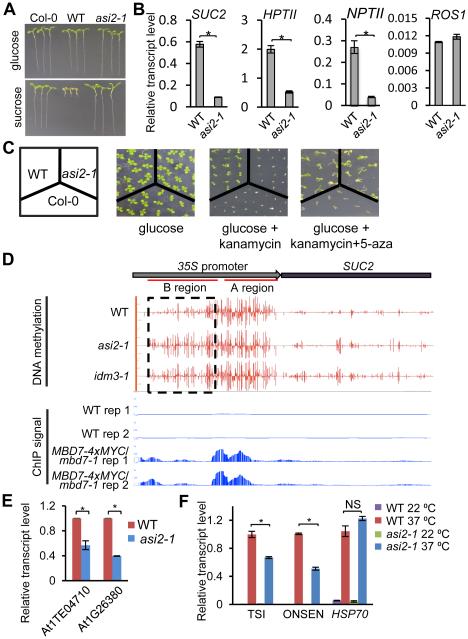
We previously developed a screen for anti-silencing mutants in Arabidopsis which employs wild-type (WT) plants that express a cauliflower mosaic virus 35S promoter-driven sucrose transporter 2 (35S::SUC2) transgene and exhibit a short-root phenotype when grown on medium containing sucrose (Figure 1A, Lei et al., 2014; Wang et al., 2013). Further, these plants express 35S promoter-driven neomycin phosphotransferase II (NPTII) and hygromycin phosphotransferase II (HPTII) transgenes and thus are resistant to kanamycin and hygromycin. We identified a recessive mutant, asi2-1, from a population of WT plants mutagenized with ethylmethane sulfonate. A si2-1 mutant plants developed long roots on sucrose-containing media due to reduced SUC2 expression (Figures 1A, 1B, and S1A) and were sensitive to hygromycin and kanamycin due to reduced expression of HPTII and NPTII (Figures 1B, 1C, S1A and S1B). We performed chromatin immunoprecipitation (ChIP) and observed reduced occupancy of RNA polymerase II (Pol II) and enrichment of the repressive chromatin mark H3K9me2 at the 35S promoter in asi2-1 mutant plants, suggesting that reduced RNA levels are caused by decreased Pol II transcription from this promoter (Figure S1C and S1D).
Figure 1. ASI2 Antagonizes Transcriptional Silencing and Prevents DNA Hypermethylation.
(A) Isolation of the asi2-1 mutant. Transgenic over-expression of the sucrose transporter gene SUC2 in Arabidopsis (ecotype Col-0) results in hypersensitivity to exogenous sucrose, as indicated by the short root phenotype in the transgenic plants (Wild type, WT). Dysfunction of ASI2 rescues the sucrose-inhibited root growth phenotype.
(B) RT-qPCR quantification of transgene transcript levels in asi2-1 in comparison to WT plants. Relative ROS1 transcript level was included as an unchanged control.
(C) ASI2 dysfunction causes a loss of kanamycin resistance in the transgenic plants. Treatment with a DNA methylation inhibitor, 5-aza, restores kanamycin resistance in the asi2-1 mutant.
(D) DNA methylation status and MBD7 enrichment at the 35S::SUC2 transgene in WT, asi2-1 and idm3-1 mutants. Shown are screenshots from IGB (Integrative Genome Browser) display of whole genome bisulfite sequencing and ChIP-seq results. Vertical bars on each track indicate DNA methylation levels or MBD7 enrichment at chromatin. Two replicates of ChIP-seq data from MBD7-4xMYC transgenic plants and from WT control plants are displayed.
(E) RT-qPCR analysis of At1TE04710 and At1G26380 in WT and asi2-1 plants.
(F) ASI1 dysfunction compromises heat-induced expression of ONSEN and TSI but not HSP70.
In (B), (E) and (F), RT-qPCR measurements of transcript levels in the asi2-1 mutant are relative to the corresponding values in the WT. All error bars indicate SD, n = 3.
*P<0.01; NS, not significant (two-tailed t-test). See also Figure S1.
Next, we analyzed DNA methylation of asi2-1 and WT plants by genomic bisulfite sequencing. We found that part of the 35S promoter (region A) is hypermethylated in WT plants, but this hypermethylation extends to the neighboring region (region B) in asi2-1 mutant plants (Figure 1D). Thus, the spread of DNA methylation at the 35S promoter may silence the transgenes in asi2-1 mutant plants. In support of this hypothesis, treatment with the DNA methylation inhibitor 5-aza-2’-deoxycytidine (5-aza) suppressed the kanamycin sensitivity of asi2-1 mutant plants (Figure 1C) and restored the expression of NPTII and SUC2 (Figure S1E).
In plants, transposable elements (TEs) and other repeats are silenced by DNA methylation and repressive histone modifications (He et al., 2011; Law and Jacobsen, 2010; Tariq and Paszkowski, 2004). We found that the expression of At1G26380, a gene in a repetitive gene cluster (Qian et al., 2012), is reduced in asi2-1 (Figure 1E). Similarly, the expression of the TE At1TE04710 is also decreased in asi2-1 (Figure 1E). Expression of some repeats, like ONSEN and TSI, can be induced by prolonged heat treatment (Ito et al., 2011). We observed reduced heat-induced expression of ONSEN and TSI in asi2-1 compared to WT plants (Figure 1F). In addition, At1G26380, At1TE04710, TSI and ONSEN display increased DNA methylation in asi2-1 (Figures S1F). Thus, the recessive asi2-1 mutation causes the DNA hypermethylation and silencing of transgenic and some endogenous loci, thereby indicating an anti-silencing role for ASI2.
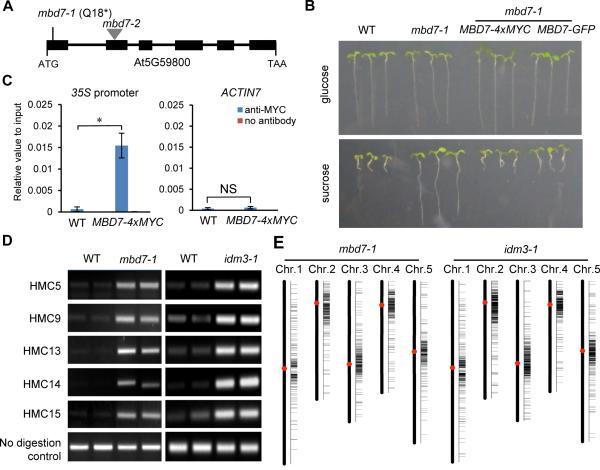
The methyl CpG-binding protein MBD7 is encoded by ASI2
Through map-based cloning, we found that asi2-1 has a C-to-T point mutation in MBD7 (AT5g59800) that creates a premature stop codon (Figure 2A). The long-root phenotype of asi2-1 could be complemented by native promoter-driven MBD7 fused with either a 4xMYC tag or a GFP tag (Figure 2B). RT-PCR assays indicated that the MYC-tagged wild type MBD7 is able to rescue the suppressed expression of 35S::SUC2 and 35S::HPTII transgenes in asi2-1 (Figure S2A). Further, we found that MYC-tagged MBD7 protein was enriched at the 35S transgene promoter by ChIP-qPCR (Figure 2C). These results confirm that the transgene silencing in asi2-1 was caused by the mutation in MBD7. We thus refer to the asi2-1 mutant as mbd7-1 hereafter. We also introduced the 35S::SUC2 transgene into a T-DNA insertion allele of MBD7 (SAIL_697_E08/mbd7-2) by crossing the T-DNA mutant with WT plants. As expected, mbd7-2 mutant plants containing the 35S::SUC2 transgene also showed a long-root phenotype (Figure S2B). These results demonstrate that MBD7 dysfunction causes the transgene silencing phenotypes in plants.
Figure 2. ASI2 Encodes the mCpG-binding Protein MBD7, and asi2-1 and idm3-1 Methylome Analysis.
(A) A diagram of the MBD7 gene showing the mutation site in mbd7-1 and the T-DNA insertion position in the mbd7-2 allele. Boxes and lines denote exons and introns, respectively.
(B) The long root phenotype in asi2-1 is caused by MBD7 mutation. Transgenic expression of MBD7 driven by its native promoter restored the short root phenotype in mbd7-1.
(C) Examination of MBD7 enrichment at the 35S transgene promoter using ChIP assay. Promoter of ACTIN7 was used as a control. All error bars indicate SD, n = 3. *P<0.01; NS, not significant (two-tailed t-test).
(D) Chop-PCR validation of hyper-DMRs identified in mbd7-1 and idm3-1. Results of two biological replicates are shown. Methylation-sensitive restriction enzymes and primers are listed in Table S1.
(E) hromosomal distribution of hyper-DMRs in mbd7-1 and idm3-1. See also Figure S2.
The mbd7-1 mutation induces DNA hypermethylation at endogenous genomic regions
Whole-genome bisulfite sequencing identified 1390 differentially methylated regions (DMRs) in mbd7-1, among which 1144 are hypermethylated and 246 are hypomethylated. Five hypermethylated cytosines (HMC) in mbd7-1 were selected for validation by Chop-PCR, an assay in which genomic DNA is digested with a methylation-sensitive restriction endonuclease and then tested as a template for PCR with primers flanking the restriction sites. We observed amplification from genomic digests of mbd7-1 but not WT DNA (Figure 2D), consistent with hypermethylation of the candidate sites in mbd7-1. The greater number of hypermethylated than hypomethylated regions in mbd7-1 suggests that MBD7 mainly antagonizes DNA methylation.
We found that about 85% (974) of the hypermethylated DMRs in mbd7-1 are located in TEs, with no obvious preference for any specific type (Figure S2C). The DNA methylation levels are increased at these TE DMRs in CG, CHG and CHH sequence contexts, and the increase is independent of TE length (Figure S2D).
In mbd7-1 mutant plants, hypermethylated DMRs are concentrated near centromeric and pericentromeric regions (Figure 2E). To test whether MBD7 preferentially regulates TEs located at these regions, we compared the number of hypermethylated DMR-associated TEs to the total number of TEs across consecutive 500-kb windows. This density distribution of hypermethylated TEs across five chromosomes showed no obvious preference for centromeric or pericentromeric TEs, but rather that the concentration of hypermethylated TEs within these regions corresponds to a higher total number of TEs (Figure S2E).
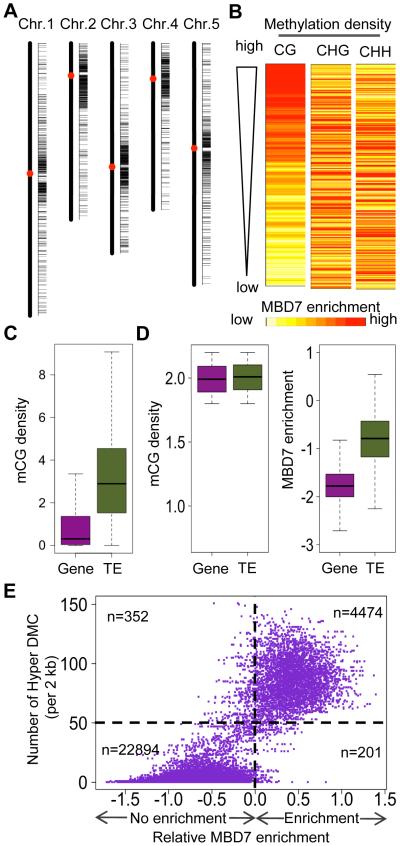
MBD7 associates with mCG-dense genomic regions
MBD7 was previously identified as a methyl CpG-binding protein (Zemach and Grafi, 2007), however its genomic targets are not known. To identify endogenous MBD7 targets, we performed ChIP using plants expressing native promoter-driven MBD7-4xMYC followed by high throughput sequencing (ChIP-seq). We identified 2452 peaks of MBD7 enrichment on chromatin; 1930 (78.7%) are within TE regions, 94 (3.8%) are within intergenic regions, 401 (16.4%) correspond to genes (202 of the genes contain annotated repeats). The pattern of chromosomal distribution of MBD7 binding peaks is similar to that of mbd7-1 hyper-DMRs (Figures 3A and 2E), and approximately 63% of the mbd7-1 hyper-DMRs overlap with MBD7 binding peaks.
Figure 3. MBD7 preferentially binds to regions with high mCG density and the binding coincides with its role in preventing DNA hypermethylation.
(A) hromosomal distribution of MBD7 binding peaks.
(B) Correlation of MBD7 enrichment with DNA methylation density. The top 1% of MBD7-enriched regions was selected for analysis. For each region, MBD7 enrichment was represented by color from light yellow (lowest) to red (highest). Methylation density was examined in the contexts of CG, CHG, and CHH.
(C) Comparison of CG methylation density between genes and TEs in the Arabidopsis genome.
(D) omparison of MBD7 chromatin enrichment (right panel) at a subset of Gene and TE loci that show similar mCG densities in the range of 1.8-2.2 (left panel).
(E) Correlation of MBD7 enrichment with DNA hypermethylation in mbd7-1 mutant plants. For each 2 kb genomic region, the relative MBD7 enrichment and number of hyper DMCs in mbd7-1 are calculated. The numbers of displayed 2 kb genomic regions with or without MBD7 enrichment and with >50 or <50 hyper DMCs are indicated.
See also Figures S3.
We examined the correlation between MBD7 binding and DNA methylation in CG, CHG, and CHH contexts. First, the genome was divided into 1-kb regions, and MBD7 enrichment was calculated for each region. The top 1% of MBD7-enriched regions was selected for further analysis. To evaluate the influences of DNA methylation level on MBD7 enrichment, we ranked these regions by CG, CHG, and CHH methylation levels, and generated a heat map of MBD7 enrichment. However, no pattern of MBD7 enrichment was evident for any of the three ranks (data not shown), suggesting that the methylation level of individual CG, CHG or CHH motifs does not determine MBD7 binding.
Methylation density is a value reflecting both methylation level and methyl cytosine density for a region, and it is equal to the sum of methylation percentages of individual cytosines in a region normalized by the region length (Baubec et al., 2013). To consider not only methylation level but also the methyl cytosine density of a region, we calculated the methylation density for 1-kb genomic regions. When these regions were ranked by mCG, mCHG, and mCHH densities, the heat map pattern indicated that MBD7 enrichment is associated with CG methylation density rather than with CHG and CHH methylation densities (Figure 3B), which is consistent with previous in vitro finding that MBD7 can bind to mCG sites but not to mCHG sites (Zemach and Grafi, 2007). The tight relationship between MBD7 enrichment and CG methylation density genome-wide (instead of the top 1%) was supported by an analysis using a previously described method (Baubec et al., 2013), where all 1-kb genomic regions were ranked from left to right by their CG methylation density, and their MBD7 enrichment and CG methylation density values are indicated by red and blue lines, respectively (Figure S3A). When the same method was used, no correlation was observed between MBD7 enrichment and methylation density in either the CHG or CHH context (data not shown).
The majority of MBD7 binding sites are in TE regions. We analyzed the CG methylation density of all TE-associated 1-kb windows (TE windows) and all gene-associated 1-kb windows (gene windows), and found that TE windows generally have higher CG methylation density than gene windows (Figure 3C), as expected. To test whether the TE-preferred binding of MBD7 is due to the higher CG methylation density of TE or due to other TE-specific features, we selected TE windows and gene windows with comparable CG methylation density and then compared their MBD7 enrichment (Figure 3D). We found a greater enrichment for MBD7 in TEs than in genes, even when they had comparable mCG densities (Figure 3D); this pattern was consistent for all the ranges of mCG density we tested (Figure S3C). This result suggests that MBD7 may not bind to all regions with a high mCG density, but instead may recognize and bind to mCGs located in TEs.
MBD7 binding coincides with its role in preventing DNA hypermethylation
To investigate the correlation of MBD7 binding with its influence on DNA methylation, we calculated MBD7 enrichment and the number of hyper differentially methylated cytosines (DMCs) in the mbd7-1 mutant for each 2-kb genomic region. Interestingly, the numbers of hyper differentially methylated cytosines (DMCs) in the mbd7-1 mutant positively correlated with MBD7 enrichment; most of the 2-kb bin genomic regions with MBD7 enrichment (4474) have higher numbers of hyper DMCs (>50 per 2 kb bin), compared to the numerous 2-kb bin genomic regions without MBD7 enrichment (22894) (Figure 3E). The result indicates that MBD7 binding to chromatin coincides with its function in preventing DNA hypermethylation.
To further understand the influence of MBD7 on DNA methylation at its binding sites, we compared the methylation levels between the WT and mbd7-1 at MBD7-binding regions. Two groups of control regions were randomly selected, and their methylation levels were also calculated. In mbd7-1, mCHG, mCHH as well as mCG levels were elevated at MBD7-binding regions but not in control regions (Figure S3C). Boxplot analysis indicated that the increase in average methylation levels at the MBD7-binding regions is due to increases in the majority of the regions rather than extremely large increases in a small number of regions (Figure S3D).
The ChIP-seq data also showed an enrichment of MBD7 protein in the 35S promoter region (Figure 1D), which is consistent with the ChIP-qPCR result (Figure 2C). In addition, the ChIP-seq data revealed MBD7 enrichment at At1G26380, At1TE04710, ONSEN and TSI regions (Figure S1F), which had increased DNA methylation in mbd7-1. These results suggest that physical binding of MBD7 at these loci attenuates the silencing of these loci (Figure 1E and 1F) by preventing DNA hypermethylation.
idm3 mutants phenocopy mbd7 mutants
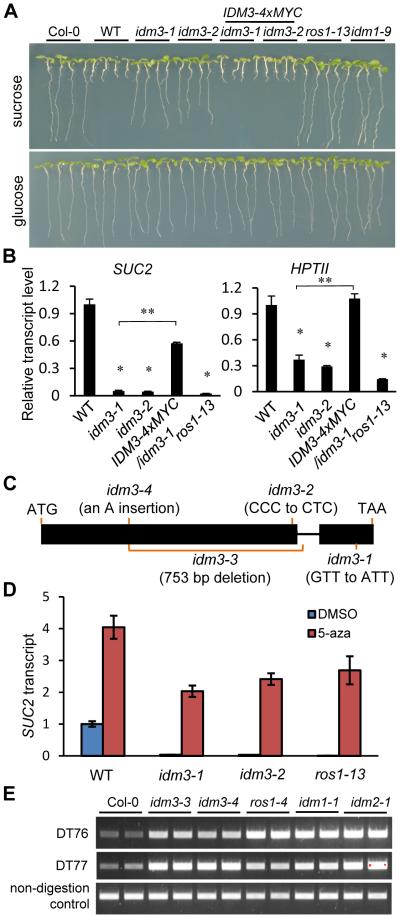
From the same mutant screen, we isolated idm3-1 (Increased DNA Methylation 3-1) and idm3-2, which showed a long-root phenotype on sucrose-containing media and reduced SUC2 and HPTII transcript levels (Figure 4A and 4B). Map-based cloning of idm3-1 and idm3-2 identified missense mutations in At1G20870 (Figure 4C). To further confirm that the mutations in At1G20870 are responsible for the anti-silencing defects in the idm3 mutants, we transformed a native promoter-driven wild type At1G20870 gene fused with 4xMYC into the idm3 mutants, and found that the root growth as well as SUC2 and HPTII transgene expression phenotypes were rescued (Figures 4A and 4B). IDM3 is predicted to encode an alpha-crystallin domain protein localized to the nucleus (Figure S4A), and is similar to IDM2 (Qian et al., 2014) (Figure S4B). Although both IDM2 and IDM3 belong to the family of small heat shock proteins, like IDM2 (Qian et al., 2014), IDM3 expression is not heat inducible (Figure S4C).
Figure 4. IDM3 Functions in Preventing Transcriptional Silencing.
(A) The idm3 mutants were isolated based on its long root phenotype when grown with exogenous sucrose. Transgenic expression of the wild type IDM3 gene rescued idm3 root growth phenotype. The ros1-13 and idm1-9 were used as controls.
(B) IDM3 dysfunction suppresses SUC2 and HPTII transgene expression. RT-qPCR results are means ± SD of three biological replicates where the fold changes are normalized to transcript levels in WT. *P<0.01 compared to WT; NS, not significant compared to WT; **P<0.01 compared to idm3-1 (two-tailed t-test).
(C) A diagram of the IDM3 gene showing the mutation sites in idm3 mutants. Boxes and lines denote exons and introns, respectively.
(D) Transcriptional silencing of 35S::SUC2 in idm3 mutants can be released by chemical inhibition of DNA methylation. Plants were treated with the DNA methylation inhibitor 5-aza or the solvent DMSO as a control. Gene expression levels were normalized to that of DMSO-treated WT plants. RT-qPCR results are means ± SD of three biological replicates, where the fold changes are normalized to transcript levels in WT.
(E) Analysis of DNA methylation levels at the DT76 and DT77 loci in idm3-3 and idm3-4 mutants by Chop-PCR.
See also Figure S4.
Genomic bisulfite sequencing of idm3-1 mutant plants revealed that DNA methylation spread to neighboring sequence in the 35S transgene promoter (Figure 1D). Consistent with the notion that increased DNA methylation was responsible for the silencing of the transgenes, expression of the 35S::SUC2 transgene was restored in idm3 mutants after treatment with the cytosine methylation inhibitor 5-aza, as revealed by RT-qPCR (Figure 4D). The 5-aza treatment also increased the expression of the 35S::SUC2 transgene in the WT (Figure 4D). The result suggests that the transgenes were already subjected to some DNA methylation-dependent silencing in WT plants, which is consistent with the high level of DNA methylation observed within region A of the 35S promoter (Figure 1D).
As in mbd7-1, there were many more hyper-DMRs (1703) than hypo-DMRs (274) in idm3-1. Approximately 47% (799) of the hyper-DMRs are also hypermethylated in mbd7-1. Chop-PCR analysis confirmed that several endogenous genomic sites were hypermethylated in idm3-1 mutant plants, as in mbd7-1 (Figure 2D). Because we suspected that the idm3-1 and idm3-2 point mutations represent weak alleles, and also because we could not find any T-DNA insertion alleles of idm3, we used the CRISPR/Cas system (Mao et al., 2013) to generate two new alleles, idm3-3 and idm3-4. In idm3-3, a 753 bp fragment was deleted from the IDM3 gene. In idm3-4, a frameshift mutation at the 5’ region of IDM3 created a premature stop codon (Figure 4C). We examined DNA methylation by Chop-PCR at two loci, DT76 and DT77 (Qian et al., 2012), in idm3-3 and idm3-4. The amplification from genomic digests of the mutants but not WT DNA indicates that these loci were hypermethylated in idm3-3 and idm3-4 mutants, as they were in ros1-4, idm1-1 and idm2-1 (Figure 4E). Thus, like MBD7, IDM3 is required to prevent transgene silencing and DNA hypermethylation.
MBD7 interacts with IDM2 and IDM3
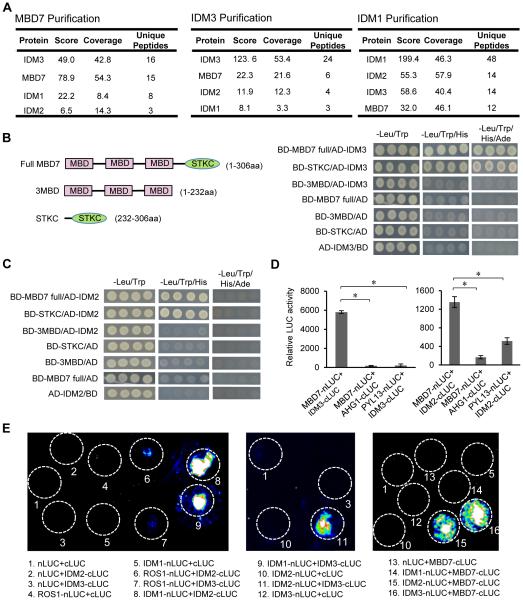
In order to understand how MBD7 functions, we identified MBD7-interacting proteins by performing immunoprecipitation (IP) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with mbd7-1 mutant plants complemented with native promoter-driven MBD7-4xMYC. WT plants not expressing MBD7-4xMYC served as controls. Of the proteins co-precipitated with anti-MYC antibodies in the MBD7-4xMYC transgenic plants but not in the control plants, four had been uncovered from the 35S::SUC2 transgene-based screen as anti-silencing factors: MBD7 itself, IDM1 (Qian et al., 2012), IDM2 (Qian et al., 2014) and IDM3 (Figure 5A). Similar IP-LC-MS/MS experiments were carried out using idm3-1 mutant plants complemented with native promoter-driven IDM3-4xMYC. In the anti-MYC immunoprecipitates, we identified not only IDM3 itself but also MBD7, IDM2 and IDM1 (Figure 5A). In addition, in anti-GFP immunoprecipitates from idm1-1 mutant plants complemented with native promoter-driven IDM1-GFP-HA, we found IDM1 itself and IDM2, IDM3 and MBD7 (Figure 5A). These results show that MBD7 is associated with IDM3, IDM2 and IDM1 in vivo.
Figure 5. MBD7 Physically Interacts with IDM2 and IDM3.
(A) Detection of proteins that associate with MBD7, IDM3 or IDM1. Proteins were detected by LC-MS/MS following immunoprecipitation of MYC-tagged MBD7, MYC-tagged IDM3 or GFP-tagged IDM1. There were two biological replicates for each of the protein purifications, one representative result is shown here.
(B) Tests of IDM3-MBD7 interaction by yeast two-hybrid assays. Full-length protein (MBD7 full) and truncated forms (3MBD and STKC) of MBD7 were fused with the GAL4 binding domain (BD). Full-length IDM3 was fused with the GAL4 activation domain (AD).
(C) Tests of IDM2-MBD7 interaction by yeast two-hybrid assays. Full-length protein (MBD7 full) and truncated forms (3MBD and STKC) of MBD7 were fused with BD. Full-length IDM2 was fused with AD.
(D) Analyses of IDM2-MBD7 and IDM3-MBD7 protein interactions by split luciferase complementation assays in Arabidopsis protoplasts. The co-transfected GUS gene was used to standardize protoplast transfection efficiency. Protoplasts expressing the tested protein and proteins unrelated to gene silencing including AHGI (ABA-hypersensitive germination 1) and PYL13 (PYR1-like protein 13) served as negative controls.
(E) Examination of protein interactions between ROS1, IDM1, IDM2, IDM3 and MBD7 by split luciferase complementation assays in tobacco (N. benthamiana) leaves. Luciferase activities were detected at 48 hours post infiltration. White circles indicate leaf region that were infiltrated with Agrobacterium strains containing the indicated constructs.
See also Figure S5.
Yeast two-hybrid (Y2H) assays were used to determine whether MBD7 interacts directly with IDM1, IDM2, and IDM3. Yeast cells expressing binding domain (BD)-MBD7 and activation domain (AD)-IDM3 were able to grow on both minus-three media (media lacking leucine, tryptophan, and histidine) and minus-four media (media lacking leucine, tryptophan, histidine and adenine) (Figure 5B), suggesting a strong interaction between MBD7 and IDM3. BD-MBD7 and AD-IDM2 co-transfected yeast cells grew on minus-three media but not on minus-four media (Figure 5C), indicating a relatively weaker interaction between MBD7 and IDM2. MBD7 did not interact with IDM1 in the Y2H assay (data not shown). The interaction between MBD7 and IDM3 was confirmed in a split luciferase complementation assay in protoplasts (Figure 5D). The split luciferase complementation assay also indicated an interaction between MBD7 and IDM2, although this interaction is much weaker than that between MBD7 and IDM3 as indicated by the relative luciferase activities (Figure 5D).
The MBD7 interactions with IDM3 and IDM2 were also confirmed using a split luciferase complementation assay in tobacco leaves (Figure 5E). Interestingly, IDM3 also interacted with IDM2 in the assay. We observed an interaction between IDM2 and IDM1 in the assay, which is consistent with previous results (Qian et al., 2014; Zhao et al., 2014). Like IDM2, IDM3 also interacted with IDM1 in the split luciferase complementation assay (Figure 5E). The Y2H and split luciferase complementation results show that MBD7 interacts with IDM3 and IDM2, and that IDM3 interacts with IDM2 and IDM1. These results are consistent with our IP-LC-MS/MS data (Figure 5A) that showed MBD7 association with IDM3, IDM2 and IDM1 in vivo.
MBD7 contains three MBD domains and a C-terminal domain known as the StkC domain (Figure 5B). We performed the Y2H assay with different deletion mutants and found that both IDM2 and IDM3 bind to the StkC domain of MBD7 but not the three MBD domains (Figures 5B and 5C). To test whether StkC, the protein interaction domain, is important for the anti-silencing function of MBD7, we separately transformed the native promoter-driven full length MBD7 coding sequence (proMBD7::MBD7-4xMYC) and native promoter-driven MBD7 containing only the three MBD domains (proMBD7::3MBD-4xMYC) into mbd7-1. Unlike the full length MBD7 protein, the three MBD domains could not rescue the root phenotype of mbd7-1 (Figure S5A), indicating that the StkC domain is required for the anti-silencing function of MBD7, at least at the transgene loci.
Our IP-LC-MS/MS results indicated an association between MBD7 and IDM1 in vivo (Figure 5A). We showed previously that IDM1 is a histone H3 acetyltransferase that catalyzes H3K18 and H3K23 acetylation (Qian et al., 2012), and that H3K18ac and H3K23ac marks are reduced at the silenced 35S promoter in idm1 mutant plants (Qian et al., 2014). ChIP-qPCR assays revealed that H3K18ac and H3K23ac levels are reduced in mbd7-1 mutant plants at the 35S promoter, as in idm1-9 mutants (Figures S5B and S5C). The results are consistent with the notion that MBD7 and IDM1 are in the same protein complex in vivo, and suggest that MBD7 is important for IDM1 function in plants.
MBD7 affects a subset of genomic regions targeted for active DNA demethylation
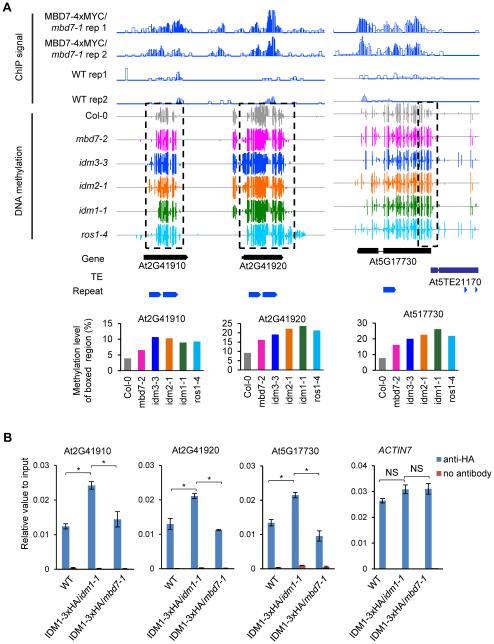
IDM1 and IDM2 have been shown to be required to prevent the hypermethylation of hundreds of genomic regions that are a subset of the targets of active demethylation by ROS1, DML2 and DML3 (Qian et al., 2012; 2014). We previously determined the methylomes of the idm1-1, idm2-1 and rdd (ros1, dml2 and dml3 triple mutant) mutants. We found that these methylomes could not be compared to those of the mbd7-1 and idm3-1 mutants because the genetic background for these mutants and their WT control (containing the 35S::SUC2 transgene) was twice subjected to Agrobacterium-mediated transformation and contains several transgenes (see Experimental Procedures) and even the WT control methylome is very different from that of Col-0 control (data not shown). We therefore determined the DNA methylomes of mbd7-2 and idm3-3, which are in the standard Col-0 background as idm1-1, idm2-1 and rdd. We found 1014 and 1725 DMRs with increased DNA methylation (hyper-DMRs) in mbd7-2 and idm3-3, respectively. Approximately 77% (776) of the hyper-DMRs in mbd7-2 overlap with the hyper-DMRs in rdd, 61% (613) overlap with the hyper-DMRs in ros1-4, 44% (441) overlap with the hyper-DMRs in idm1-1, 46% (461) overlap with the hyper-DMRs in idm2-1, and 42% (425) overlap with the hyper-DMRs in idm3-3. Several examples of shared hyper-DMRs in the mbd7-2, idm1-1, idm2-1, idm3-3 and ros1-4 mutants are shown in Figures 6A and S6A-D, which also display ChIP-seq results showing MBD7 enrichment levels in these regions. Furthermore, we examined IDM1 occupancy at some of these loci using ChIP against 3xHA tagged IDM1. The ChIP assay indicated that IDM1 protein was enriched in the chromatin at the tested loci in WT plants, but the enrichment was substantially reduced in mbd7-1 mutant plants (Figure 6B and S6E). These results suggest that IDM1 binding to target loci is MBD7-dependent.
Figure 6. Shared Hyper-DMRs between mbd7-2 and Other Anti-silencing Mutants and the Effect of mbd7 on IDM1 Binding to Chromatin.
(A) Levels of DNA methylation and MBD7 chromatin enrichment at several shared hyper-DMRs (dashed boxes) in the mbd7-2, idm3-3, idm2-1, idm1-1 and ros1-4 mutants. Total DNA methylation levels of boxed regions in each mutant were quantified from the whole-genome bisulfite sequencing data and shown in the bottom panel.
(B) Effect of mbd7 on IDM1 protein enrichment at the shared hyper-DMRs. ChIP against IDM1-3xHA was performed in WT, IDM1-3xHA/idm1-1 and IDM1-3xHA/mbd7-1. Promoter of ACTIN7 was used as a control region. All error bars indicate SD, n = 3. *P<0.01; NS, not significant (two-tailed t-test).
See also Figure S6.
Our results suggest that MBD7 affects only a specific subset of genomic regions that require IDM1 or IDM2 for DNA demethylation. To understand how MBD7 may achieve this specificity, we analyzed the CG methylation density of the different groups of hyper-DMRs. In idm1, idm2 and idm3 mutants, the hyper-DMRs that overlap with those in mbd7-2 have higher CG methylation density than the non-overlapping hyper-DMRs. Simulations with randomly selected regions do not show a significant difference (Figure S6F). The results suggest that MBD7 functions together with IDM1 and IDM2 preferentially at genomic regions with a high CG methylation density, consistent with the requirement of high mCG density for MBD7 binding. Indeed, analyses of the MBD7 ChIP-seq data showed that MBD7 is more highly enriched at the dense mCG/overlapping hyper-DMRs than at the non-overlapping hyper-DMRs (Figure S6G). No significant difference was found using the control ChIP-seq data from WT plants (Figure S6G).
DISCUSSION
We have found that MBD7 and IDM3 are two cellular anti-silencing factorsthat inhibit DNA hypermethylation at some genomic regions and prevent transcriptional gene silencing. DNA methylation is a conserved epigenetic mark which silences TEs and other invasive elements (He et al., 2011; Law and Jacobsen, 2010). Many plant and animal genomes have abundant TEs, such that the genes are islands in the sea of TEs (Bennetzen and Wang, 2014). Methylation at TEs may spread to and silence adjacent, transcriptionally active genes. Active DNA demethylation is one mechanism for preventing DNA methylation spread to protect nearly genes. For example, the EPF2 gene, which controls the size of the stomatal stem cell population in leaf epidermis, is close to a methylated TE in Arabidopsis, and active DNA demethylation is required to prevent methylation spreading and transcriptional silencing of EPF2 (Yamamuro et al., 2014). In the DNA demethylase ros1 mutants, EPF2 is silenced by DNA methylation that spread from the proximal TE, resulting in an over-production of stomatal lineage cells (Yamamuro et al., 2014). Many imprinted genes in plants require the DNA demethylase DME for DNA demethylation and expression in the endosperm because these genes evolved to have TEs in or near their regulatory sequences (Gehring et al., 2009). Our findings reveal that MBD7 associates with methylated DNA and recruits other anti-silencing factors to create a permissible chromatin environment for binding of DNA demethylases such as ROS1. Thus, our results suggest that MBD7 and the IDM proteins limit the spread of DNA methylation by promoting active DNA demethylation.
Active DNA demethylation is also necessary for pruning the DNA methylation landscape of many TEs and is thus important for preventing over-silencing of TEs (Zhu et al., 2007). In addition, it has been known for a long time that DNA methylation can cause transcriptional silencing of transgenes in plants (Matzke et al., 1989). It is important to understand how transcriptional silencing of transgenes can be avoided or prevented in order to keep transgenic traits stable in the agricultural biotechnology industry.
The key enzymes for active DNA demethylation have been identified in recent years. In vertebrates, active DNA demethylation is initiated by deamination or oxidation of 5mC by AID/APOBECs or TET enzymes, respectively followed by the DNA glycosylase TDG or MBD4 (Rai et al., 2008; Kohli and Zhang, 2013), whereas in plants, active DNA demethylation is initiated by the ROS1/DME family of 5mC DNA glycosylases (Zhu, 2009). How the DNA demethylation enzymes are targeted to specific genomic loci is poorly understood. In plants, the histone acetyltransferase IDM1 is required for targeting ROS1 to a subset of genomic regions for demethylation (Qian et al., 2012). Although it is not known how the H3K18 and H3K23 acetylation marks created by IDM1 affect ROS1 targeting, this regulation may be considered an “acetylation switch”, analogous to the model proposed for targeting of the chromatin remodeling complex SWR1 in yeast (Ranjan et al., 2013). Like IDM1, IDM2 regulates the demethylation of a similar subset of genomic loci targeted by the ROS1 family of demethylases (Qian et al., 2014). IDM2/ROS5 is a nuclear alpha-crystallin domain (ACD) protein that interacts with IDM1and affects its H3K18 acetylation activity (Qian et al., 2014; Zhao et al., 2014). Interestingly, we have now identified IDM3, an ACD protein that is closely related to IDM2 and is also required to prevent DNA hypermethylation and gene silencing.
MBD7 is an MBD protein that contains three MBD domains and a C-terminal Stkc domain. Consistent with its ability to bind methylated CpGs in vitro (Zemach and Grafi, 2007), our ChIP-seq assays revealed that genome-wide binding of MBD7 correlates with the density of methylated CG but not methylated CHG or CHH sites. Our results suggest that MBD7 does not bind to all regions with a high mCG density, but instead may preferentially recognize and bind to mCGs located in TEs. It is possible that in addition to high mCG density, TE features such as heterochromatic histone marks may also contribute to MBD7 binding. We showed that MBD7 binding is coincident with the role of MBD7 in preventing DNA hypermethylation. Therefore, instead of reading the DNA methylation signal to cause silencing, MBD7 interprets the DNA methylation signal to avoid DNA methylation spread or hypermethylation to prevent silencing.
MBD7 interacts directly with both IDM2 and IDM3. Like IDM2 (Qian et al., 2014; Zhao et al., 2014), IDM3 also interacts with IDM1. In addition, IDM2 and IDM3 interact with each other. Our IP-LC-MS/MS results show that MBD7 is associated with IDM1 as well as with IDM2 and IDM3 in vivo. The IP-LC-MS/MS results indicated that the amounts of IDM1 and IDM2 that were co-immunoprecipitated with MBD7 or IDM3 were low, compared to the amounts of IDM3 or MBD7 (Figure 5A). The results suggest that while MBD7 and IDM3 may exist in a tight complex, IDM2 and IDM1 are more loosely or transiently associated with the complex. In addition to having three MBD motifs, MBD7 also contains a Stkc domain that is conserved in plant MBD7 orthologs but is not found in other proteins. Our results show that the Stkc domain is necessary for MBD7 function in anti-silencing in plants, consistent with the Stkc domain being responsible for mediating the interaction of MBD7 with IDM2 and IDM3.
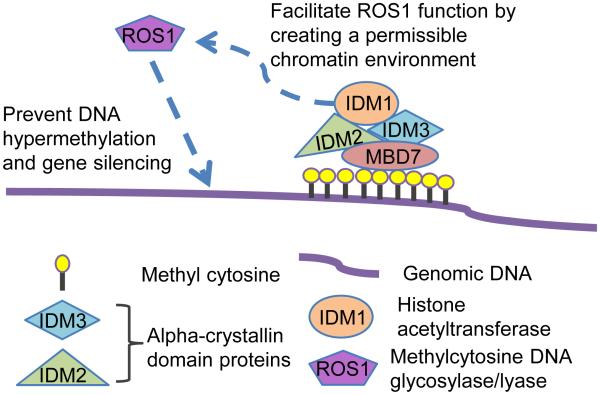
Our results suggest a model in which MBD7 binds to methylated TEs and other repeats through its MBD motifs, and uses its Stkc domain to bind to IDM3 and IDM2, thus bringing IDM1 to the methylated DNA (Figure 7). The H3K18ac and H3K23ac marks created by IDM1 then allow ROS1 and related DNA demethylases to be recruited to restrict methylation spread or to prevent hypermethylation by active demethylation (Figure 7). Consistent with this model, IDM1 recruitment to target genomic regions is dependent on MBD7 (Figures 6B and S6E), and H3K18ac and H3K23ac marks are reduced in mbd7 mutant plants, as in idm1 plants, at the 35S promoter (Figures S5B and S5C). It is interesting that IDM1 also contains an MBD domain that can bind methylated CG in vitro (Qian et al., 2012). It seems that the single MBD domain of IDM1 is not sufficient to bring IDM1 to some of its target genomic regions, and the targeting requires MBD7 that contains multiple MBD domains. Nevertheless, the single MBD domain may help anchor IDM1 to the genomic sites once it is recruited by MBD7.
Figure 7. A Working Model of the functions for MBD7 and IDM3 in Anti-silencing.
MBD7 binds to highly methylated genomic regions through its MBD motifs, and uses its Stkc domain to bind to IDM3 and IDM2, thus bringing IDM1 to the methylated DNA. The H3K18ac and H3K23ac marks created by IDM1 then allow ROS1 and related DNA demethylases to be recruited to curb methylation spread or to prevent hypermethylation by active demethylation.
Our results suggest that MBD7 and the three IDM proteins form a previously unknown anti-silencing complex that promotes DNA demethylation. Protein complexes that promote DNA methylation and transcriptional gene silencing have been well studied (Matzke and Mosher, 2014). In contrast, nothing has been known in any system about protein complexes that function in regulating DNA demethylation. Our results suggest that MBD7 binds to highly methylated genomic regions through its MBD motifs, and uses its Stkc domain to bind to IDM3 and IDM2, thus bringing IDM1 to the methylated DNA (Figure 7). The H3K18ac and H3K23ac marks created by IDM1 then allow ROS1 and related DNA demethylases to be recruited to restrict methylation spread or to prevent hypermethylation by active demethylation (Figure 7). MBD7 may also recruit histone modification enzymes other than IDM1 to help target the DNA demethylases since IDM1 only affects a subset of genomic regions demethylated by ROS1 and related 5mC DNA glycosylases. It is possible that at some genomic regions MBD7 may affect DNA methylation by preventing the establishment or maintenance of DNA methylation rather than by facilitating active DNA demethylation. On the other hand, not all genomic regions subjected to active DNA demethylation are affected by MBD7, indicating the presence of MBD7-independent mechanisms for targeting the DNA demethylases.
EXPERIMENTAL PROCEDURES
Plant materials, mutant screening and map-based cloning
Wild-type (WT) in this study refers to transgenic plants expressing the phosphate starvation responsive AtPT2 promoter driven firefly luciferase reporter gene and 35S::NPTII (Karthikeyan et al., 2002) that were later transformed with a construct containing the CaMV 35S promoter-driven sucrose transporter 2 (35S::SUC2) and 35S::HPTII transgenes (Lei et al., 2011). An EMS-mutagenized pool of plants was generated and screened for mutants with a long-root phenotype (Wang et al., 2013). M2 seedlings were grown vertically on ½ MS plates with 2% sucrose and 1% agar. WT plants exhibit severely suppressed root growth on this medium, and we screened for mutants with long-root phenotype among 7-day-old seedlings. Genetic mapping and gene cloning was performed as described previously (Lei et al., 2014). Details of plant materials and gene cloning are provided in Supplemental Experimental Procedures.
RNA analysis and Chop-PCR
RNA analysis and Chop-PCR assays were carried out according to Lei et al (2014). Detailed procedures are described in the Extended Experimental Procedures. Primer information is included in Table S1.
Whole-genome bisulfite sequencing and data analysis
Fourteen-day-old seedlings were used for extraction of genomic DNA. Bisulfite conversion, library construction, and deep sequencing were performed by the Beijing Genomics Institute (BGI) in Shenzhen, China. DMRs were identified according to Qian et al. (2012) with some modifications, which are detailed in Supplemental Experimental Procedures.
Chromatin immuno-precipitation (ChIP) assay
The ChIP assay was performed as described (Wierzbicki et al., 2008). Dynabeads (Invitrogen, cat# 10003D) were used for pre-clearing and antibody binding. The antibodies were anti-H3K9me2 (Abcam, cat# ab1220), anti-H3K18ac (Abcam, cat# ab1191), anti-H3K23ac (Millipore, cat#07-355), anti-MYC (Millipore, cat#05-724), and anti-Pol II (Abcam, cat#ab5408).
MBD7 ChIP-seq and data analysis
proMBD7::gMBD7-4xMYC transgenic plants were used for ChIP-seq, with WT plants as controls. ChIP was performed according to a previously published protocol (Wierzbicki et al., 2008). ChIP samples were sent to the Genomics Core Facilities of the Shanghai Center for Plant Stress Biology, SIBS, CAS (Shanghai, China) for library construction and Illumina sequencing.
MBD7 binding Peaks were called with SICER (Zang et al., 2009), which compared the two replicates of MBD7-Myc ChIP samples with the two replicates of wild-type control. The MBD7 protein enrichments were calculated according to Baubec et al. (Baubec et al., 2013) as follows: Enrichment = log2( 8+ n_ChIP ) − log2( 8 + n_Input * N_ChIP/N_Input ), where n_ChIP and n_Input represent the number of mapped ChIP and input tags in the corresponding 1-kb bin, and N_ChIP and N_Input are the sums of all mapped tags. In Figure 3E, the relative enrichment value is calculated as: log2(MBD7 enrichment value) − log2(WT enrichment value). Detailed methods are described in Supplemental Experimental Procedures.
Yeast two-hybrid assay
Coding sequences of IDM2, IDM3, MBD7 and truncated MBD7 (3MBD or STKC) with stop codon were cloned into pDEST22 or pDEST32. Yeast two-hybrid assays were performed as described (Bai et al., 2013).
IP and LC-MS/MS analysis
For immunoprecipitation (IP), 5 g of floral tissues for each epitope-tagged transgenic line were used. Dynabeads (Thermo Fisher Scientific) conjugated with MYC antibody (Millipore, cat#05-724) were used for IP. For affinity purification as described in Law et al (2010), the protein samples were subjected to LC-MS/MS analysis as detailed in Supplemental Experimental Procedures.
Split luciferase complementation assays
Split luciferase complementation assays were performed in tobacco leaves and Arabidopsis protoplasts, the details of which are provided in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 and by the Chinese Academy of Sciences (to J.-K. Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All high-throughput sequencing data generated in this study were submitted to NCBI’s Gene Expression Omnibus (GSE58789, GSE58787 and GSE59712). Additional datasets used in this study are GSE33071 (Qian et al., 2012) and GSE49421 (Qian et al. 2014).
REFERENCES
- Bai G, Yang DH, Zhao Y, Ha S, Yang F, Ma J, Gao XS, Wang ZM, Zhu JK. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 2013;83:651–664. doi: 10.1007/s11103-013-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Ivanek R, Lienert F, Schübeler D. Methylation-dependent and-independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Wang H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014;65:505–530. doi: 10.1146/annurev-arplant-050213-035811. [DOI] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D'Urzo MP, Damsz B, Raghothama KG. Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 2002;130:221–233. doi: 10.1104/pp.020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, La H, Lu K, Wang P, Miki D, Ren Z, Duan CG, Wang X, Tang K, Zeng L, et al. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc. Natl. Acad. Sci. U S A. 2014;111:527–532. doi: 10.1073/pnas.1320106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 2011;156:1116–1130. doi: 10.1104/pp.110.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu J-K. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant. 2013;6:2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Primig M, Trnovsky J, Matzke A. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. Embo J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. A Histone Acetyltransferase Regulates Active DNA Demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Miki D, Lei M, Zhu X, Zhang H, Liu Y, Li Y, Zhao Y, Lang Z, Wang J, et al. Regulation of Active DNA Demethylation by an α-Crystallin Domain Protein in Arabidopsis. Mol. Cell. 2014;55:361–371. doi: 10.1016/j.molcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Wang X, Duan C-G, Tang K, Wang B, Zhang H, Lei M, Lu K, Mangrauthia SK, Wang P, Zhu G, et al. RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc. Natl. Acad. Sci. U S A. 2013;110:15467–15472. doi: 10.1073/pnas.1315399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Miki D, Zheng Z, Ma J, Wang J, Yang Z, Dong J, Zhu JK. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat. Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Grafi G. Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci. 2007;12:80–85. doi: 10.1016/j.tplants.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xie S, Li X, Wang C, Chen Z, Lai J, Gong Z. REPRESSOR OF SILENCING5 Encodes a Member of the Small Heat Shock Protein Family and Is Required for DNA Demethylation in Arabidopsis. Plant Cell. 2014;10:2660–2675. doi: 10.1105/tpc.114.126730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.