Abstract
Interleukin 17 (IL-17) is a proinflammatory cytokine that promotes the expression of different cytokines and chemokines via the induction of gene transcription and the post-transcriptional stabilization of mRNAs. Here we show that IL-17 increases the half-life of the Zc3h12a mRNA via interaction of the adaptor protein CIKS with the DEAD box protein DDX3X. Interleukin 17 stimulation promotes the formation of a complex between CIKS and DDX3X, this interaction requires the helicase domain of DDX3X but not its ATPase activity. DDX3X knock-down decreases the IL-17-induced stability of Zc3h12a without affecting the stability of other mRNAs. IKKε, TRAF2 and TRAF5 were also required to mediate the IL-17-induced Zc3h12a stabilization. DDX3X directly binds the Zc3h12a mRNA after IL-17 stimulation. Collectively, our findings define a novel, IL-17-dependent mechanism regulating the stabilization of a selected mRNA.
Introduction
Interleukin-17 (IL-17A) is the signature cytokine produced by CD4+ T helper-17 cells, and belongs to a molecular family composed of six members (IL-17A to F), which are structurally unrelated to other cytokines (1, 2). IL-17 is a proinflammatory cytokine that induces transcription and stabilization of different mRNAs encoding for other inflammatory proteins, such as cytokines, chemokines and metalloproteinases, to amplify the inflammatory response. Although IL-17 is required for host defense against bacterial and fungal infection, it has also been linked to the development of various autoimmune and inflammatory diseases, including rheumatoid arthritis, inflammatory bowel disease, and Systemic Lupus Erythematosus (3–6). IL-17A and the other members of the IL-17 family signal through its binding to heterodimeric receptors composed of members of the IL-17 receptor family (7). Connection to IκB kinase and Stress-activated protein kinase (CIKS) (a.k.a. Traf3ip2 or Act1) is an adaptor protein required for signaling by these receptors(8, 9). After IL-17 receptor triggering, CIKS is recruited to the receptor via a homotypic interaction between its SEFIR domain and the SEFIR domain of the receptor. CIKS, in turn interacts with TRAF6 and this interaction is required for NF-κB and JNK activation, and subsequent transcription of proinflammatory genes (10–12). Other members of the TRAF family (TRAF2 and TRAF5) have been demonstrated to interact with CIKS, but this interaction seems to be dispensable for NF-κB activation, rather it controls the IL-17-induced mRNA stability (13). For this function, IKKε (a.k.a. IKKi) has been rather claimed as essential molecule. After stimulation with IL-17, IKKε forms a complex with CIKS, indeed, mouse embryonic fibroblasts isolated from IKKε KO mice failed to stabilize the IL-17-induced cytokine mRNA(s) (14). Although increased transcription is a requisite for the induced expression of IL-17-target genes, regulation of the half-life of corresponding mRNAs is also critical in determining the magnitude of their expression. Indeed, highly unstable mRNAs require IL-17 to be stabilized during inflammatory responses, and to effectively express the encoded proteins (15). This in turn has fostered the notion that mRNA stabilization is the primary function of IL-17. Stabilization of mRNAs encoding cytokines and chemokines involves regions in the 5’- and 3’-untranslated regions (UTR) of the message, which are specifically recognized by proteins whose function is controlling exonucleolytic degradation of the RNA (16). RNA helicases modulate almost every aspect of RNA metabolism from transcription to translation, and are classified in superfamilies and families based on sequence and structural features (17). DEAD box proteins form the largest helicase family and are characterized by the presence of an Asp-Glu-Ala-Asp (DEAD) motif (18, 19). DDX3X is an ubiquitously expressed member of this family, it consists of 662 amino acids and contains a central core helicase domain. DDX3X, as most of the members of the helicase family, appears to be involved in almost every step of RNA metabolism, and a role for DDX3X in cell cycle control and apoptosis was also proposed (20–24). Recently, DDX3X has also been demonstrated to have a positive role in Interferon (IFN) induction: i) by binding to PolyI:C and to viral RNA in solution; ii) as a component of the IPS-1 and TBK1/IKKε complex; iii) via direct binding to the IFN-β promoter (25– 27).
ZC3H12a (a.k.a. MCP-1–induced protein 1 (MCPIP1), or Regnase-1), is a LPS-inducible gene, and contains a highly conserved Nedd4-BP1, YacP nuclease/deubiquitinase (NYN/DUB) domain with intrinsic RNase and DUB activities at the N terminus, a single CCCH-type ZF domain with RNA-binding potential in the middle region, and a proline-rich (PRR) domain for protein oligomerization at the C-terminus (29, 30). The RNase and DUB activities of ZC3H12a are involved in various biological functions, such as cellular RNA decay and negative regulation of cellular inflammation. The RNase activity of ZC3H12a can directly degrade certain mRNAs of cytokines, such as IL-6 and IL-12p40, via AU-rich element–independent mechanisms (30, 31). The DUB function of ZC3H12A inhibits LPS-, IL-1β–, and TNF-α–mediated NF-κB– and JNK-signaling pathways by removing ubiquitin moieties of TNFR-associated factors (TRAFs), including TRAF2, TRAF3, and TRAF6 (32). Zc3h12a -deficient mice exhibited severe immune syndrome disorders characterized by severe anemia, autoimmune response, and severe inflammation, and most mice died within 12 wk of birth (30).
Although the role of the above mentioned RNA-binding proteins in regulating the half-life of mRNA is well recognized, the high number of proteins involved in such effect, and the signaling pathways downstream of extracellular stimuli (including IL-17) involved are poorly understood. For example, it has been recently demonstrated that the splicing-regulatory factor SF2 (ASF) prolongs the half-life of the chemokine cxcl1 mRNA, after IL-17 stimulation (13). However, the role of SF2 seems to be restricted to specific RNAs and does not involve all the IL-17-induced mRNAs, further suggesting that multiple, RNA-specific mechanisms exist. Here we demonstrate that the DEAD-box helicase DDX3X is mediating the stabilization of the Zc3h12a mRNA. DDX3X interacts with CIKS in an IL-17 dependent manner, and this interaction requires the helicase domain of DDX3, and both the N-terminus and the C-terminus of CIKS. DDX3X knock-down decreases the IL-17-induced stability of Zc3h12a without affecting the stability of other mRNAs. This effect is independent on NF-κB activation, while it depends upon IKKε, TRAF2 and TRAF5. Collectively, our findings unveil a new-, IL-17-dependent mechanism regulating the stabilization of a selected mRNA.
Material and Methods
Reagents, cell lines, and constructs
Recombinant IL-17 and TNF-α were from Peprotech, and were used at 200ng/ml and 2000U/ml, respectively. Anti-M2 (Flag) was from Sigma-Aldrich. Anti-DDX3X was produced in rabbit using a recombinant fragment spanning the amino acids 1–222 of mouse DDX3X as antigen. Anti-TRAF6 (sc-7221), anti-TRAF2 (sc-876), anti-TRAF5 (sc-7220), were from Santa Cruz Biotechnology. Anti-IKKε (D61F9) was from Cell Signaling. Anti-CIKS was produced in rabbit using a recombinant peptide spanning the amino acids 382–574 of CIKS as antigen.
HEK293, wild type mouse embryonic fibroblast (MEF), and CIKS−/− MEF (28) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (Sigma-Aldrich), antibiotics (100 µg/mL penicillin, 100 µg/mL streptomycin), and 1 mM L-glutamine (Invitrogen).
The cDNA encoding CIKS and its mutants were previously described (8, 33). The cDNA encoding DDX3X was a gift of Dr Tilmann Bürckstümmer (Haplogen GmbH, Wien, Austria). DDX3X mutants were generated by PCR and cloned in pCDNA3.1-HA or FLAG (Invitrogen).
Transfection and immunoprecipitation
Lipofectamine-mediated transfections were performed according to the manufacturer’s instructions (Invitrogen). All transfections included supplemental empty vector to ensure that the total amount of transfected DNA was kept constant in each dish culture. For immunoprecipitation of transfected proteins, HEK293 cells (3 × 106) were transiently transfected and 24 h after transfection, cells were lysed in Triton X-100 lysis buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, and Complete Protease Inhibitor mixture). After additional 15 min on ice, cell extracts were centrifuged for 10 min at 14,000 × g at 4 °C, and supernatants were incubated with anti-FLAG antibodies bound to agarose beads (M2, Sigma) for 3 hours, at 4°C. Immunoprecipitates were washed five times with Triton X-100 lysis buffer and subjected to SDS-PAGE.
RNA-interference (RNAi)
Cells were transfected with small interfering RNA oligonucleotides (20 nM final concentration) and Interferin (PolyPlus), according to manufacturer’s instructions. The siRNA sequences used are listed below:
Mouse DDX3X: 5′-GAUGCUGGCUCGUGAUUUCU-3′
Mouse TRAF2: 5′-CGACAUGAACAUCGCAAGC-3′
Mouse TRAF5: 5'-AAGCCAGUGACCAGAGAUUAGUU-3'
To knock-down mouse IKKε and TRAF6, we used the esiRNA#EMU029351 (Sigma) and esiRNA #EMU046421 (Sigma), respectively. The scrambled control was from Thermo Scientific (siRNA ON-TARGETplus Non-targeting Pool #D-001810-10-05). Forty-eight or seventy-two hours after transfection, cells were collected for RNA or protein extraction.
Lentivirus production and infections
FLAG-CIKS, CIKS E17A and CIKSΔUbox mutant cDNAs were subcloned into pWPT lentiviral vector at BamHI/SalI sites. The constructs were sequenced to confirm correct DNA sequence and orientation. Subconfluent 293T lentivirus packaging cells were cotransfected with either pWPT-GFP or pWPT-CIKS and pMD2G and pCMV-R8.91 by calcium phosphate precipitation. After 24h, medium was changed, and supernatant was harvested after 48 and 72 hours. Lentiviral supernatant, cleared of cell debris, was concentrated by centrifugation at 23000 rpm for 90 min, at 4°C in a Beckman SW28 swinging bucket rotor. For transduction, CIKS−/− MEFs were plated on 12-well plate, and infected with lentiviruses in the presence of 10% foetal bovine serum and polybrene (8µg/ml final concentration) (Sigma).
Coimmunoprecipitation and mass spectrometry analysis
CIKS−/− MEFs reconstituted with FLAG-CIKS or with empty vector were left untreated or treated with IL-17 at different time points. After stimulation, cells were washed with ice-cold PBS, and then lysed with lysis buffer (20 mM HEPES pH7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 10 mM NaF, 1 mM Na3VO4) freshly supplemented with protease inhibitors cocktail (Roche). Nuclear and cellular debris were removed by centrifugation at 14,000 × g for 30 min, at 4°C. For identification of the CIKS-interacting proteins, 100 mg of cell extract from CIKS−/− and FLAG-CIKS-reconstituted MEFs were incubated with 500 µl of M2 beads (Sigma), which were preventively washed with 0.1 M glycine pH 3.0, to eliminate non-covalently bound antibody. After 3 hours incubation at 4°C, beads were washed four times with lysis buffer, and twice with high salt (1 M NaCl) lysis buffer. Proteins were eluted from the beads by using 500 µl of 3×FLAG peptide (Sigma) (200µg/ml final concentration). Eluted proteins were separated by SDS-PAGE on a 9–16% gradient gel, and stained with colloidal Coomassie G-250 (34). Gel slices (11 in number) were excised from the lanes corresponding to the bait and mock samples, S-alkylated and in gel digested with trypsin (35). After desalting, peptide mixtures were analyzed by nLC-ESI-LIT-MS/MS; a LTQ XL mass spectrometer (Thermo, San Jose, CA, USA) equipped with a Proxeon nanospray source connected to an Easy-nanoLC (Proxeon, Odense, Denmark) was used to this purpose. Peptide mixtures were separated on an Easy C18 column (100 × 0.075 mm, 3 µm) (Proxeon) by using a gradient of acetonitrile in aqueous 0.1% formic acid, at flow rate of 300 nL/min. Spectral acquisition was performed as previously reported (36). Two technical replicates were performed for each gel slice. Proteome Discoverer platform v.1.3 (Thermo) was used to search raw mass data against an updated Uniprot database of mouse sequences (2011/10) with both Sequest (Thermo, USA) and Mascot (Matrix Science, UK) algorithms. nLC-ESI-LIT-MS/MS data were searched by using experimental parameters previously reported (36). Candidate proteins with more than 2 assigned peptides with an individual MASCOT score >25, or filtered by Sequest Xcorr (>1.8 for +1, >2.0 for +2, >2.2 for +3 and higher charges), and with a significant threshold p < 0.05, were further considered for protein identification. CIKS-interacting proteins were then identified by subtracting the components ascertained within the bait with that ones from the corresponding mock.
Cell stimulation, RNA isolation and Real-Time PCR
To measure mRNA stability, MEFs were stimulated with TNF-α (2000U/ml) for 60 minutes, and then after removing the TNF-α, with IL-17 (200 ng/ml) plus actinomycin D (5µg/ml) for different time points. Total RNA was extracted by using TRIZOL reagent according to manufacturer’s instructions (Invitrogen). Real-Time RT-PCR was carried out with cDNAs reverse-transcribed from total RNA by using GoTaq qPCR Master Mix (Promega) and CFX Manager software (Bio-Rad). Experimental ΔΔCt values were normalized to GAPDH. Statistical analysis has been performed by using the student’s t-test.
The primers used were:
GAPDH: Fw: 5'-ATGGTGAAGGTCGGTGTGAAC-3' Rev: 5'-CCATGTAGTTGAGGTCAATGAAG-3'
Zc3h12a: Fw: 5'-AACTGGTTTCTGGAGCGAGG-3' Rev: 5'-CGAAGGATGTGCTGGTCTGT-3'
Cxcl1: Fw: 5'-AGCCACCCGCTCGCTTCTCT-3' Rev: 5'-GTCCCGAGCGAGACGAGACCA-3'
IKKε: Fw: 5'-CCACGTCTTTTCCCTACCCC-3' Rev: 5'-GATGGCAATCGTGTTGTGGG-3'
DDX3X: Fw: 5'-GTAGAGGCAGCCTTTGCTCA-3' Rev: 5'-ATAACGCCCTTTGCTTGCTG-3'
CXCL2: Fw: 5'-CCCAGACAGAAGTCATAGCCA -3' Rev: 5'-CAGGTACGATCCAGGCTTCC -3'
CCL2: Fw: 5'-AGCTGTAGTTTTTGTCACCAAGC-3' Rev: 5'-GTGCTGAAGACCTTAGGGCA-3'
CXCL5: Fw: 5'-AGAAGGAGGTCTGTCTGGATCCA-3' Rev: 5'-CGAGTGCATTCCGCTTAGCTTTC-3'
IL6: Fw: 5'-AAAGCCAGAGTCCTTCAGAGAGA-3' Rev: 5'-GGTCCTTAGCCACTCCTTCTGTG-3'
LCN2: Fw: 5'-ACAGAGCTACAATGTGCAAGTG-3' Rev: 5'-CAGCTCCTTGGTTCTTCCATACA-3'
TRAF2: Fw: 5’-GCCTTTCCAGATAACGCTGC -3’ Rev 5’-TCGTGGCAGCTCTCGTATTC-3’
TRAF5: Fw: 5’-CGCACCTGTCCCTGTACTT-3’ Rev 5’-AGGCAATGTTCATCTCGCCA-3’
TRAF6: Fw: 5’-ATCCATAAGGGATGCAGGGC-3 Rev: 5’-GGCACTTTACCGTCAGGGAA-3’
RNA immunoprecipitation
To detect the Zc3h12a mRNA in the DDX3X immunoprecipitate, HeLa cells (3 × 106) were transiently transfected with FLAG-DDX3X by using Lipofectamin. Twenty-four hours after transfection, cells were treated with TNF-α (2000U/ml) plus IL-17 (200 ng/ml), and then lysed in Triton X-100 lysis buffer containing RNasin 1U/µl. After an additional 15 min on ice, cell extracts were centrifuged for 10 min at 14,000×g at 4 °C, and supernatants were incubated with anti-FLAG antibodies bound to agarose beads (M2, Sigma) for 3 h at 4°C. Immunoprecipitates were washed five times with Triton X-100 lysis buffer containing RNasin 1U/µl. RNA was extracted by using TRIZOL reagent according to manufacturer’s instructions (Invitrogen). Immunoprecipitated-RNA was reverse-transcribed by using the GoScript Reverse Transcriptase (Promega). PCR was carried out with cDNAs by Expand High Fidelity PCR System (Roche). To amplify the 5'-region of the human Zc3h12a mRNA, we used the following primers:
Fw: 5'-GTCTGAGCTATGAGTGGCCC-3'; Rev: 5'-GGCTGTGGCTGTCCTCAAAT-3'
To amplify the 3’-portion of the human Zc3h12a mRNA, we used the following primers:
Fw: 5'-TGAGGCCCAAACCGTCTTTT-3'; Rev: 5'-GCCTCATTGGCGTAGAGGAC-3'
Experimental PCR conditions were: 95°C 30”; 60°C 30”; 68°C 1’, 40 cycles. The PCR products were analyzed by 1% agarose gel electrophoresis.
RESULTS
CIKS interacts with DDX3X
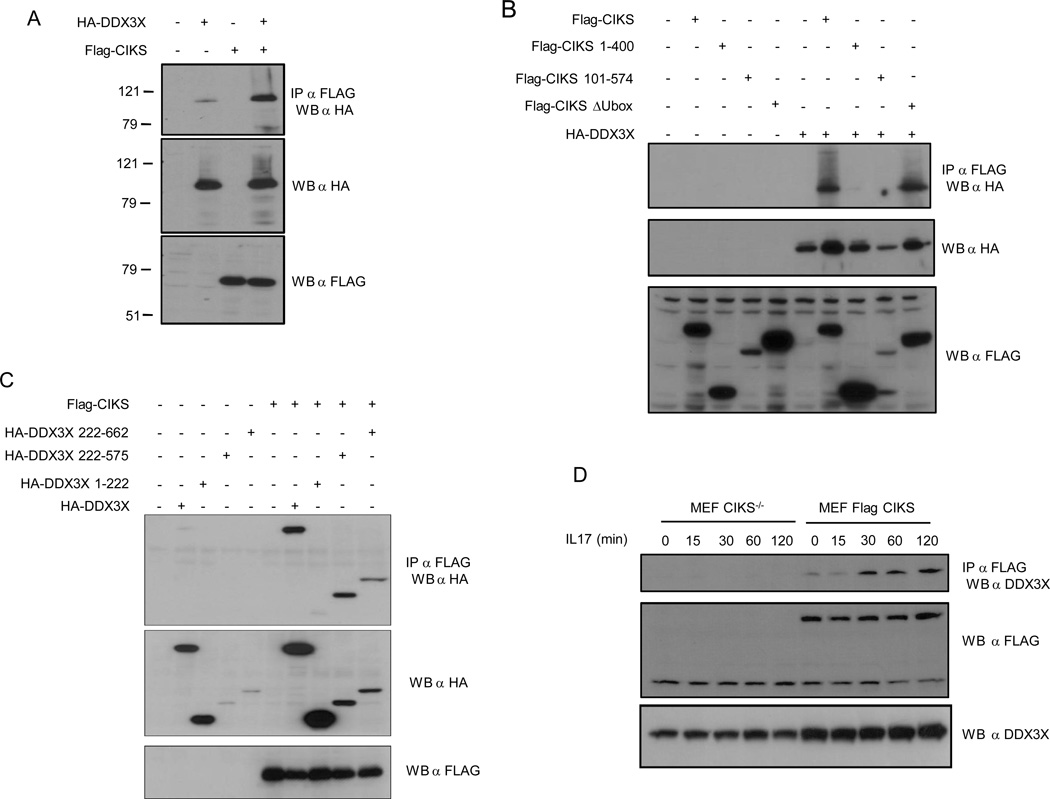
To search for CIKS-interacting proteins, we reconstituted CIKS−/− mouse embryonic fibroblasts (MEFs) with FLAG-tagged CIKS, immunoprecipitated FLAG-CIKS, and analysed the products by mass spectrometry. Among the proteins recovered from the reconstituted cells, but not from the CIKS−/− counterpart (control), we identified the DEAD-box RNA helicase DDX3X. We confirmed this interaction by co-immunoprecipitation (Fig. 1A), and we mapped the sites on both CIKS and DDX3X responsible for the interaction. As shown in Fig. 1B, both the N-terminus and the C-terminus of CIKS were required for a proper interaction with DDX3X. Indeed, deletion of the first 100 amino acids, or the last 100 aminoacids of CIKS sequence, resulted in a loss of the interaction between CIKS and DDX3X. Internal deletions, such as the one comprising the U-box domain (amino acids 271–334) (CIKS ΔUbox) did not affect complex formation. Next, a searching for the DDX3X domains responsible for a proper interaction with CIKS was performed, we found that the helicase domain of DDX3X was required for interaction with CIKS (Fig. 1C). Worth noting a DDX3X mutant (K230A) unable to bind ATP was proved to retain the ability to interact with CIKS (data not shown). We also found that the interaction between CIKS and endogenous DDX3X was dependent upon IL-17 stimulation. In fact, as shown in Fig. 1D, treatment of reconstituted CIKS MEF with IL-17 enhanced the interaction of endogenous DDX3X with CIKS.
Figure 1. Physical interaction between CIKS and DDX3X.
(A) Anti-FLAG immunoprecipitates (IP) of HEK293 cell extracts transfected with FLAG-CIKS and HA-DDX3X were Western-blotted with anti-HA antibodies to detect coimmunoprecipitated DDX3X (Top panel). (Middle and Bottom panels) Western blots with anti-HA (anti-DDX3X) and anti-FLAG (anti-CIKS) antibodies on whole-cell extracts are shown, respectively. (B) Mapping of the DDX3X-binding domain on CIKS. HEK293 cells were transfected with HA-DDX3X and FLAG-CIKS or different FLAG-CIKS deletion mutants. Cell extracts were immunoprecipitated with anti-FLAG antibodies (CIKS) followed by Western blot (WB) anti-HA (DDX3X). The presence of -HA and -FLAG proteins in the whole cell extract is shown in the middle and bottom panels, respectively. (C) Mapping the CIKS-binding domain on DDX3X. HEK293 cells were transfected with FLAG-CIKS and HA-DDX3X or different HA-DDX3X deletion mutants. Cell extracts were immunoprecipitated with anti-FLAG antibodies (CIKS) followed by Western blot (WB) anti-HA (DDX3X). The presence of -HA and -FLAG proteins in the whole cell extract is shown in the middle and bottom panels, respectively. (D) Interaction between CIKS and endogenous DDX3X. CIKS−/− and reconstituted FLAG-CIKS MEFs, were treated for the indicated period of time with IL-17 (200ng/ml). Cell extracts were immunoprecipitated with anti-FLAG antibodies and Western blotted (WB) with anti-DDX3X antibodies.
All together these data demonstrated that CIKS interacted with DDX3X in an IL-17-dependent manner.
DDX3X mediates mRNA stability
Since CIKS is essential for the IL-17-mediated biological responses, we sought to investigate if DDX3X down-regulation was interfering with some of the IL-17-mediated functions, such as gene expression and mRNA stability. To this purpose, we knocked-down DDX3X expression in MEFs by using siRNA (Fig. 2A and B), and in these cells we evaluated: NF-κB and AP-1 activation after IL-17 stimulation. No differences were observed between parental and DDX3X-interfered cells both in terms of NF-κB and AP-1 activation (Supplementary figure 1).
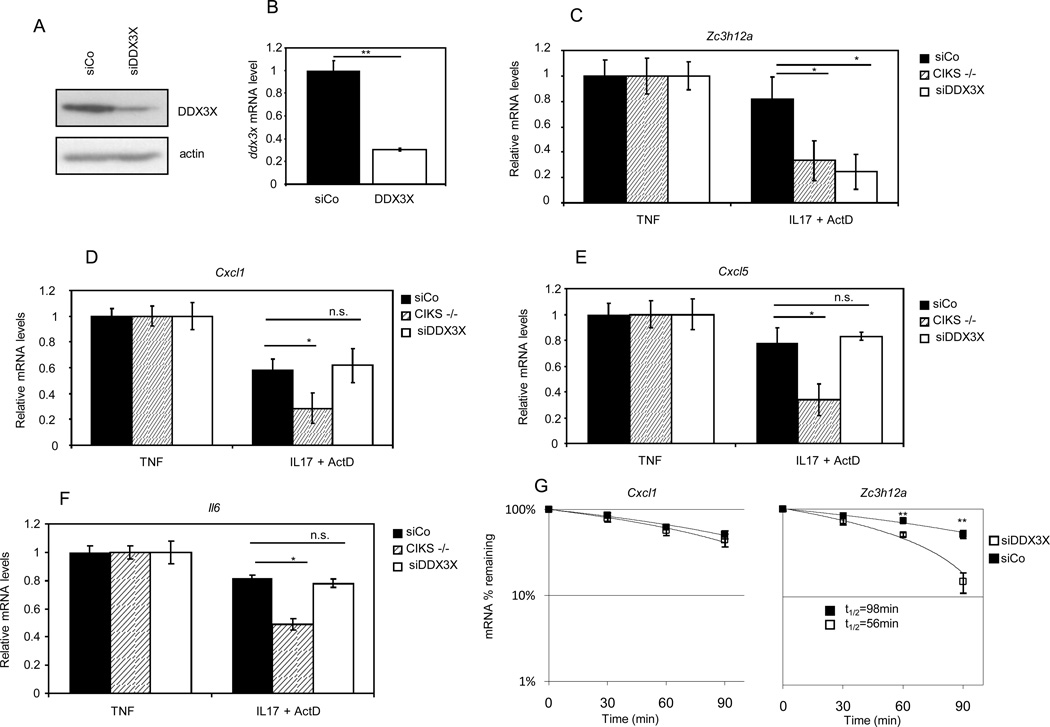
Figure 2. DDX3X controls the half-life of the Zc3h12a mRNA.
Control MEF (siCo), DDX3X-depleted MEF (siDDX3X) and CIKS−/− MEF were treated with TNF-α (2000U/ml) for 60 minutes, and then with IL-17 (200ng/ml) and actinomycin D (5µg/ml). After additional 90 min RNA was extracted and the abundance of the Zc3h12a (C), Cxcl1 (D), Cxcl5 (E), Il-6 (F) was measured by Real-Time PCR. (G) Time course analysis of Cxcl1 and Zc3h12a abundance after treatment with TNF-α (60 minutes) followed by the combined addition of IL-17 and actinomycin D. Expression of DDX3X protein (A) and mRNA (B) in control (siCo) and DDX3X interfered (siDDX3X) MEFs is shown. Data are representative of three independent experiments. Statistical analysis by unpaired Student’s t-test: * p<0.05, ** p<0.01. N.S. not statistically significant.
Next, we investigated the ability of DDX3X to mediate the IL-17-induced stabilization of some short-lived mRNAs induced by TNF-α. We treated CIKS−/− MEFs, reconstituted MEFs, and DDX3X knocked-down MEFs with TNF-α for 60 minutes to induce transcription of target genes. Then, we treated cells with actinomycin D (to block de novo transcription) plus IL-17 to induce mRNA stabilization. After 90 minutes cells were harvested, RNA extracted, and analysed by Real-Time PCR. As shown in Fig. 2C, the level of Zc3h12a was significantly decreased in DDX3X knocked-down cells compared to MEFs treated with a control siRNA. The ability of DDX3X to stabilize mRNA was relatively selective for Zc3h12a, as the levels other mRNAs known to be stabilized by IL-17, including Cxcl1, Cxcl5 and Il6, was not affected in the absence of DDX3X (Fig. 2D–F). Fig. 2G illustrates a time course of mRNA stability for Cxcl1 and Zc3h12a, in cells knocked-down for DDX3X expression. The half-life of Zc3h12a was decreased by almost 50%. These results suggested that DDX3X is involved in the stabilization of selected mRNAs after IL-17 treatment.
IKKε forms a complex with CIKS and DDX3X and is required for mRNA stabilization
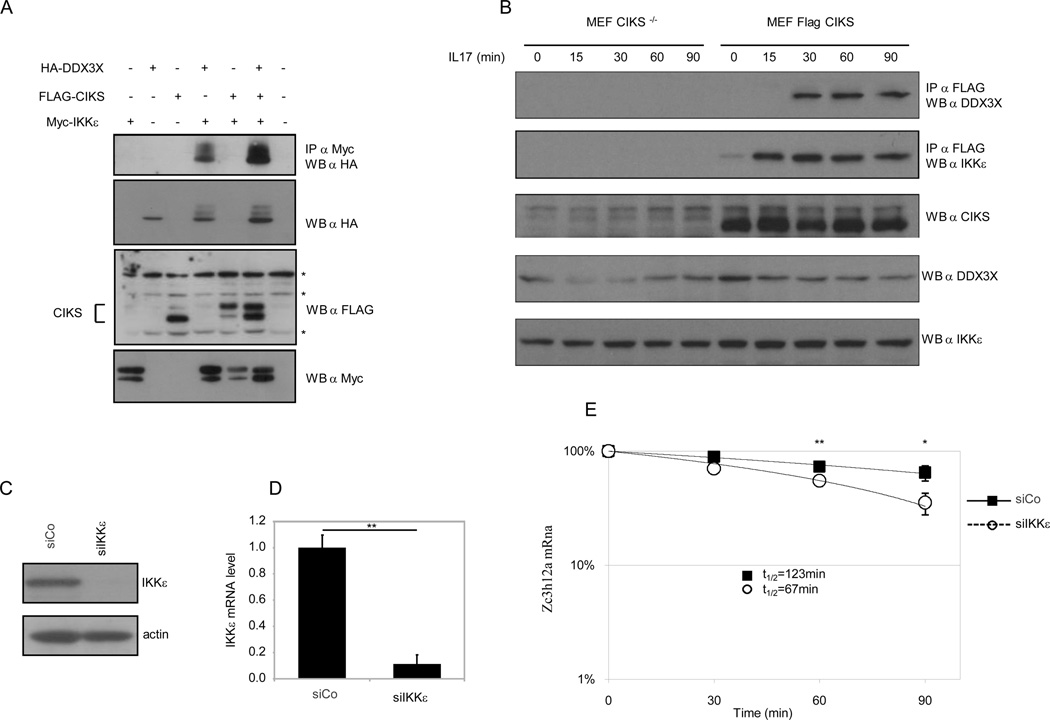
In order to gain insights into the mechanism underlying the DDX3X-based stabilization of Zc3h12a, we investigated if IKKε, which has already been shown to be involved in mRNAs-protecting machineries, was present in a complex containing CIKS and DDX3X. We transfected HEK293 cells with the indicated expression vector (Fig. 3A), immunoprecipitated Myc-tagged IKKε and looked for co-immunoprecipitating DDX3X. As shown in Fig. 3A, IKKε was interacting with DDX3X, suggesting the formation of a complex between CIKS, IKKε and DDX3X. To investigate if the formation of such a complex was dependent upon IL-17 stimulation, we treated CIKS−/− and reconstituted MEF with IL-17, immunoprecipitated FLAG-CIKS and analyzed the presence of IKKε and DDX3X in the immunoprecipitate. As shown in Fig. 3B both IKKε and DDX3X was co-immunoprecipitating with CIKS in an IL-17-dependent manner. It was possible to detect IKKε 15’ minutes after stimulation, while DDX3X appeared in the complex at a later time point, thus suggesting that IL-17 treatment induces the formation of a complex containing CIKS, IKKε and DDX3X.
Figure 3. IKKε interacts with DDX3X and controls the Zc3h12a mRNA stability.
(A) Anti-Myc immunoprecipitates (IP) of HEK293 cell extracts transfected with Myc-IKKε, FLAG-CIKS and HA-DDX3X were Western-blotted with anti-HA antibodies to detect coimmunoprecipitated DDX3X (Top panel). Western blots with anti-HA (DDX3X) and anti-FLAG (CIKS) and anti-Myc (IKKε) antibodies on whole-cell extracts are shown. * Indicates non-specific cross-reactive bands.
B) Interaction between CIKS and endogenous IKKε and DDX3X. CIKS−/− and reconstituted FLAG-CIKS MEFs, were treated for the indicated period of time with IL-17 (200ng/ml). Cell extracts were immunoprecipitated with anti-FLAG antibodies and Western blotted (WB) with anti-IKKε and with anti-DDX3X antibodies. Expression of IKKε protein (C) and mRNA (D) in control (siCo) and IKKε-interfered (siIKKε) MEFs. (E) Zc3h12a mRNA level in control (siCo) and IKKε-interfered MEFs (siIKKε), after treatment with TNF-α (60 minutes) followed by combined addition of IL-17 and actinomycin D. Data are representative of three independent experiments. Statistical analysis by unpaired Student’s t-test: * p<0.05, ** p<0.01.
Next, we knocked-down expression of IKKε in MEF (Fig 3 C, D), and we evaluated the stability of the Zc3h12a mRNA. Cells were treated with TNF-α for 60 min, and then treated with actinomycin D plus IL-17. As shown in Fig. 3E, the half life of Zc3h12a was significantly decreased in IKKε knocked-down cells, compared to MEFs treated with a control siRNA, thus suggesting that the IL-17-induced Zc3h12a stabilization requires IKKε.
TRAF2 and TRAF5, but not TRAF6, are involved in Zc3h12a stability
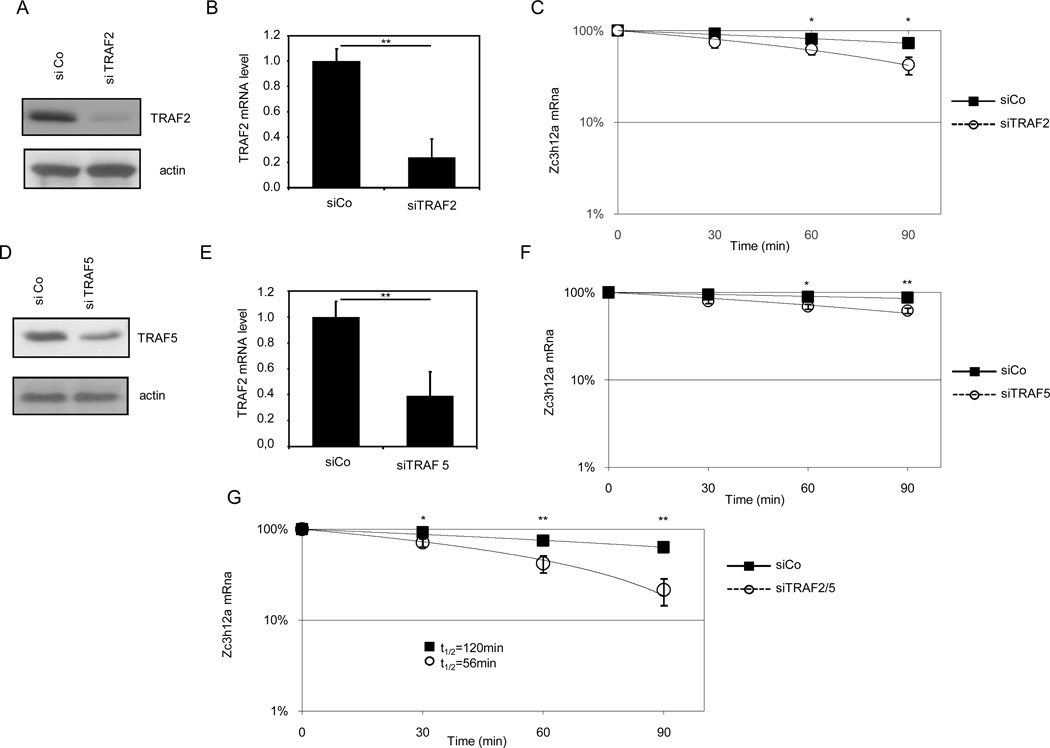
TRAF2 and TRAF5 are already known to be involved in the stabilization of mRNAs induced by IL-17. In order to investigate if TRAF2, TRAF5 or both were also involved in the stabilization of Zc3h12a, we knocked-down their expression in CIKS-reconstituted MEF (Fig 4A, B, D, E), and we analyzed the stability of the Zc3h12a mRNA after IL-17 treatment. As shown in Fig. 4C, in the absence of TRAF2, IL-17 treatment slightly affected Zc3h12a stabilization. Similarly, in the absence of TRAF5, Zc3h12a stabilization was partially affected after IL-17 stimulation (Fig. 4F). As TRAF2 and TRAF5 are known to have redundant functions (37), we knocked-down expression of both proteins by siRNA. As shown in Fig. 4G, knocking down expression of both molecules resulted in a loss of mRNA accumulation in interfered cells, when compared to the scrambled-treated counterpart. These results suggest that either TRAF2 or TRAFA5 (or both) are required for stabilization of Zc3h12a.
Figure 4. TRAF2 and TRAF5 are required for IL-17-induced stabilization of the Zc3h12a mRNA.
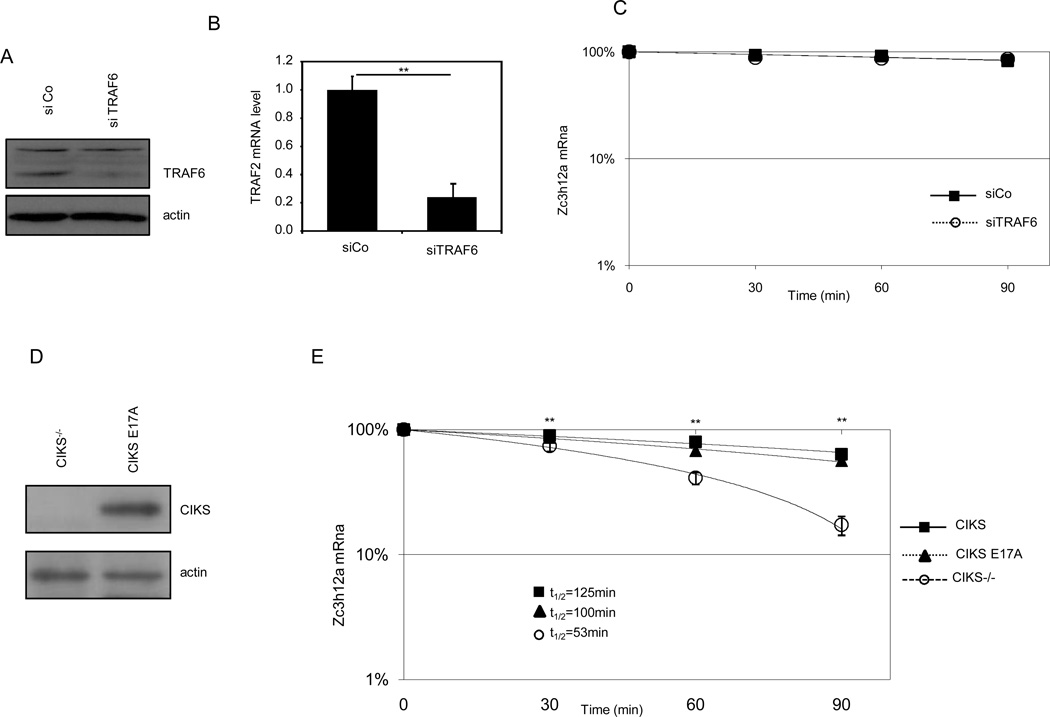
Expression of TRAF2 (A, B) and TRAF5 (D, E) in control (siCo), TRAF2 (siTRAF2) and TRAF5 (siTRAF5) interfered MEFs. Zc3h12a mRNA level in control (siCo), TRAF2 (siTRAF2) (C), TRAF5 (siTRAF5) (F) and double interfered (siTRAF2/5) (G) MEFs, after treatment with TNF-α (60 minutes) followed by combined addition of IL-17 and actinomycin D for the indicated period of time. Data are representative of three independent experiments. Statistical analysis by unpaired Student’s t-test: * p<0.05, ** p<0.01.
We also investigated if the ability of DDX3X to stabilize the Zc3h12a mRNA was dependent upon the ability of CIKS to bind TRAF6 and to mediate NF-κB activation. We knocked-down TRAF6 expression in CIKS-reconstituted MEFs, and we analyzed the stability of Zc3h12a, after IL-17 treatment. As shown in Fig. 5 A, B, C, TRAF6 knock-down did not affect the stability of the Zc3h12a. Similarly, we reconstitute CIKS−/− MEFs with the point mutant CIKS E17A, unable to bind TRAF6 and to activate NF-κB (38, D.S. and A.L. unpublished observation). As shown in Fig. 5D, E, CIKS−/− MEFs reconstituted with CIKS or CIKS E17A had the same levels of the Zc3h12a mRNA, thus suggesting that TRAF6 and NF-κB activation were not part of the mechanism regulating mRNA stability.
Figure 5. TRAF6 is not required for IL-17-induced stabilization of the Zc3h12a mRNA.
Expression of TRAF6 (A, B) in control (siCo), and TRAF6-interfered (siTRAF6) MEFs. (C) Zc3h12a mRNA level in control (siCo), and siTRAF6 MEFs, after treatment with TNF-α (60 minutes) followed by combined addition of IL-17 and actinomycin D. (D) Expression of CIKS E17A in CIKS−/− MEFs reconstituted with CIKSE17A. (E) Zc3h12a mRNA level in CIKS−/− MEFs, reconstituted with CIKS or CIKS E17A after treatment with TNF-α (60 minutes) followed by combined addition of IL-17 plus actinomycin D for the indicated period of time. Data are representative of three independent experiments. Statistical analysis by unpaired Student’s t-test: ** p<0.01.
IL-17 modulates the binding of DDX3X to Zc3h12a
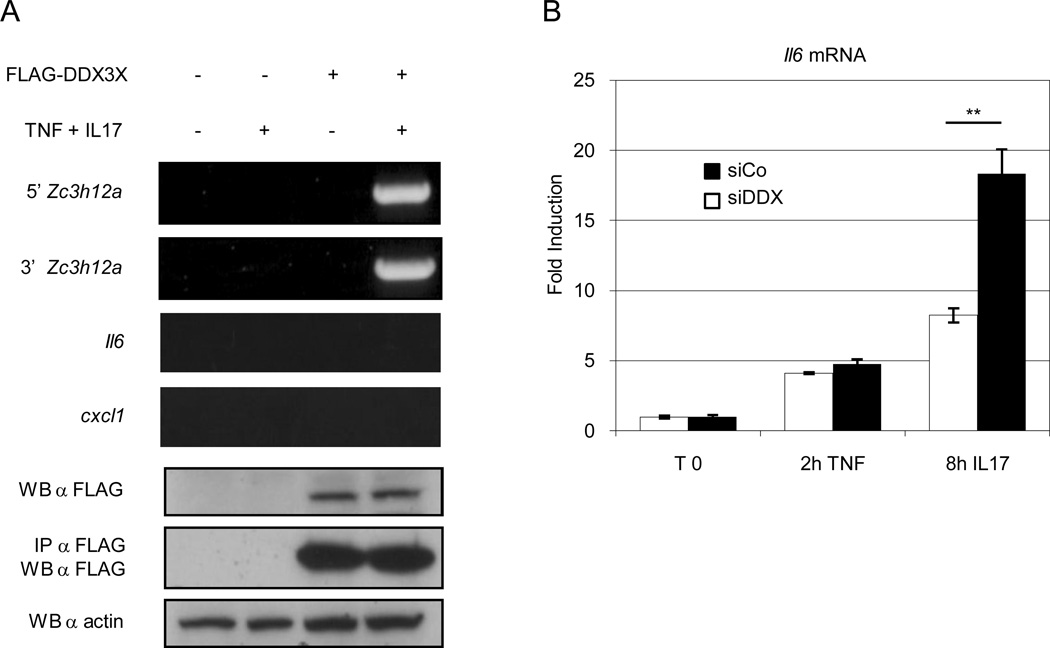
To demonstrate whether DDX3X binds the Zc3h12a mRNA, we ectopically expressed FLAG-DDX3X in HeLa cells. Cells were treated with TNF-α plus IL-17, lysed, and DDX3X was immunoprecipitated by using anti-FLAG antibody. RNA was extracted from the immunoprecipitate, reverse transcribed, and the presence of Zc3h12a was detected by PCR. As shown in fig. 6A it was possible to amplify the Zc3h12a mRNA in the DDX3X immunoprecipitate after TNF plus IL-17 stimulation. It was not possible to amplify other mRNAs known to be stabilized by IL-17, such as il6 and Cxcl1, thus demonstrating that DDX3X specifically binds Zc3h12a. In order to investigate the functional consequence of the decreased level of zc3h12a in the absence of DDX3X, we knocked-down DDX3X expression in MEF, treated the cells with TNF-α plus IL-17 and evaluated the level of il6, one of the known target of the RNase activity of ZC3H12a. As shown in fig 6B, the level of il6 was significantly increased in DDX3X knocked-down MEF, suggesting that the DDX3X-mediated zc3h12a stabilization is one of the mechanisms used to down-regulate immune responses.
Figure 6. IL-17 modulates the binding of DDX3X to the Zc3h12a mRNA.
A) HeLa cells were transfected with an expression vector encoding FLAG-DDX3X or with an empty vector. Twenty-four hours after transfection, cells were treated with TNF-α (2000U/ml) plus IL-17 (200 ng/ml) for two hours, and DDX3X was immunoprecipitated by using anti-FLAG antibodies. RNA was extracted from the immunoprecipitates, reverse transcribed, and Zc3h12a, Il6 and cxcl1 were amplified by PCR. The presence of transfected DDX3X in the whole cell lysate and in the immunoprecipitates is shown. B) Il6 mRNA level in control (siCo) and DDX3X-interfered MEFs (siDDX) after treatment with TNF-α (2 hours) or TNF-α (2 hours) followed by IL-17 for additional 8 hours. Statistical analysis by unpaired Student’s t-test ** p<0.01.
DISCUSSION
In this manuscript we report that the helicase family member DDX3X contributes to the regulation of mRNA stability induced by IL-17. We demonstrate that DDX3X is interacting with CIKS, an essential component of the IL-17 signaling pathway, and that this interaction is dependent upon IL-17 stimulation. CIKS-DDX3X interaction finally stabilizes a selected mRNA. In fact, DDX3X selectively enhances the half life of the Zc3h12a mRNA by directly binding the mRNA in an IL-17-dependent manner, possibly protecting it from degradation. The DEAD-box RNA helicase DDX3X is a multifunctional protein involved in different cellular processes linked to RNA metabolism and gene expression (20). Despite its involvement in almost every step of mRNA metabolism, this is the first evidence linking DDX3X to the stabilization of a selected mRNA following IL-17 stimulation. DDX3X has also been demonstrated to be required for translation of selected mRNAs, by directly binding the 5’-end of such RNAs and facilitating the recruitment of eIF4F complex to start translation (39). It is then possible that binding of DDX3X to the Zc3h12a mRNA is important to modulate both the stability of the Zc3h12a mRNA and its translation. It is not yet clear how DDX3X increases the half life of Zc3h12a. One possible explanation is that, after receptor triggering, DDX3X binds the mRNA displacing other factors controlling the mRNA decay, and recruits the translational machinery to start translation. ZC3H12a regulates its own mRNA stability by binding to its 3’UTR, thus the physical interaction that here we show between DDX3X and the Zc3h12a mRNA might suggest the displacement of ZC3H12a binding as one possible mechanism for its stabilization (40).
DDX3X has also been involved in viral sensing pathway at different levels: by i) enhancing the IPS-1 function; ii) recruiting TBK1 and IKKε; iii) directly binding to the IFN-β promoter; iv) directly binding to viral RNA (25–27).
These results suggest that DDX3X may be an important component of the innate immunity, and that it may use the adaptor protein CIKS to signal the interferon response. However, at least in our experimental system, we were not able to detect an interaction between CIKS and DDX3X after stimulation with different substances mimicking viral nucleic acids, such as Poly(I:C) (both low and high molecular weight) and Poly (dA:dT). In addition, we have not detected a significant variation in IRF3/7 phosphorylation in CIKS−/− MEFs treated with Poly(I:C) and Poly(dA:dT), compared to WT MEFs. These data prompt us to envisage a scenario in which DDX3X may function in various signaling pathways by regulating different immune responses: determining mRNA stability in the IL-17 pathway (via interaction with CIKS), and type I interferon production in the innate immune system (via interaction with TBK1/IKKε).
In this manuscript, we also demonstrated that the DDX3X-mediated stabilization of the Zc3h12a mRNA requires IKKε, TRAF2 and TRAF5. This finding is not surprising given that TRAF2, TRAF5 and IKKε are required to stabilize other mRNAs, such as Cxcl1 (13–14). Notably, mRNA stability was completely abrogated only in TRAF2/TRAF5 double deficient cells, suggesting a redundant function of these two molecules in the IL-17 pathway. The functional redundancy of TRAF2 and TRAF5 is not restricted to the IL-17 pathway. For instance, one of the two proteins may compensate the absence of the other one in TNF-α, signaling pathways. In this line, the phenotype of the TRAF2/TRAF5 double KO mice is way more complex that the phenotype of the single KO (37). However, in our experimental system, TRAF2 knock-down was more effective in blocking the IL-17 mediated stabilization of Zc3h12a, thus suggesting that TRAF2 is the regulator physiologically relevant. It remains to be determined if after IL-17 stimulation DDX3X forms a complex with TRAF2 and/or TRAF5, and we are currently investigating this hypothesis.
We also present evidence that CIKS, IKKε and DDX3X form a complex after IL-17 stimulation. IKKε and DDX3X are recruited in the complex with a different kinetic. In fact, while it is possible to detect IKKε in the CIKS immunoprecipitate immediately after IL-17 stimulation, DDX3X appeared in the complex at a later time point. It is possible to speculate that after IL-17 stimulation IKKε is first recruited to CIKS and then DDX3X is recruited to the complex to regulate its biological function. To this regard, it has been recently reported that IKKε phosphorylates DDX3X and that this phosphorylation modulated the interaction between DDX3X and IRF3 (41). Therefore, it is likely that the phosphorylation of DDX3X may affect its function also in the IL-17 pathway. We are currently testing if the ability of DDX3X to bind Zc3h12a is affected by IKKε. It has been demonstrated that IKKε phosphorylates CIKS and this event is required for the formation of the CIKS-TRAF2-TRAF5 complex that mediates mRNA stability, while the formation of the CIKS-TRAF6 complex, regulating activation of NF-κB, is not affected (14). Our data confirm and expand this observation, as also the DDX3X-mediated mRNA stabilization relies on a similar mechanism and is independent of NF-κB activation. In fact, DDX3X-mediated Zc3h12a mRNA stabilization was not affected in CIKS−/− MEFs reconstituted with the CIKS mutant E17A unable to bind TRAF6 and to activate NF-κB. Similarly, the Zc3h12a mRNA stability was also retained in CIKS−/− MEFs reconstituted with a CIKS mutant lacking the Ubox domain, unable to activate NF-κB (Suppl. Fig 2). On this basis, it is then possible to speculate that the CIKS-TRAF2-TRAF5 axis may activate different effector mechanisms, each regulating the stability of selected mRNA. The use of different machineries to stabilize different RNAs may reflect the different biological functions exerted by the different RNA products, CXCL1 being pro-inflammatory, and ZC3H12a being anti-inflammatory.
ZC3H12a (a.k.a. Regnase-1 or MCPIP-1) is a novel RNAase with CCCH-type zinc-finger domain expressed mainly by immune cells. It has been shown that Zc3h12a expression is rapidly induced either by LPS or by MCP-1, and it negatively regulates mRNA stability of pro-inflammatory cytokines such as TNF-α, IL-1 and IL-6 (42). It is also able to de-ubiquitinate TRAF2, TRAF3 and TRAF6, and its over-expression can inhibit NF-κB and AP-1 activity (32). However, ZC3H12a−/− macrophages are still able to induce NF-κB and AP-1 dependent genes upon LPS stimulation, thus suggesting that the major role of ZCH12a is to regulate the level of different cytokine genes by affecting their mRNA stability rather than regulate their expression via NF-κB or AP-1 function (43). At present it is still unknown whether ZC3H12a targets mRNA alone or in combination with other protein(s). Our data suggest that DDX3X might be one of these protein partners.
The increased half-life of Zc3h12a induced by DDX3X could also play a major role for the stabilization of different cytokine mRNAs induced by IL-17 to finely tune the inflammatory response. It has been reported that ZC3H12a is essential for the inhibition of unwanted T-cell mediated immune response and our data are in line with these observations (43). In fact, the lack of ZC3H12a stabilization in the DDX3X knocked-down MEF results in increased level of the pro-inflammatory cytokine IL6. In this regard it is worth noting that the relative abundance of il6 mRNA does not change in DDX3X knockdown cells at early time point after IL-17 stimulation (fig. 2F), but it is greatly increased at later time point (8 hours after stimulation, fig 6B). This apparent discrepancy may be simply explained by the fact that the stability of il6 mRNA is not directly regulated by DDX3X, rather it is regulated by ZC3H12a, that is, in turn, a direct target of the DDX3X-mediated mRNA stabilization activity. Then, the CIKS/DDX3X -mediated stabilization of Zc3h12a mRNA upon IL-17 stimulation might be part of a regulatory pathway involved in both innate and adaptive immunity to avoid sustained expression of pro-inflammatory cytokines aimed to suppress inflammatory response.
In brief, we report for the first time the association between CIKS and DDX3X, and we provide evidence that DDX3X mediates the stability of selected mRNA induced by IL-17. The possibility to manipulate this interaction may be an important point to control over activation of the immune response.
Supplementary Material
Acknoledgments
Authors thank Dr. Tilmann Bürckstümmer, Haplogen GmbH, Wien, Austria, for providing DDX3X expression vector.
This work was partially supported by the Intramural Research Programs of the National Institute of Allergy and Infectious Disease, National Institutes of Health.
References
- 1.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 2.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9(8):556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziolkowska M, Koc A, Luszczykiewicz G, Ksiezopolska-Pietrzak K, Klimczak E, Chwalinska-Sadowska H, Maslinski W. High levels of IL-17 in rheumatoid arthritis patients: IL-15 triggers in vitro IL-17 production via cyclosporin A-sensitive mechanism. J. Immunol. 2000;164:2832–2838. doi: 10.4049/jimmunol.164.5.2832. [DOI] [PubMed] [Google Scholar]
- 4.Doreau A, Belot A, Bastid J, Riche B, Trescol-Biemont MC, Ranchin B, Fabien N, Cochat P, Pouteil-Noble C, Trolliet P, Durieu I, Tebib J, Kassai B, Ansieau S, Puisieux A, Eliaou JF, Bonnefoy-Bérard N. Interleukin 17 acts in synergy with B cell-activating factor to influence B cell biology and the pathophysiology of systemic lupus erythematosus. Nat Immunol. 2009;10(7):778–785. doi: 10.1038/ni.1741. [DOI] [PubMed] [Google Scholar]
- 5.Kolls JK. Th17 cells in mucosal immunity and tissue inflammation. Semin. Immunopathol. 2010;32:1–2. doi: 10.1007/s00281-010-0198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11(10):763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 7.Chang SH, Dong C. Signaling of interleukin-17 family cytokines in immunity and inflammation. Cell Signal. 2011;23(7):1069–1075. doi: 10.1016/j.cellsig.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leonardi A, Chariot A, Claudio E, Cunningham K, Siebenlist U. CIKS, a connection to Ikappa B kinase and stress-activated protein kinase. Proc Natl Acad Sci U S A. 2000;97(19):10494–10499. doi: 10.1073/pnas.190245697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Commane M, Nie H, Hua X, Chatterjee-Kishore M, Wald D, Haag M, Stark GR. Act1, an NF-kappa B-activating protein. Proc Natl Acad Sci U S A. 2000;97(19):10489–10493. doi: 10.1073/pnas.160265197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J. Biol. Chem. 2006;281:35603–35607. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 11.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang QW, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, Li X. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8(3):247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 12.Sønder SU, Saret S, Tang W, Sturdevant DE, Porcella SF, Siebenlist U. IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem. 2011;286(15):12881–12890. doi: 10.1074/jbc.M110.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF) Nature Immunol. 2011;12(9):853–860. doi: 10.1038/ni.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulek K, Liu C, Swaidani S, Wang L, Page RC, Gulen MF, Herjan T, Abbadi A, Qian W, Sun D, Lauer M, Hascall V, Misra S, Chance MR, Aronica M, Hamilton T, Li X. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat Immunol. 2011;12(9):844–852. doi: 10.1038/ni.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartupee J, Liu C, Novotny M, Li X, Hamilton T. IL-17 enhances chemokine gene expression through mRNA stabilization. J. Immunol. 2007;179:4135–4141. doi: 10.4049/jimmunol.179.6.4135. [DOI] [PubMed] [Google Scholar]
- 16.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 17.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linder P, Fuller-Pace FV. Looking back on the birth of DEAD-box RNA helicases. Biochim Biophys Acta. 2013;1829(8):750–755. doi: 10.1016/j.bbagrm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 20.Schroder M. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem Pharmacol. 2010;79:297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 21.Sun M, Song L, Li Y, Zhou T, Jope RS. Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ. 2008;15:1887–1900. doi: 10.1038/cdd.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, Wu Lee YH. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene. 2006;25:1991–2003. doi: 10.1038/sj.onc.1209239. [DOI] [PubMed] [Google Scholar]
- 23.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok V, Winnard P, Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, Patel AH, Raman V. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27(28):3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 25.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3). helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40(4):940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 26.Soulat D, Bürckstümmer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, Hantschel O, Bennett KL, Decker T, Superti-Furga G. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 2008;27(15):2135–2146. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27(15):2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claudio E, Sønder SU, Saret S, Carvalho G, Ramalingam TR, Wynn TA, Chariot A, Garcia-Perganeda A, Leonardi A, Paun A, Chen A, Ren NY, Wang H, Siebenlist U. The adaptor protein CIKS/Act1 is essential for IL-25-mediated allergic airway inflammation. J Immunol. 2009;182(3):1617–1630. doi: 10.4049/jimmunol.182.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jura J, Skalniak L, Koj A. Monocyte chemotactic protein-1-induced protein-1 (MCPIP1). is a novel multifunctional modulator of inflammatory reaction. Biochimica et Biophysica Acta. 2012;1823:1905–1913. doi: 10.1016/j.bbamcr.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 30.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 31.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B, Jura J. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FASEB J. 2009;276:7389–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 32.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207(13):2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mauro C, Vito P, Mellone S, Pacifico F, Chariot A, Formisano S, Leonardi A. Role of the adaptor protein CIKS in the activation of the IKK complex. Biochem Biophys Res Commun. 2003;309(1):84–90. doi: 10.1016/s0006-291x(03)01532-8. [DOI] [PubMed] [Google Scholar]
- 34.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25(9):1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 35.D'Ambrosio C, Arena S, Salzano AM, Renzone G, Ledda L, Scaloni A. A proteomic characterization of water buffalo milk fractions describing PTM of major species and the identification of minor components involved in nutrient delivery and defense against pathogens. Proteomics. 2008;8(17):3657–3666. doi: 10.1002/pmic.200701148. [DOI] [PubMed] [Google Scholar]
- 36.Scippa GS, Rocco M, Ialicicco M, Trupiano D, Viscosi V, Di Michele M, Arena S, Chiatante D, Scaloni A. The proteome of lentil (Lens culinarisMedik.). seeds: discriminating between landraces. Electrophoresis. 2010;31(3):497–506. doi: 10.1002/elps.200900459. [DOI] [PubMed] [Google Scholar]
- 37.Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, Hashimoto H, Mak TW, Yagita H, Okumura K, Yeh WC, Nakano H. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem. 2001;276(39):36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 38.Sønder SU, Paun A, Ha HL, Johnson PF, Siebenlist U. CIKS/Act1-mediated signaling by IL-17 cytokines in context: implications for how a CIKS gene variant may predispose to psoriasis. J Immunol. 2012;188(12):5906–5914. doi: 10.4049/jimmunol.1103233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Décimo D, Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31(18):3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM, Akira S. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-. 1. Nat Immunol. 2011;12(12):1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 41.Gu L, Fullam A, Brennan R, Schroder M. Human DEAD box helicase 3 couples IκB kinase ε to interferon regulatory factor 3 activation. Mol. Cell Biol. 2013;33(10):2004–2015. doi: 10.1128/MCB.01603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uehata T, Akira S. mRNA degradation by the endonuclease Regnase-1/Zc3H12a/MCPIP-1. Biochim Biophys Acta. 2013;1829(6–7):708–713. doi: 10.1016/j.bbagrm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, Tsujimura T, Rakugi H, Isaka Y, Takeuchi O, Akira S. Malt1-induced cleavage of regnase-1 in CD4(+). helper T cells regulates immune activation. Cell. 2013;153(5):1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.