Abstract
Progesterone (P4) has emerged as an important hormone regulating mammary stem cell populations. In breast cancer, P4 and synthetic analogs increase the number of stem-like cells within luminal estrogen receptor (ER) and progesterone receptor (PR) positive breast cancers. These cells gain expression of de-differentiated cell markers CD44 and cytokeratin 5 (CK5), lose luminal markers ER and PR, and are more therapy resistant. We previously described that P4-downregulation of microRNA (miR)-29a contributes to the expansion of CD44high and CK5+ cells. Here we investigated P4-downregulation of miR-141, a member of the miR-200 family of tumor suppressors, in facilitating an increase in stem-like breast cancer cells. miR-141 was the sole member of the miR-200 family P4-downregulated at the mature miRNA level in luminal breast cancer cell lines. Stable inhibition of miR-141 alone increased the CD44high population, and potentiated P4-mediated increases in both CD44high and CK5+ cells. Loss of miR-141 enhanced both mammosphere formation and tumor initiation. miR-141 directly targeted both PR and Stat5a, transcription factors important for mammary stem cell expansion. miR-141 depletion increased PR protein levels, even in cells lines where PR expression is estrogen-dependent. Stat5a suppression via siRNA or a small molecule inhibitor reduced the P4-dependent increase in CK5+ and CD44high cells. These data support a mechanism by which P4-triggered loss of miR-141 facilitates breast cancer cell de-differentiation through deregulation of PR and Stat5a, two transcription factors important for controlling mammary cell fate.
Keywords: breast cancer, cytokeratin 5, microRNA, miR-141, progesterone, Stat5
Introduction
A major role of progesterone (P4) in the mammary gland is to promote lobulo-alveolar development during specific stages such as puberty, pregnancy, and lactation (1). In murine models, this process involves the P4-dependent expansion of mammary stem cells (MaSCs) that in turn rapidly generate additional lobular epithelial cells (2, 3). This occurs through a paracrine mechanism whereby progesterone receptor (PR)+ cells in the luminal compartment signal through RANKL to expand basal-located stem cells (4, 5). In humans, P4 increases progenitor cells in recapitulated breast epithelial acini structures (6). RANKL is P4-regulated in human breast (7), and thus this expansion could also occur through inter-cellular signaling. Expansion of pre-malignant stem cell populations may explain increased breast cancer incidence among women taking hormone therapies containing progestin (8). We previously demonstrated in estrogen receptor (ER)+ and PR+ breast cancer cells that P4 treatment leads to the emergence of cells expressing cytokeratin 5 (CK5) (9, 10), a marker of stem and progenitor cells in the normal breast and breast cancer (11-13). CK5+, compared to the bulk CK5− tumor cells, are endocrine and chemo-therapy resistant, and have enhanced mammosphere forming potential (11, 14). Progestin exposure of existing breast cancers may thus negatively affect tumor progression.
miRNAs are small regulatory RNA molecules that regulate expression of specific target genes by base-pairing to their mRNAs and interfering with their translation and/or inducing their degradation. miRNA maturation is regulated by several processing steps. The primary miRNA transcript (pri-miRNA) is usually RNA Pol II generated and can include multiple miRNA sequences from a cluster. The pri-miRNA is then processed to its precursor hairpin form (pre-miRNA) by the ribonuclease III protein Drosha, exported out of the nucleus, and cleaved into its mature form by the ribonuclease III enzyme Dicer (15). miRNA species are in general downregulated in cancer compared to normal adult tissue (16). Loss of the miR-200 family (miR-200abc, miR-141, miR-429) in particular leads to epithelial-mesenchymal transition (EMT) in normal and cancer cells (17). Restoration of miR-200c is sufficient to revert mesenchymal breast cancer cells into a more epithelial phenotype (17-21). Progestins regulate multiple miRNAs in breast cancer cells, the majority (~70%) of which are downregulated (22-24). providing a mechanism to indirectly control cellular differentiation. We previously demonstrated that progestin suppression of miR-29 family members facilitates induction of the transcription factor KLF4 and potentiates the expansion of CD44+ and CK5+ breast cancer cells (24). miR-141 is also progestin-downregulated in breast cancer cells (22), and we therefore speculated that it may contribute to expansion of dedifferentiated breast cancer cells.
The cancer stem cell (CSC) theory posits that a subset of pre-existing CSCs, the putative origins of malignancies, perpetuate tumors through indefinite self-renewal and replicative potentials (25). Breast CSCs, compared to bulk tumor cells, are relatively resistant to conventional drugs and are mostly ER−PR−, thus avoiding endocrine therapies (14, 25, 26). Notably, a CSC state may be acquired either spontaneously or through environmental signals during tumor progression (27, 28). Progesterone is one such factor that controls mammary stem cell plasticity; this could be partially facilitated through miRNA regulation. We therefore tested whether miR-141 suppression could contribute to P4-mediated cellular dedifferentiation. Here we demonstrate that miR-141 inhibition enhanced the P4-mediated increase in CD44high and CK5+ cells. miR-141 directly targeted PR and Stat5a, a transcription factor and P4-regulated gene (29, 30) that regulates the mammary luminal progenitor population (31). Stat5a inhibition reduced the P4-mediated increase in CK5+ and CD44high populations. These findings emphasize that hormonal alteration of miRNA levels contributes to the regulation of breast CSCs via transcription factors that promote mammary stem cell populations.
Results
P4 downregulates mature miR-141 in luminal breast cancer cell lines
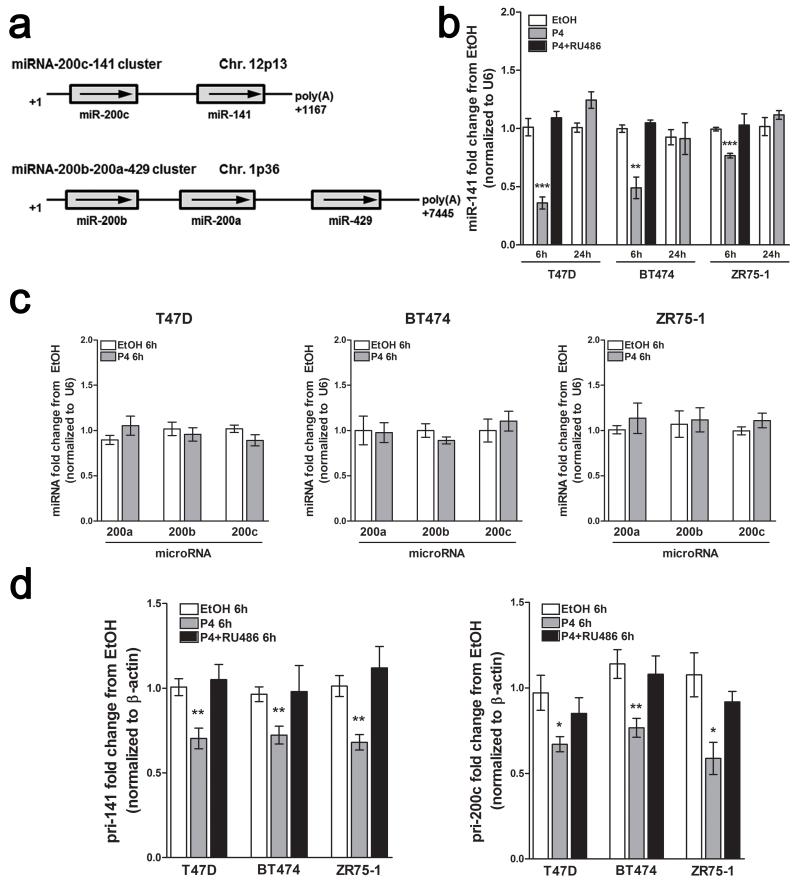
The miR-200 family is organized on two different chromosomes; the miR-141 gene is downstream of miR-200c on chr12p13, with the remaining members (miR-200b-200a-429) clustered on chr1p36 (Figure 1a). To determine whether miR-141 expression correlates with a particular breast cancer subtype, we assessed miR-141 expression in multiple breast cancer cell lines. miR-141 was expressed at detectable levels in luminal and basal-like triple negative (TN) cell lines, and was absent in mesenchymal-like TN cell lines (Supplementary Figure 1), where the miR-200c/141 cluster is silenced by CpG methylation (32).
Figure 1.
P4 downregulates mature miR-141 in a PR-dependent manner. (a) Chromosomal organization of the miR-200 family; the miR-200c-141 and miR-200b-200a-429 clusters are located on different chromosomes. (b) P4 downregulates mature miR-141. Using quantitative reverse-transcriptase PCR (qRT-PCR), miR-141 levels decrease by 6 h 100 nM P4 treatment in luminal breast cancer cell lines (T47D, BT474, and ZR75-1), and restore to original levels after 24 h 100 nM P4 treatment; downregulation of miR-141 is blocked by the PR antagonist RU486. Indicated cells were treated with vehicle, 100 nM P4, or P4 plus 1 μM RU486 for 6 h and miR-141 levels measured by qRT-PCR. Data represent fold change in miRNA levels relative to vehicle (EtOH) treatment normalized to U6 levels. Asterisks indicate statistical significance in comparison to 6h EtOH treatment. (c) miR-200abc levels do not significantly change with 6 h 100 nM P4 treatment as compared to EtOH treated cells. Data represent fold change in miRNA levels measured by qRT-PCR relative to EtOH treatment normalized to U6 levels. (d) P4 downregulates the primary miR-141 and miR-200c transcripts. Cells were treated as indicated and pri-miRNAs measured by qRT-PCR. Data represent fold change in pri-miRNA levels relative to EtOH treatment normalized to β-actin mRNA levels. For all panels, bars are mean +/− SEM of biological triplicates. Significance is indicated as * P<0.05, ** P<0.01, *** P<0.0001. In all experiments, BT474 and ZR75-1 cells were pretreated with 10 nM E2 for 48 h to induce PR expression prior to P4 treatment. All qPCR experiments were performed in biological triplicate, and repeated twice independently.
Global miRNA profiling of T47D breast cancer cells treated with vehicle or the synthetic progestin medroxyprogesterone acetate (MPA) identified miR-141 downregulation at 6 h post-treatment (22). To confirm P4-mediated regulation of miR-141, luminal breast cancer cell lines T47D, BT474, and ZR75-1 were treated with vehicle (ethanol) or 100 nM P4 for 6 or 24 h, and mature miR-141 levels measured by qPCR. BT474 and ZR75-1 cells were pre-treated with 10 nM 17β-estradiol (E2) for 48 h to induce PR levels. In all three cell lines, mature miR-141 levels significantly decreased at 6 h post P4 treatment, and returned to pre-treatment levels at 24 h (Figure 1b). Co-treatment with the progestin antagonist RU486 for 6 h blocked miR-141 downregulation, suggesting a PR-dependent mechanism. E2 alone had no effect on miR-141 levels (Supplementary Figure 2). In contrast, mature miR-200a, miR-200b, and miR-200c levels did not significantly change with 6 h P4 treatment (Figure 1c). An expanded time course confirmed 6 h as the maximal miR-141 reduction point, with no changes in the other family members (Supplementary Figure 3).
Since miR-141 is located downstream of miR-200c on the miR-200c/-141 cluster (32), but P4 decreases only mature miR-141, we sought to determine if P4 regulates the pri-miRNA transcript. qPCR with primers specific to pri-miR-141 or pri-miR-200c (provided in Supplementary Figure 4a) found that both transcripts decreased at 6 h post P4 treatment, and were blocked by RU486 (Figure 1d). An extended time course (0-12 h) confirmed maximal downregulation of pri-miR-141 and pri-miR-200 at 6 h (Supplementary Figure 4b). There is emerging evidence for differential processing of miRNAs located in the same cluster (33), with separate transcription units observed for miR-141 and miR-200c (32). These data suggest differential biogenesis of miR-141 and miR-200c following P4 treatment; selective underexpression of miR-141 has been previously observed in prostate cancer (34).
Stable inhibition of miR-141 potentiates the P4-mediated expansion of CD44high and CK5+ cells
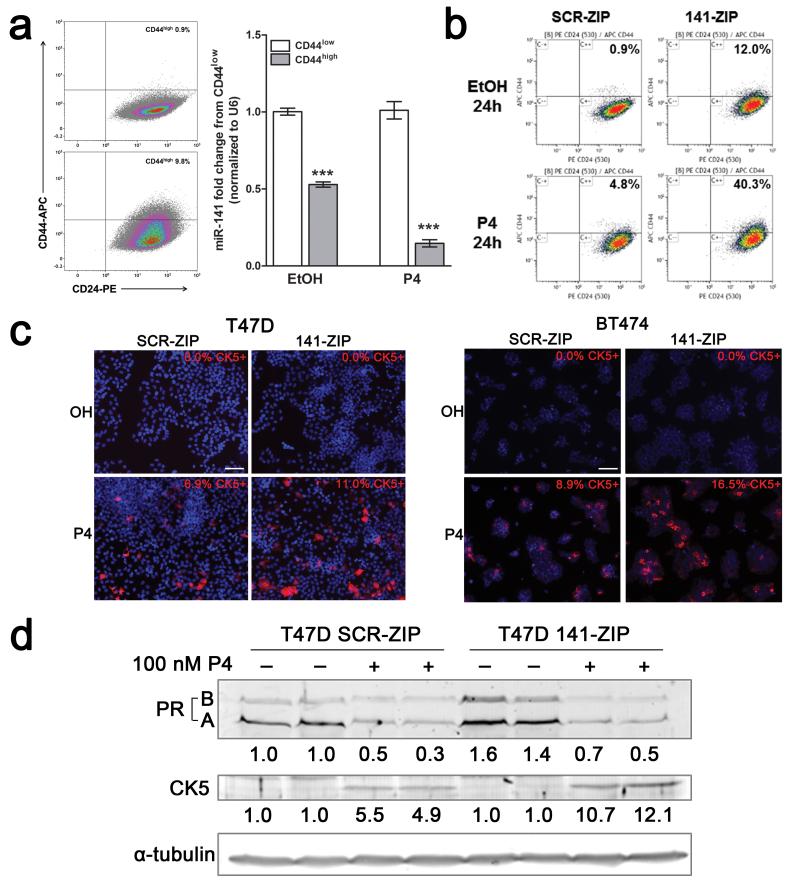
We evaluated CD44high and CK5+ populations as both are reported markers of CSCs and are P4-regulated (9, 24, 35). While there is considerable overlap between the two populations, a distinct CD44highCK5− population exists (9). Since miR-141 is underexpressed in CD44highCD24−/low breast CSCs isolated from patient tumors (36), we sought to determine if miR-141 levels are changed in the P4-induced CD44high population. T47D cells were treated with vehicle or P4 for 24 h then fluorescence-activate cell sorting (FACS) performed to isolate the CD44high and CD44low populations. Expression of mature miR-141 was significantly reduced in both the endogenous and P4-induced CD44high populations (Figure 2a). While this could indicate direct expansion of the CD44high population, conversion is more probable given a cell doubling time of 36 h and maximal downregulation of miR-141 at 6 h.
Figure 2.
Stable inhibition of miR-141 increases the CD44high and CK5+ populations in response to P4. (a) CD44high cells express significantly less miR-141 than CD44low cells. T47D cells were treated with vehicle or 100 nM P4 for 24 h, immunolabeled with APC-CD44 and PE-CD24, and the CD44high and CD44low fractions collected by FACS. Representative flow density plots are shown, with percentage of CD44high cells indicated. Graph depicts qRT-PCR for miR-141 from triplicate samples of CD44high and CD44low cells. Bars are mean +/− SEM. Significance is indicated as *** P<0.0001. (b) T47D cells that stably inhibit miR-141 (141-ZIP) or with negative control (SCR-ZIP) were treated with either EtOH or 100 nM P4 for 24 h, immunolabeled, and CD44 expression measured by flow cytometry. Representative flow density plots are shown, with percentage of CD44high cells indicated. (c) T47D (left) and BT474 (right) 141-ZIP cells express more CK5+ cells in response to P4 treatment than SCR-ZIP cells. CK5 expression (red) was determined by immunofluorescence in EtOH- or P4-treated T47D SCR-ZIP or 141-ZIP cells. Representative images show DAPI (4′,6-diamidino-2-phenylindole (blue)) and CK5+ expression. The percentage of CK5+ cells indicated was measured in 6 fields and at least 300 cells per condition in biological triplicates. Scale bars, 100 μm. (d) T47D 141-ZIP cells express more CK5 protein after P4 treatment than SCR-ZIP cells. PR and CK5 expression was determined by Western blot; α-tubulin was used as loading control. Fold change of total PR and CK5 in P4-treated SCR-ZIP or 141-ZIP compared to EtOH-treated SCR-ZIP cells is indicated; quantification is normalized to α-tubulin.
To determine if miR-141 suppression exacerbates the P4-induced expansion of CD44high and CK5+ populations, we generated stable control (SCR-ZIP) and miR-141-inhibited (141-ZIP) cell lines. SCR-ZIP and 141-ZIP T47D cells were treated with vehicle or P4 for 24 h and the CD44high and CK5+ populations measured by flow cytometry, or immunofluorescence and Western blot, respectively. P4 treatment led to a 5-fold increase in the CD44high population (5%) in SCR-ZIP cells (Figure 2b), as previously described (24). Vehicle treated 141-ZIP compared to SCR-ZIP cells had a 12 fold increase in the CD44high population (12%). P4 treated 141-ZIP compared to SCR-ZIP had a 8 fold increase in the CD44high population (40%) (Figure 2b). CK5+ cells were absent in vehicle treated SCR-ZIP and 141-ZIP T47D and BT474 cells. The observed P4-induced CK5+ cells in SCR-ZIP cell lines were potentiated ~2-fold in 141-ZIP cell lines (Figure 2c). By western blot, total PR protein levels increased 1.5 fold in 141-ZIP cells, and still underwent ligand-dependent downregulation. CK5 protein levels increased >2 fold in P4-treated 141-ZIP compared to SCR-ZIP cells (Figure 2d). These results indicate that downregulation of miR-141 alone contributes to expanding the CD44high population, and potentiates the P4-mediated increases in the CD44high and CK5+ populations.
Stable inhibition of miR-141 increases the mammosphere and tumor initiating capacity of breast cancer cells
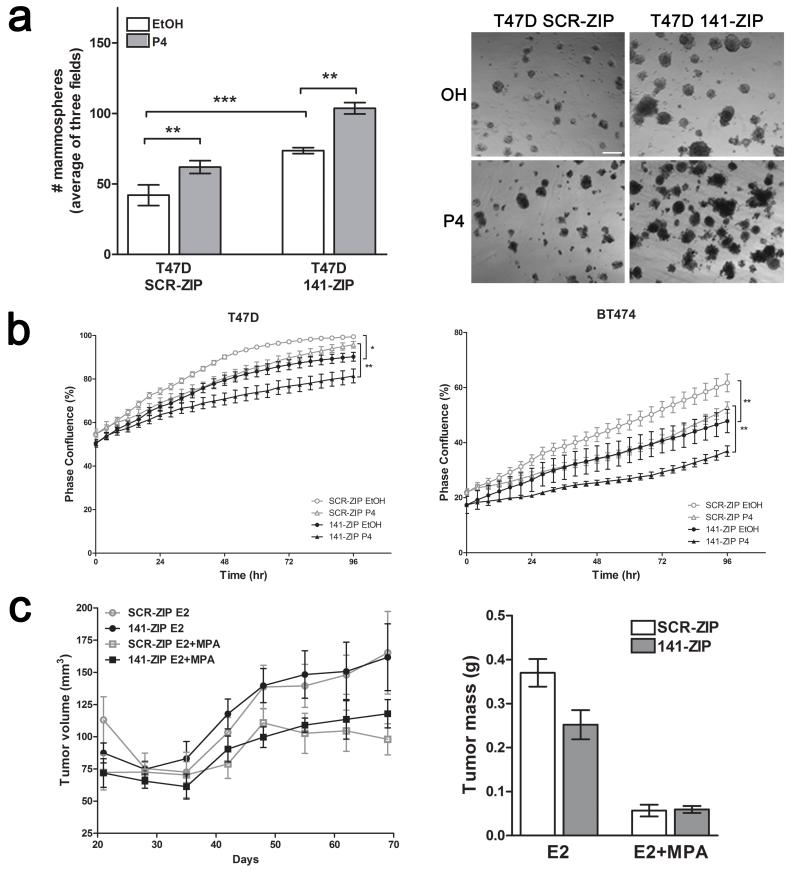
To test whether miR-141 inhibition affects breast cancer cell self-renewal, we performed mammosphere-formation assays using T47D SCR-ZIP and 141-ZIP cells. P4 treatment alone increased mammosphere formation of SCR-ZIP cells (Figure 3a). 141-ZIP compared to SCR-ZIP cells had increased mammosphere formation in both control and P4 treated samples. Thus, inhibition of miR-141 alone increases the sphere-forming capability of luminal breast cancer cells, potentially due to increasing the CD44high population, and enhances the P4-mediated increase in mammosphere formation, potentially due to increased CD44high and CK5+ cells.
Figure 3.
Stable inhibition of miR-141 increases mammosphere formation and growth of progestin treated tumors. (a) T47D 141-ZIP cells form significantly more mammospheres than SCR-ZIP cells. A total of 1000 cells were plated in mammosphere media, in triplicate, with EtOH or 100nM P4, and mammospheres counted 14 days later. Measurements were taken digitally and mammospheres were quantified using six pictures per well. Left: Mean number of mammospheres >75 μm for T47D SCR-ZIP and 141-ZIP with EtOH or P4 treatment. Bars indicate mean +/− SEM. Significance is indicated as ** P<0.01, *** P<0.0001. Right: Representative images of T47D SCR-ZIP and 141-ZIP treated with EtOH or P4. Scale bar, 250 μm. (b) 141-ZIP cells show reduced proliferation. T47D and BT474 (E2 pretreated for 48 h) SCR-ZIP and 141-ZIP were treated with EtOH or P4 in sextuplicate and tracked using the IncuCyte™ live-cell imaging system every 4 h for 96h total. Graphs indicate percent phase confluence over time; significance is indicated as *P<0.05, **P<0.01. (c) Seven ovariectomized Female NOD/SCID mice (n=7 per group) were injected with 1×106 T47D SCR-ZIP and 141-ZIP cells in the left and right fourth mammary fat pad, respectively. Mice were implanted subcutaneously with either E2 alone or in combination with MPA (E2 + MPA) at the time of cell injection and tumor size measured using digital calipers weekly after injection. Tumor volumes are plotted versus the number of days of incubation in E2-treated and E2+MPA-treated mice (left panel). Upon euthanasia, tumors were excised and weighed (right panel). Data represent mean +/− SEM.
We next evaluated the ability of SCR-ZIP and 141-ZIP cells to initiate tumors; cells were pre-treated for 24 h with P4 in vitro then injected bilaterally into the fourth mammary fat pads of female nude mice at dilutions of 102 to 104. Measured at 5 and 6 weeks post-implantation, 141-ZIP cells initiated tumors more efficiently compared to SCR-ZIP cells (Table 1). These data show that loss of miR-141 enhances tumor-initiating ability, likely due to amplified CD44high and CK5+ populations.
Table 1.
Tumor-initiating capacity of miR-141ZIP compared to SCRZIP T47D cells
|
Number of cells injected per mammary fat
pad |
Number of tumors per number of
injected fat pads a |
|
|---|---|---|
|
| ||
| Week 5 after implantation | ||
| SCRZIP | 141ZIP | |
| 1 × 104 | 2/5 | 5/5 |
| 1 × 103 | 3/10 | 6/10 |
| 1 × 102 | 1/10 | 4/10 |
| Tumor-initiating frequency (95% CI) | (1/7894) | (1/712) |
| Tumor-initiating range (95% CI) | (1/19965 - 1/3121) | (1/1440 - 1/352) |
| P-value | 5.47 × 10−6 | |
| Week 6 after implantation | ||
|---|---|---|
| SCRZIP | 141ZIP | |
| 1 × 104 | 3/5 | 5/5 |
| 1 × 103 | 3/10 | 7/10 |
| 1 × 102 | 2/8b | 5/8b |
| Tumor-initiating frequency (95% CI) | (1/4924) | (1/495) |
| Tumor-initiating range (95% CI) | (1/11721 - 1/2069) | (1/1003 - 1/245) |
| P-value | 3.07 × 10−6 | |
Abbreviation: CI, confidence interval. Limiting dilution analysis of the tumor-initiating frequency of T47D-141ZIP cells as compared with SCRZIP negative control cells.
Shown as number of tumors per number of injected fat pads.
Two animals were euthanized for health reasons between week 5 and week 6, and were excluded from the analysis. Data were analyzed using ELDA software.
To determine whether the observed differences in sphere and tumor formation could be due to differential cell growth rate, we measured proliferation of SCR-ZIP and 141-ZIP in vitro and in vivo. For in vitro experiments, SCR-ZIP and 141-ZIP T47D and BT474 cells were plated in sextuplicate in 96 well plates, treated with vehicle or 100 nM P4 (T47D), or E2 and E2+P4 (BT474) and proliferation measured via the Incucyte kinetic live cell imaging system over 4 days. In two luminal breast cancer lines, 141-ZIP compared to SCR-ZIP cells had significantly reduced proliferation in the absence or presence of P4 (Figure 3b).
To evaluate tumor growth in vivo, we injected 1×106 T47D SCR-ZIP and 141-ZIP cells in the fourth mammary fat pad of NOD/SCID mice supplemented with either E2 alone or E2 + MPA. SCR-ZIP and 141-ZIP cells were implanted on opposing mammary glands for internal comparison. There was no statistical size difference between SCR-ZIP and 141-ZIP tumors in the E2-treated group (Figure 3c). Tumors in E2+MPA supplemented mice were overall smaller as previously observed (24). 141-ZIP compared to SCR-ZIP tumors were not statistically different in volume or mass, except for one data point at 69 days post-implantation (Figure 3c). We conclude that miR-141 inhibition decreases short-term cell proliferation, and has no overall effect on long-term tumor growth. Therefore its enhancement of sphere and tumor initiation is not a function of increased growth capacity.
PR is a direct target of miR-141 and increases with miR-141 inhibition
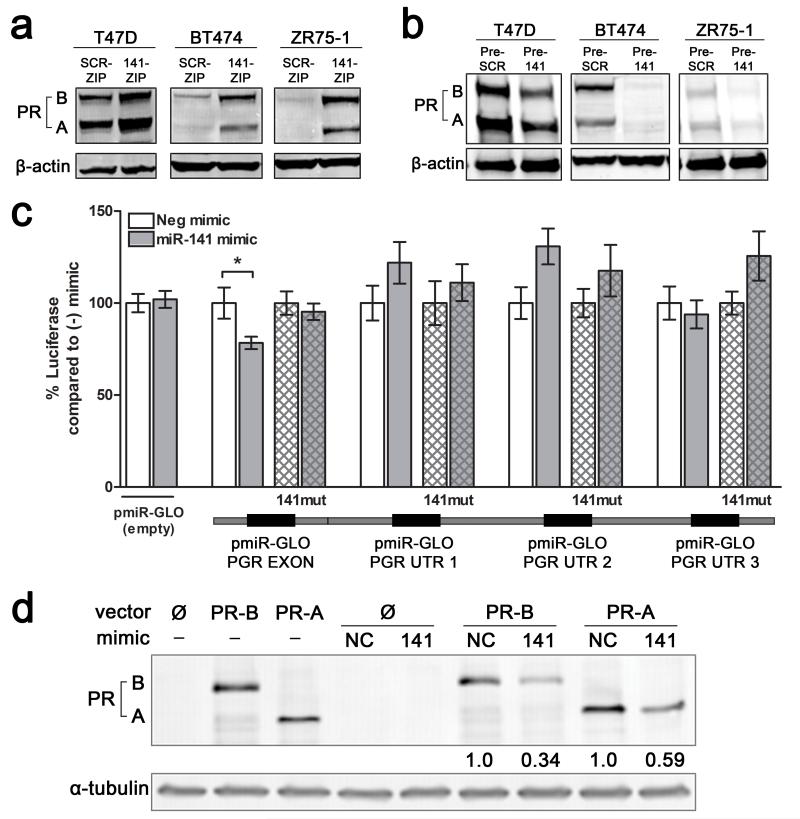
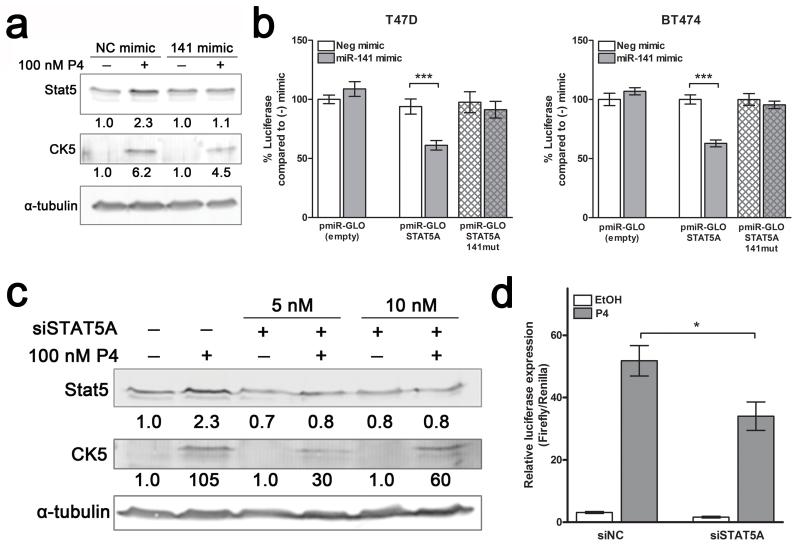
To determine relevant targets of miR-141 that may be involved in promoting the expansion of stem-like cells, we focused on transcription factors important in mammary gland differentiation. Since miR-141 is predicted to target the PGR gene, which encodes both isoforms of PR (PR-B, PR-A), we first analyzed the effect of miR-141 manipulation on PR expression. PR protein expression significantly increased in three different luminal breast cancer cell lines (T47D, BT474, and ZR75-1) with miR-141 inhibition (141-ZIP) (Figure 4a; Figure 2d). Conversely, PR expression was decreased in the same three cell lines when miR-141 was overexpressed using a lentiviral vector carrying its precursor sequence (Pre-141) or a scrambled control (Pre-SCR) (Figure 4b).
Figure 4.
miR-141 regulates PR expression levels in luminal breast cancer cell lines and directly targets the PR transcript. (a) Stable inhibition of miR-141 increases PR expression. PR expression was measured by Western blot in untreated T47D, BT474, or ZR75-1 cells with stable inhibition of miR-141 (141-ZIP) or scrambled control (SCR-ZIP). PR-A and PR-B isoforms are indicated. β-actin was used as loading control. (b) Stable overexpression of miR-141 decreases endogenous PR expression. PR expression measured by Western blot in untreated T47D, BT474, or ZR75-1 cells with stable overexpression of miR-141 (Pre-141) or control (Pre-SCR). β-actin was used as loading control. (c) miR-141 directly targets PR through a binding site in the last exon. Predicted miR-141 binding sites in the PR 3′UTR and last exon are outlined below the graph. Regions of the 3′UTR as indicated were cloned singly downstream of luciferase in the pMIR-GLO vector and each site was mutagenized to abolish miR-141 binding. Each luciferase construct or its mutagenized counterpart was transfected into T47D cells with either 50 nM negative control (−) or miR-141 (141) mimic and luciferase activity measured after 48 h. Data represents relative luciferase activity normalized to constitutively active Renilla contained on the same vector. Experiments were repeated twice. Bars are mean +/− SEM; * P<0.05. (d) Plasmids encoding PRB (hPR1) and PRA (hPR2) were transiently transfected into HEK293 cells alone or with negative control (NC) or miR-141 mimics. PR protein levels were measured by Western blot. Fold change of PR compared to NC mimic is indicated; quantification is normalized to α-tubulin.
To test if miR-141 directly targets the PR transcript, we analyzed four predicted miR-141 binding sites (Figure 4c); three within the 3′UTR as identified through Targetscan (http://www.targetscan.org/) and one in the last exon predicted based on Argonaute HITS-CLIP analysis and corresponding seed match with prediction algorithms (37). These sequences were each placed separately downstream of a luciferase reporter gene and luciferase activity measured in the presence of control or miR-141 mimics. MiR-141 mimic significantly decreased luciferase activity with the coding site (PGR EXON), but not the 3′UTR sites, and mutation of the predicted coding miR-141 binding site rescued the decrease (Figure 4c; hatched bars). These results indicate direct targeting of PR through a miR-141 site in the last exon, which is present in transcripts for both PR-A and PR-B isoforms (38).
To further test this in context, PR-negative HEK293 cells were transiently transfected with plasmids expressing PR-A and PR-B that contain ~998 bp of the 3′UTR (38). These contain the exonic binding site but none of the three predicted 3′UTR binding sites. Expression of both PRA and PRB protein was reduced by miR-141 mimic but not control (Figure 4d). These data confirm that PR expression levels can be directly altered by miR-141 targeting of PR transcripts. The heightened PR expression observed with miR-141 inhibition may be a contributing mechanism that helps potentiate the P4-mediated expansion of CK5+ and CD44high populations.
Stat5a is a direct target of miR-141 and is important for the P4-mediated increase in CK5+ cells
Stat5a (signal transducer and activator of transcription 5A) is a progestin-regulated gene in the normal mammary gland that dictates luminal cell fate and is necessary for full lobular-alveolar development (39, 40). Stat5a is also progestin-regulated in luminal breast cancer (30, 41), and is a predicted target of miR-141. We therefore investigated Stat5a regulation by miR-141 and its involvement in the P4 expansion of stem-like cells. Treatment with a miR-141 mimic blocked P4-mediated Stat5 upregulation (Figure 5a). To determine if miR-141 directly targets Stat5a, we placed the predicted miR-141 binding site sequence downstream of a luciferase reporter gene and measured luciferase activity in the presence of control or miR-141 mimics. miR-141 mimic significantly decreased luciferase activity for the Stat5a miR-141 site construct in both T47D and BT474 cells; mutation of the predicted miR-141 binding site blocked this decrease (Figure 5b). We also analyzed a predicted miR-141 site in Stat5b and found no significant regulation (Supplementary Figure 5). These data support that miR-141 directly and specifically regulates Stat5a in breast cancer cells.
Figure 5.
Stat5a is a direct target of miR-141 and contributes to the P4 expansion of CK5+ cells. (a) P4 increases Stat5 expression, which is partially blocked by miR-141. Western blot analysis of Stat5 and CK5 protein expression in T47D cells treated with EtOH (−) or 100 nM P4 (+), and with negative control mimic (NC) or miR-141 mimic. Fold change of Stat5 and CK5 compared to EtOH/NC mimic is indicated; quantification is normalized to α-tubulin (loading control). (b) Stat5a is directly targeted by miR-141. The STAT5A 3′-UTR was cloned downstream of luciferase in the pMIR-GLO vector and the predicted binding site was mutagenized to abolish miR-141 binding ability. The construct (solid bars) or its mutagenized counterpart (patterned bars) was transfected into T47D or BT474 cells with either 50 nM negative control or miR-141 mimic and luciferase activity measured after 48 h. Data represents relative luciferase activity normalized to constitutively active Renilla contained on the same vector. Experiments were repeated twice. Bars are mean +/− SEM; *** P<0.0001. (c) siSTAT5A blocks P4-mediated upregulation of Stat5. Western blot of Stat5 and CK5 protein expression of T47D cells treated with EtOH or 100 nM P4, with or without 5 or 10 nM siSTAT5A. α-tubulin was used as loading control. Fold changes in Stat5 and CK5 relative to vehicle with negative control siRNA are indicated. (d) siSTAT5A inhibits P4 induction of CK5+. T47D cells stably expressing a luciferase reporter driven by the CK5 gene (KRT5) promoter were plated at 10,000 cells per well in 96-well plates. After 24 h, cells were transfected with SV40-Renilla vector and 5 nM negative control (siNC) or On-Target pool siSTAT5A using dual transfection reagent. Cells were treated for an additional 24 h with vehicle (EtOH) or 100 nM P4, and luciferase activity measured. Renilla was used to normalize luciferase data for transfection efficiency. Bars are mean +/− SEM. *P<0.05.
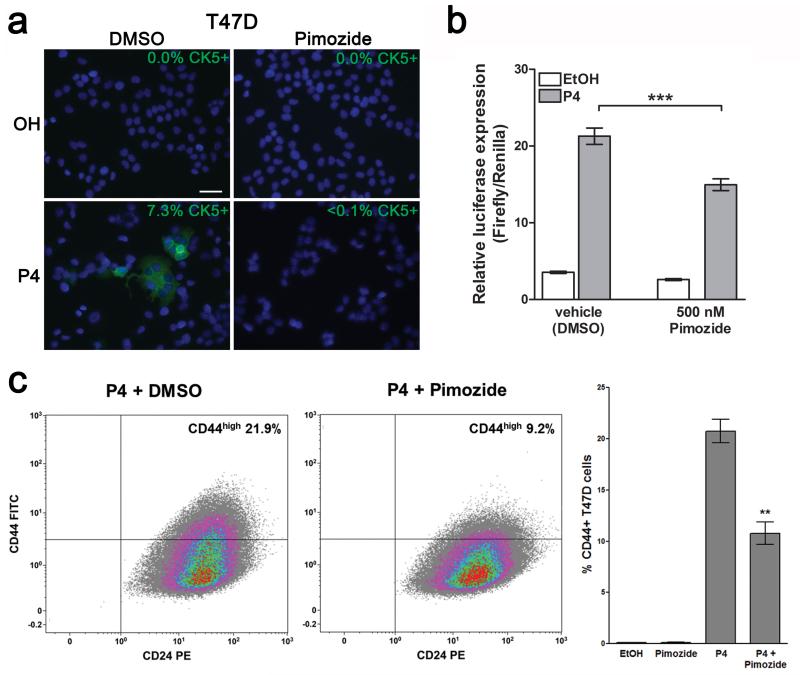
To determine whether Stat5a upregulation plays a functional role in the P4-induced expansion of CK5+ cells, we employed two methods to block Stat5a. First, we used siRNA to reduce Stat5a protein levels; P4-mediated increases in Stat5 and CK5 expression were both blocked in the presence of siSTAT5A (Figure 5c). We then analyzed the effects of siSTAT5A on CK5 promoter activity using T47D cells stably expressing a luciferase reporter gene driven by the human KRT5 (CK5) promoter (11). The P4-mediated increase in CK5 expression was significantly inhibited in the presence of siSTAT5A (Figure 5d). Second, we analyzed the effects of the Stat5 inhibitor Pimozide on the P4-mediated increase in CK5 expression. Pimozide is a Stat5 inhibitor that acts through inhibition of phospho-Stat5 production, but has no effect on NF-kB, Stat3, or Stat1 (42). Treatment of T47D cells with 500 nM Pimozide for 24 h inhibited P4 induction of CK5+ cells, measured by immunofluorescence for CK5 (Figure 6a) and CK5-promoter-luciferase activity (Figure 6b). Likewise, Pimozide reduced the P4 induction of CD44high cells by half (Figure 6c). Taken together, these results suggest that loss of miR-141 contributes to positive regulation of Stat5a, which in turn contributes to the P4-dependent expansion of CK5+ and CD44high breast cancer cells.
Figure 6.
The Stat5a inhibitor Pimozide reduces the P4-mediated expansion of CK5+ and CD44high cells. (a) Pimozide blocks P4-mediated induction of CK5+ cells. T47D cells were treated with EtOH or P4 for 24 h, with vehicle or 500 nM of the Stat5 inhibitor Pimozide, and immunofluorescence for CK5 (green) performed. The percentage of CK5+ cells indicated was measured in 6 fields and at least 300 cells per condition in biological triplicates. Scale bar, 50 μM. (b) Pimozide inhibits P4 induction of CK5. Stable CK5-promoter-luciferase-expressing T47D cells were plated at 10,000 cells per well in 96-well plates. After 24 h, cells were transfected with SV40-Renilla vector. Cells were treated for an additional 24 h with EtOH (vehicle) or P4, and DMSO (vehicle) or 500 nM Pimozide in eight wells per group. Luciferase activity was measured and normalized using Renilla; bars are mean +/− SEM. ***P<0.0001. Experiments were repeated twice. (c) Pimozide partially inhibits P4 induction of CD44high cells. T47D cells were treated with 100 nM P4 + vehicle (DMSO) or P4 + 500 nM Pimozide for 24 h, immunolabeled with FITC-CD44 and PE-CD24, and analyzed using flow cytometry. Representative flow analyses of cells −/+ Pimozide are shown. Graph depicts percentage of CD44high cells in triplicate samples. Bars represent mean +/− SEM. Significance is indicated as ** P<0.01.
Discussion
Progestins have emerged as crucial regulators of stem cells in the normal mammary gland and in breast cancer. In normal tissue, this regulation occurs both temporally during peak P4 levels, and spatially, as luminal-located cells signal to basally located MaSCs to expand (2, 3). Our group was first to describe that progestins increase a population of CK5+ stem-like cells in breast cancer (9, 10). Luminal breast cancer cells appear to directly convert from CK5− to CK5+ based on tracing studies using a CK5 promoter reporter (11, 43). Interestingly, transformed human mammary epithelial cells can convert to a CD44+CD24−/low phenotype spontaneously or via cytokine signals (27, 28). The P4-mediated expansion of stem-like cells, while necessary for normal breast function, is detrimental in breast cancer and may cause increased treatment resistance and recurrence. Sustained progestin use in hormone replacement therapy is hypothesized to increase breast tumorigenesis through expansion of pre-malignant stem cells (8). Since breast cancer cells lose compartmentalization and become more reliant on autocrine signaling (44), we focused on intracellular P4 signaling events that could potentially influence cell phenotype. In this article we describe a mechanism by which P4 suppression of miR-200 family member miR-141 cooperates to increase stem-like breast cancer cells.
We demonstrate that miR-141 is the only miR-200 family member temporally downregulated at the mature level in response to progestins in breast cancer cells. Although the primary transcript for the miR-141/200c cluster is downregulated, only mature miR-141 levels decrease (Figure 1). There is emerging evidence supporting differential processing, through multiple mechanisms including differential processing, maintenance of miRNA stability, and direct or indirect degradation (45-49). A prime example is context dependent processing of miR-18a within the miR-17 cluster of intronic miRNAs through the RNA binding protein hnRNP A1 (33). miR-200c is sometimes more abundant than miR-141 due to differential splicing that creates independent transcription units (32). This could also explain why it is less sensitive to transient downregulation of the primary transcript. Notably, miR-141, but not miR-200c, is underexpressed in CD44highCD24−/low prostate CSCs, further supporting differential regulation of miRNAs located in the same cluster (34). miR-141 is constitutively underexpressed in CD44high cells, as previously reported (36), and is transiently downregulated by P4 in the total cell population. This indicates its loss helps set in motion the gain of CD44high and CK5+ cells (Figure 2). This is similar to a feedback loop described in hepatocarcinoma cells; several miRNAs and HNF4a set oncogenesis into motion that cannot be reversed once initiated, even when miRNAs are restored to normal levels (50).
Transcription factors involved in controlling cell fate decisions are common miRNA targets. In breast cancer these include miR-200 or miR-205 targeting of EMT and stem cell promoting transcription factors Zeb1/2, Suz12, TWIST1, and BMI1 (17-21, 51, 52). Here we demonstrate miR-141 directly targets two transcription factors involved in regulating mammary cell differentiation: PR and Stat5a. P4 repression of miR-141 may allow for increased PR translation to recover from its ligand-dependent downregulation. The enhanced number of P4-induced CD44high and CK5+ cells with miR-141 inhibition could be partially due to more robust PR signaling. Stable inhibition of miR-141 was in fact sufficient to induce PR expression in breast cancer cell lines that typically require E2/ER-dependent PR expression (Figure 4). We also demonstrate that miR-141 targets both PR-A and PR-B at a site within the last coding exon mapped by genome-wide analysis of miRNA binding sites in luminal breast cancer cells (37).
The two highly conserved Stat5 transcription factors (transcribed from two separate genes, STAT5A and STAT5B) are both positively-regulated by progestins in breast cancer (30). We demonstrate that Stat5a is specifically targeted by miR-141 in breast cancer cells. Stat5a is also targeted by miR-141 in the bovine mammary gland (53). Selective deletion of Stat5 genes determined that Stat5a, but not Stat5b, is required for mammary development (39). Furthermore, Stat5a-null mammary epithelial cells show impaired differentiation, and are unable to expand the luminal progenitor population, which produces mature ER+PR+ alveolar cells (40, 54). Notably, CK5 is a major marker of luminal progenitor cells, the putative origin of BRCA1 breast cancers (12). We demonstrate that knockdown and inhibition of Stat5a reduces P4-mediated expansion of CK5+ and CD44high breast cancer cells (Figures 5&6). Stat5 is activated by another P4-regulated gene, prolactin receptor (PRLR). Prolactin (PRL) has been shown to reduce the P4 increase in CK5+ breast cancer cells, potentially though blocking induction of the transcriptional repressor BCL6 (55). Other reports found PRL antagonism could reduce clonogenicity and improve chemotherapy treatment of breast cancer cells and primary tumor xenografts (56), and could block PRL/Stat5 induction of basal stem-like populations in prostate cancer (57). Thus, interplay between PR-Stat5a-PRLR signaling in regulating the breast cancer CK5+ population requires further investigation, and could be a source for therapeutic intervention.
Both PR and Stat5a are positive prognostic indicators in breast cancer, and predict better response to tamoxifen-based endocrine therapy (58, 59). Loss of Stat5 signaling measured by nuclear phopho-Stat5 correlates with worse prognosis (60). Conversely, both PR- and Stat5a-null animals exhibit reduced mammary tumorigenesis in murine models (1, 39); this could be due to reduced stem/progenitor cell populations. In breast cancer cells, P4 upregulates Stat5a transcriptionally through its promoter and post-transcriptionally by depleting its repressor miR-141. Progestin regulation of stem-like cancer cell populations is complex; our data demonstrate miRNAs are utilized to modulate expression of mammary cell fate transcription factors. Downregulation of miR-141 is sufficient for increasing the CD44high population, as we previously demonstrated with miR-29 (24), and serves to amplify the progestin signal in increasing the CD44high/CK5+ populations. This luminal to basal/stem-like cell switch likely involves convergence of multiple signaling factors, including miRNAs as we demonstrate here. Ultimately, there may be opportunities for small molecule manipulation of breast cancers to maintain cells in a more treatment-vulnerable state.
Materials and Methods
Cell lines
Luminal breast cancer cell lines (T47D, BT474, and ZR75-1) were obtained from the University of Colorado Cancer Center Tissue Culture core. T47D cells were maintained in minimal Eagle’s medium, 5% fetal bovine serum (FBS), 1× NEAA, 1×10−9M insulin, 0.1 mg/mL sodium pyruvate, and 2 mM L-glutamine. BT474 and ZR75-1 cells were maintained in RPMI, 10% FBS, and 2.05 mM L-glutamine.
Reagents
On-Target pooled siRNAs, miRNA mimics/inhibitors, and transfection reagents were obtained from Thermo-Fisher Scientific (Pittsburgh, PA, USA); pmiR-GLO luciferase vector and Dual Luciferase Reporter assay were obtained from Promega (Madison, WI, USA). shRNA vectors were from the University of Colorado Functional Genomics Facility (Boulder, CO, USA). Primary antibodies used included: Stat5 (SC-835, SC-836; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), CK5 (mouse NCL-L-CK5 and rabbit 2290-1; Vector Laboratories/Epitomics, Burlingame, CA, USA), PR (Dako, Carpinteria, CA, USA), α-tubulin and β-actin (Sigma-Aldrich, St. Louis, MO, USA). For western blot, secondary antibodies included: IRDye 800CW Goat-Anti-Mouse IgG (926-32210) and IRDye 680LT Goat-Anti-Rabbit IgG (926-68021)(Li-Cor Biosciences, Lincoln, NE, USA). For Western blot imaging, the Odyssey Infrared Imaging System (Li-Cor Biosciences). For immunocytochemistry, secondary antibodies: AlexaFluor488 goat anti-mouse (A11029) and goat anti-rabbit (A11008), AlexaFluor594 goat anti-mouse (A11032) and goat anti-rabbit (A11037) (Invitrogen/Life Technologies, Grand Island, NY, USA). Hormones (17β-estradiol, progesterone, and MPA) were purchased from Sigma. Pimozide (573110) was purchased from EMD Millipore (Billerica, MA, USA).
RNA extraction and quantitative reverse-transcriptase PCR (qRT-PCR)
Total RNA from cultured cell lines was isolated using Trizol; total RNA from flow-sorted cells was isolated using the RNAqueous-Micro kit (Ambion, Austin, TX, USA) following manufacturer instructions. 10,000-50,000 cells were collected in provided lysis buffer. Analysis of mature miR-141, miR-200abc, and RNU6B (used for normalization) used TaqMan MicroRNA Assays, TaqMan MicroRNA RT kit, and TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Carlsbad, CA, USA). Primers for pri-miR-141 and pri-200c were designed to flank their respective stem-loop, and analysis was performed using the Verso cDNA Synthesis kit and ABsolute Blue Sybr Green with Fluorescein (Thermo-Fisher Scientific). β-actin was used for normalization. The relative mRNA or miRNA levels were calculated using the Pfaffl method (61).
Flow cytometry
Cells were labeled with antibodies CD44-APC, CD44-FITC, CD24-PE (BD Biosciences, Franklin Lakes, NJ, USA) at a concentration of 1×106 cells per ml in PBS+0.5% BSA, and were subjected to either flow cytometry analysis on a Gallios or fluorescence-activated cell sorting analysis on a Moflo XDP 100 (Beckman-Coulter, Indianapolis, IN, USA). Analysis was performed using Kaluza Analysis Software (Beckman-Coulter).
Stable cell lines
Lentiviral vectors were used to stably inhibit miR-141 by expressing complementary sequences to the mature miRNA (pMIRZIPs, System Biosciences, Mountain View, CA, USA). Lentiviral vectors containing the precursor sequence for miR-141 were used for overexpression (pMIRNAs, System Biosciences). A scrambled non-silencing vector was used as the negative control. Stably-expressing cells were selected using GFP-based cell sorting (for pMIRNA vectors) or puromycin (miR-ZIP vectors).
Mammosphere assay
Cells were seeded in non-adherent 6-well plates at 1000 cells/well in 2 mL of complete Mammocult medium (Stemcell Technologies, Vancouver, BC, Canada). After 24h, cells were treated with EtOH or 100 nM P4 for 10 days. Mammospheres were photographed, four fields/well, and mammospheres larger than 75 um in size were counted.
Proliferation assay
Real-time imaging (IncuCyte™, ESSEN BioScience Inc, Ann Arbor, Michigan, USA) was used to measure cell proliferation using non-labeled phase confluence. BT474 and T47D SCR-ZIP and 141-ZIP cells were plated at 5000 cells/well in 96-well plates. BT474 cells were pre-treated with E2 for 48 hours. Cells were treated in sextuplicate with EtOH or P4. Confluence measurements were started immediately after treatment, and taken every 4h for a total of 96h.
Tumor growth and limiting dilution formation in vivo
Tumor xenografts were developed by injecting the indicated amount of cells in 100% Matrigel Basement Membrane Matrix (BD Biosciences) into the fourth mammary fad pad of ovariectomized female NOD/SCID mice (for tumor growth) or nu/nu mice (for limiting dilution experiments) (Jackson Labs, Bar Harbor, ME, USA). Silastic pellets containing either 17β-estradiol alone (1 mg) or in combination with MPA (10 mg) were implanted subcutaneously at time of tumor cell injection. Tumors were measured weekly using a digital caliper, and tumor volume estimated using the formula (lw^2)/2. At termination of the experiment, mice were euthanized and tumors were excised and weighed. These experiments were performed under an approved University of Colorado Institutional Animal Care and Use Committee protocol.
Immunocytochemistry
Immunocytochemistry was performed as described (62), with additions of EtOH/10 nM P4 and/or DMSO/500 nM Pimozide for 24 h. Images were collected using a Nikon TiE microscope equipped with a digital camera and NIS Elements software. Adobe Photoshop CS5 was used to perform linear adjustments to brightness/contrast, assemble pictures into multipanel figures, and convert images from RGB to CMYK.
Luciferase assays
For 3′UTR-targeting experiments, portions of the 3′UTRs of PGR and STAT5A/B were cloned into the pmiR-GLO vector (Promega). Sites cloned from PGR are as follows (numbering based off +1 as first base of the 3′UTR): PGR_EXON, 357–+1; PGR_UTR_1, 2997–3961; PGR_UTR_2, 5764–6710; PGR_UTR_3, 8258–9073. Mutagenesis was performed by altering each predicted miR-141 site to 5′-GTCACCA. The site cloned from STAT5A is contained within bases 990-1255; mutagenesis altered the miR-141 binding site to 5′-CAGTGTT. Sites cloned from STAT5B are as follows: STAT5B_1, 1-379; STAT5B_2, 1329-1604. Mutagenesis altered STAT5B_1 to 5′-TCACAAT; STAT5B_2 was altered to 5′-GTCACAA. Cells were plated into 96 well plates at 104 cells/well. Cells were transfected in octuplicate with dual transfection reagent (Invitrogen) using 10ng of pmiR-GLO empty vector, or vectors containing most common sequence or mutated predicted miR-141 binding sites. Cells were lysed after 24 h and luciferase assays performed using the Dual Luciferase Reporter luciferase assay (Promega).
For K5P-Luc, T47D cells (11) were plated at 104 cells per well and transfected with siNC or siSTAT5A. After 24 h, EtOH or P4 was added for 24 h, cells lysed and luciferase activity measured. For Pimozide experiments, 500 nM Pimozide or vehicle (DMSO) was added in combination with EtOH or P4 for 24 h. pRL-SV40 Renilla vector (Promega) was the transfection efficiency control.
Statistical analyses
Statistics were done using Graphpad Prism version 5.0 for Windows 7 (La Jolla, CA, USA). Two-tailed Student’s t-tests or ANOVA followed by Bonferroni post hoc tests were used, as well as paired t-tests for tumor growth. P<0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank the University of Colorado Cancer Center Flow Cytometry and Tissue Culture Cores supported by P30CA046934 and the University of Colorado Department of Pathology Sequencing Core for their technical assistance and services. This work was supported by Department of Defense BCRP grants W81XWH-11-1-0210 (CAS, JKR) and W81XWH-11-1-0101 (DMC) and NIH R01 CA140985 (CAS). PK was supported by NIH K08 CA164048.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Author contributions: JFS and PH performed most of the studies. DMC performed experiments in Fig 2a,b and Fig 5c,d. PP performed experiments in Fig 3a. PK provided the PR exonic binding site for miR-141 (Fig 4c) and technical advice. JFS, DMC, BMJ, JKR, and CAS contributed intellectual design and interpretation of results. JFS wrote the manuscript. BMJ, JKR, CAS provided editorial assistance. All authors read and approved the final manuscript.
References
- 1.Ismail PM, Amato P, Soyal SM, DeMayo FJ, Conneely OM, O’Malley BW, et al. Progesterone involvement in breast development and tumorigenesis--as revealed by progesterone receptor “knockout” and “knockin” mouse models. Steroids. 2003 Nov;68(10-13):779–87. doi: 10.1016/s0039-128x(03)00133-8. [DOI] [PubMed] [Google Scholar]
- 2.Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010 Jun 10;465(7299):798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 3.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010 Jun 10;465(7299):803–7. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Suarez E, Jacob AP, Jones J, Miller R, Roudier-Meyer MP, Erwert R, et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature. 2010 Nov 4;468(7320):103–7. doi: 10.1038/nature09495. [DOI] [PubMed] [Google Scholar]
- 5.Schramek D, Leibbrandt A, Sigl V, Kenner L, Pospisilik JA, Lee HJ, et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature. 2010 Nov 4;468(7320):98–102. doi: 10.1038/nature09387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham JD, Mote PA, Salagame U, van Dijk JH, Balleine RL, Huschtscha LI, et al. DNA replication licensing and progenitor numbers are increased by progesterone in normal human breast. Endocrinology. 2009 Jul;150(7):3318–26. doi: 10.1210/en.2008-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanos T, Sflomos G, Echeverria PC, Ayyanan A, Gutierrez M, Delaloye JF, et al. Progesterone/RANKL is a major regulatory axis in the human breast. Sci Transl Med. 2013 Apr 24;5(182):182ra55. doi: 10.1126/scitranslmed.3005654. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz KB, Sartorius CA. Progestins in hormone replacement therapies reactivate cancer stem cells in women with preexisting breast cancers: a hypothesis. J Clin Endocrinol Metab. 2008 Sep;93(9):3295–8. doi: 10.1210/jc.2008-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horwitz KB, Dye WW, Harrell JC, Kabos P, Sartorius CA. Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5774–9. doi: 10.1073/pnas.0706216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sartorius CA, Harvell DM, Shen T, Horwitz KB. Progestins initiate a luminal to myoepithelial switch in estrogen-dependent human breast tumors without altering growth. Cancer Res. 2005 Nov 1;65(21):9779–88. doi: 10.1158/0008-5472.CAN-05-0505. [DOI] [PubMed] [Google Scholar]
- 11.Axlund SD, Yoo BH, Rosen RB, Schaack J, Kabos P, Labarbera DV, et al. Progesterone-inducible cytokeratin 5-positive cells in luminal breast cancer exhibit progenitor properties. Horm Cancer. 2013 Feb;4(1):36–49. doi: 10.1007/s12672-012-0127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009 Aug;15(8):907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 13.Bocker W, Moll R, Poremba C, Holland R, Van Diest PJ, Dervan P, et al. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab Invest. 2002 Jun;82(6):737–46. doi: 10.1097/01.lab.0000017371.72714.c5. [DOI] [PubMed] [Google Scholar]
- 14.Kabos P, Haughian JM, Wang X, Dye WW, Finlayson C, Elias A, et al. Cytokeratin 5 positive cells represent a steroid receptor negative and therapy resistant subpopulation in luminal breast cancers. Breast Cancer Res Treat. 2011 Jul;128(1):45–55. doi: 10.1007/s10549-010-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013 Jan-Feb;48(1):51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005 Jun 9;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008 May;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 18.Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011 Apr 18;13(2):R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009 May;8(5):1055–66. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007 Sep 1;67(17):7972–6. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 21.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008 Apr 1;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cochrane DR, Jacobsen BM, Connaghan KD, Howe EN, Bain DL, Richer JK. Progestin regulated miRNAs that mediate progesterone receptor action in breast cancer. Mol Cell Endocrinol. 2012 May 15;355(1):15–24. doi: 10.1016/j.mce.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cochrane DR, Spoelstra NS, Richer JK. The role of miRNAs in progesterone action. Mol Cell Endocrinol. 2011 Sep 17; doi: 10.1016/j.mce.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Cittelly DM, Finlay-Schultz J, Howe EN, Spoelstra NS, Axlund SD, Hendricks P, et al. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2013 May 16;32(20):2555–64. doi: 10.1038/onc.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 Nov 1;414(6859):105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 26.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009 Aug 18;106(33):13820–5. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7950–5. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1397–402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards DP, Leonhardt SA, Gass-Handel E. Novel mechanisms of progesterone antagonists and progesterone receptor. J Soc Gynecol Investig. 2000 Jan-Feb;7(1 Suppl):S22–4. doi: 10.1016/s1071-5576(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 30.Richer JK, Lange CA, Manning NG, Owen G, Powell R, Horwitz KB. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J Biol Chem. 1998 Nov 20;273(47):31317–26. doi: 10.1074/jbc.273.47.31317. [DOI] [PubMed] [Google Scholar]
- 31.Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, et al. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009 Oct 15;23(20):2382–7. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter HI, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res Notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007 Jul;14(7):591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012 Jul 1;72(13):3393–404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003 Apr 1;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009 Aug 7;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pillai MM, Gillen AE, Yamamoto TM, Kline E, Brown J, Flory K, et al. HITS-CLIP reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res Treat. 2014 Jul;146(1):85–97. doi: 10.1007/s10549-014-3004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, et al. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990 May;9(5):1603–14. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997 Jan 15;11(2):179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 40.Vafaizadeh V, Klemmt P, Brendel C, Weber K, Doebele C, Britt K, et al. Mammary epithelial reconstitution with gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation. Stem Cells. 2010 May;28(5):928–38. doi: 10.1002/stem.407. [DOI] [PubMed] [Google Scholar]
- 41.Santos SJ, Haslam SZ, Conrad SE. Estrogen and progesterone are critical regulators of Stat5a expression in the mouse mammary gland. Endocrinology. 2008 Jan;149(1):329–38. doi: 10.1210/en.2007-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson EA, Walker SR, Weisberg E, Bar-Natan M, Barrett R, Gashin LB, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011 Mar 24;117(12):3421–9. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid BG, Jerjian T, Patel P, Zhou Q, Yoo BH, Kabos P, et al. Live multicellular tumor spheroid models for high-content imaging and screening in cancer drug discovery. Curr Chem Genomics Transl Med. 2014;8(Suppl 1):27–35. doi: 10.2174/2213988501408010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obr AE, Edwards DP. The biology of progesterone receptor in the normal mammary gland and in breast cancer. Mol Cell Endocrinol. 2012 Jun 24;357(1-2):4–17. doi: 10.1016/j.mce.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006 Aug 15;20(16):2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009 Sep 24;461(7263):546–9. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 47.Grosshans H, Chatterjee S. MicroRNAses and the regulated degradation of mature animal miRNAs. Adv Exp Med Biol. 2010;700:140–55. [PubMed] [Google Scholar]
- 48.Schwab R, Speth C, Laubinger S, Voinnet O. Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep. 2013 Jul;14(7):615–21. doi: 10.1038/embor.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011 Jul;39(13):5692–703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011 Dec 9;147(6):1233–47. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012 Dec 14;287(51):42695–707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010 Sep 10;39(5):761–72. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Liu H, Jin X, Lo L, Liu J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genomics. 2012;13:731. doi: 10.1186/1471-2164-13-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vafaizadeh V, Klemmt PA, Groner B. Stat5 assumes distinct functions in mammary gland development and mammary tumor formation. Front Biosci. 2012;17:1232–50. doi: 10.2741/3983. [DOI] [PubMed] [Google Scholar]
- 55.Sato T, Tran TH, Peck AR, Girondo MA, Liu C, Goodman CR, et al. Prolactin suppresses a progestin-induced CK5-positive cell population in luminal breast cancer through inhibition of progestin-driven BCL6 expression. Oncogene. 2013 May 27; doi: 10.1038/onc.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howell SJ, Anderson E, Hunter T, Farnie G, Clarke RB. Prolactin receptor antagonism reduces the clonogenic capacity of breast cancer cells and potentiates doxorubicin and paclitaxel cytotoxicity. Breast Cancer Res. 2008;10(4):R68. doi: 10.1186/bcr2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rouet V, Bogorad RL, Kayser C, Kessal K, Genestie C, Bardier A, et al. Local prolactin is a target to prevent expansion of basal/stem cells in prostate tumors. Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15199–204. doi: 10.1073/pnas.0911651107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamashita H, Nishio M, Ando Y, Zhang Z, Hamaguchi M, Mita K, et al. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr Relat Cancer. 2006 Sep;13(3):885–93. doi: 10.1677/erc.1.01095. [DOI] [PubMed] [Google Scholar]
- 59.Clark GM, McGuire WL. Prognostic factors in primary breast cancer. Breast Cancer Res Treat. 1983;3(Suppl):S69–72. doi: 10.1007/BF01855130. [DOI] [PubMed] [Google Scholar]
- 60.Wagner KU, Rui H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J Mammary Gland Biol Neoplasia. 2008 Mar;13(1):93–103. doi: 10.1007/s10911-008-9062-z. [DOI] [PubMed] [Google Scholar]
- 61.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001 May 1;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012 Jul 24;135(2):415–32. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.