Abstract
Background
The presence of enterohepatic Helicobacter species (EHS) is commonly noted in mouse colonies. These infections often remain unrecognized but can cause severe health complications or more subtle host immune perturbations and therefore can confound the results of animal experiments. The aim of this study was to isolate and characterize a putative novel EHS that has previously been detected by PCR screening of specific-pathogen-free mice.
Materials and Methods
Biochemical analysis of enzyme activities (API campy), morphologic investigation (Gram-staining and electron microscopy) and genetic analyses (16SrRNA and 23SrRNA analyses, DNA fingerprinting, restriction fragment polymorphisms, and pulsed-field gel electrophoresis) were used to characterize isolated EHS. Genomic DNA fragments were sequenced to develop a species-specific PCR detection assay.
Results
Scanning electron microscopy revealed the presence of spiral-shaped EHS, which varied in length (2.5–6 µm) and contained single monopolar or single bipolar sheathed flagella. The bacteria were grown under anaerobic conditions, preferably on agar plates containing serum or blood. The 16SrRNA, genetic, and biochemical analyses indicated the identification of a novel EHS species, named Helicobacter magdeburgensis. We also examined the genome content using pulsed-field gel electrophoresis. Based on the pattern produced by two restriction enzymes, BamIII and KspI, the genome size was determined to be about 1.7–1.8 Mbp.
Conclusion
We isolated and characterized a novel EHS species, H. magdeburgensis, morphologically, biochemically, and genetically. These results are important for future studies on the prevalence and pathophysiologic relevance of such infections. Our PCR assay can be used to detect and discriminate H. magdeburgensis from other Helicobacter species.
Keywords: Enterohepatic Helicobacter, genome, pulsed-field gel electrophoresis, RAPD fingerprinting, electron microscopy
As Helicobacter pylori was the first bacterium cultivated from human gastric biopsy specimens in 1982, it has become apparent that Helicobacter spp. exhibit a broad host spectrum and can be isolated from the gastrointestinal tracts of humans, non-human primates, cats, dogs, cheetahs, ferrets, rodents, cows, sheep, pigs, dolphins, and birds. Members of the genus Helicobacter are helical curved, spiral or fusiform, Gram-negative bacteria with or without helical periplasmic fibers [1]. One of the best characterized species, H. pylori, was originally described as a member of the genus Campylobacter [2,3], but it was subsequently placed in its own genus based on 16SrRNA sequence analysis data [4–6]. There are currently 24 internationally recognized Helicobacter species in the genus [7], and numerous putative new “Candidatus Helicobacter” species are described in the literature. The sequence analysis of 16S rRNA of over 225 Helicobacter spp. isolated from mammals and birds indicates the genus to be phylogenetically diverse, potentially containing over 30 additional taxa [8].
Helicobacter spp. have widths of about 0.3–0.6 µm and lengths ranging from 1 to 5 µm. These microorganisms are highly motile by means of single or multiple flagella [1]. Optimum temperature for growth is from 37 to 42 °C. These bacteria mostly grow under microaerobic conditions and have a respiratory type of metabolism. Helicobacter spp. are oxidase-producing and most strains encode for a catalase. All gastric Helicobacter spp. described to date produce copious amounts of urease. Although strains of several species are capable of growing on simple nutritional agar media, the majority require media supplemented with blood or serum, and some species may be exacting (e.g. Helicobacter bizzozeronii, Helicobacter felis and Helicobacter salomonis) [9]. Detailed biochemical, morphologic, and physiologic aspects have been previously reported for species of this genus. Most Helicobacter spp. can be discriminated from the neighboring genera Campylobacter, Arcobacter, Wolinella, Sulfurospirillum, and Thiovulum by the presence of sheathing around the flagellar apparatus. However, two Helicobacter species (Helicobacter pullorum and Helicobacter rodentium) possess unsheathed flagella and therefore resemble species assigned to other related genera. Genotypic data, such as 16S rRNA sequence analysis, are the major features to unequivocally differentiate the genus Helicobacter from all other genera.
The description of a new Helicobacter species or subspecies is based on features used for assigning the new taxon to the genus and on characteristics used to differentiate the new taxon from existing taxa of the genus. For critical comparisons with other species, controls consisting of type or reference strains of the appropriate taxa should be tested [1]. The use of standardized, well-described tests, and methods, such as defined phenotypic test procedures, the inoculum size, composition of the gaseous atmosphere, period of incubation, and composition of the basal growth medium has been recommended [10–16]. Putative new species of uncultured organisms for which molecular sequence data are available (such as 16S rRNA sequence) may qualify for assignment to the provisional taxonomic status Candidatus [17]. In accordance with the study by Murray & Stackebrandt [18], 16S rRNA sequence data are not sufficient to assign Candidatus status. Therefore, morphotype, Gram-reaction, and other preliminary metabolic data should also be collected. With culturable Helicobacter spp., it is preferred that five isolates from different sources be studied, and that multiple sequences of the putative new “Candidatus Helicobacter” species cluster together in phylogenetic analyses [1]. This convention has been used for “Candidatus Helicobacter suis” [19] and “Candidatus Helicobacter bovis” [20], and may be appropriate for other uncultured Helicobacter spp., such as other gastric spiral organisms described by Solnick et al. [21].
Our groups are interested in the investigation and characterization of novel enterohepatic Helicobacter spp. (EHS), which are an emerging group of microaerobic, motile pathogens carrying flagella with variable styles in number and locations [22,23]. EHS are known to persistently colonize multiple animal species. They can be isolated from the lower intestine, hepatobiliary system, and diarrheic feces and are potentially associated with chronic inflammation and epithelial cell hyperproliferation leading to neoplasic disease [24,25]. Helicobacter hepaticus, considered the prototype of all known EHS [22], induces chronic active hepatitis and hepatocellular carcinoma in A/JCr and B6C3F1 mice strains as well as typhlocolitis in A/JCr inbred mice. This species is also used experimentally to induce cholesterol gallstones, inflammatory bowel disease (IBD), and in certain strains of mice induces colon cancer [26,27]. Because lesions caused by EHS in mice often mimic those seen in humans with cholecystitis, their possible role in hepatobiliary disease in humans has been proposed [28]. In addition, the possible zoonotic origin of important clinical manifestations in humans and the health status of mice housed in research facilities have recently attracted the attention of scientists [29]. In contrast to H. pylori, almost nothing is known about potential virulence factors in EHS. It is known, however, that cytolethal distending toxin (CDT), a well-recognized toxin first described by Johnson and Lior [30], is encoded in the genomes of several Helicobacters species [31].
To evaluate the prevalence of EHS infections in mouse strains harbored in our specific-pathogen-free (SPF) facilities, we tested 40 mouse lines that were permanently living in nine colony rooms using a group-specific PCR, which detects all Helicobacter species currently known [32]. When Helicobacter-negative and infected mice shared the same cage, transmission of the infection occurred within two weeks at very high frequency (100%). Furthermore, we found that mice from commercial breeding facilities may carry undetected Helicobacter infections [32]. We also showed that infection with EHS may occur and spread frequently in mice under SPF conditions, and despite extensive safety precautions. Our recent PCR analyses also indicated a high prevalence of rather uncommon Helicobacter species, which may be a consequence of current routine procedures for health screening of SPF mice. Here we describe a novel EHS isolated from SPF mice. We propose to name this BHS Helicobacter magdeburgensis.
Materials and Methods
Laboratory Mice
Mice from various mouse lines (BALB/c, C3H, and C57BL/6) that were contaminated with putative novel Helicobacter species were separated in individually ventilated cages. Fecal samples of each isolated mouse were then tested for presence of Helicobacter DNA as described previously [32]. Subsequently, the infection was transferred from individual infected animals to several PCR-proven Helicobacter-free C57BL/6 mice by harboring the mice in the same cage for 2 weeks. After transfer of the infection was detected by PCR [32], Helicobacter-infected mice were then used for bacterial isolation and culturing.
Bacterial Isolation
Helicobacter-infected mice were euthanized and organs were immediately dissected in a laminar flow hood. Ileum and the entire large intestine were cut into small pieces with sterile scissors, incubated with BHI (brain heart infusion) medium (5 mL per gram material), and shaken for 20 min at 37 °C in 50 mL Falcon tubes at 1000× g. The mixture was then centrifuged for 10 min at 2000× g to remove larger particles like cells and intestinal debris. The supernatant was removed and passed through sterile filter paper (Whatman) to further remove cell debris. Bacteria were then cultured in different amounts (100, 50, 25 or 5 µL) on different agar plates (Helicobacter pylori selective agar plates (HP), GC agar plates with 10% horse serum, Campylobacter selective plates, Muller-Hinton agar plates, and Columbia agar plates containing 5% sheep blood). These plates were incubated for 2, 3, 4, and 7 days. The Campygen™, Anaerogen™ from Oxoid, Anaerocult® from Merck gas generating systems, and one anaerobic chamber (5% N2, 4.5% CO2 and 3% H2) were used for incubation at 37–42 °C. Single bacterial colonies were picked and grown for further analysis.
Bacterial Control Strains and Growth Conditions
Helicobacter pylori, Helicobacter MIT 96-1001, Helicobacter typhlonicus, Helicobacter hepaticus, Helicobacter bilis ATCC 51630, Helicobacter mustelae, and Campylobacter jejuni strains were included as controls and grown under standard conditions. Briefly, H. pylori were cultivated on HP agar plates under microaerophillic conditions, and all other strains were cultivated on Columbia 5% sheep blood agar plates in an anaerobic chamber at 37 °C for 48 hours [9,10].
DNA Isolation and Purification
Bacteria were harvested with a sterile cotton swab and suspended in 100 µL of lysis buffer (50.0 mm Tris-HCl [pH 7.6], 1 mm EDTA, 0.5% Tween-20, 20 mg of proteinase K per mL) and incubated at 58 °C for 2 hours. The proteinase K was inactivated by conventional phenol/chloroform extraction. Purified DNA was then precipitated with 2.5 volume of 96% ethanol and washed with 70% ethanol.
Amplification and Sequencing of a 1.6-kb PCR Product of the 16S rRNA Gene
For amplification of the complete 16S rRNA gene, primers C70 (5′-AGA GTT TGA TYM TGG C-3′, forward) and B37 (5′-TAC GGY TAC CTT GTT ACG A-3′, reverse) were used [28]. Amplicons were then purified and sequenced directly by using the amplification primers C70 and B37, as well as internal primers C97-20: 5′- GGC TAT GAC GGG TAT CCG GC-3′ (forward), H5A: 5′-CGC GTG GAG GAT GAA GG-3′ (forward), C98: 5′-GAT TTT ACC CCT ACA CCA-3′ (reverse), H2: 5′-TCG CAA TGA GTA TTC CTC TT-3′(reverse), and H3A-20: 5′-GCC GTG CAG CAC CTG TTT TC-3′ (reverse) [24]. The gene has the NCBI GenBank accession number EF990624.
16S rRNA Sequence Analysis
16S rRNA sequence data were entered and aligned using the program RNA, which is set for data entry, editing, sequencing alignment, secondary structure comparison, similarity matrix generation, and dendrogram construction and is written in Microsoft QuickBASIC [5]. The database used contains approximately 400 Helicobacter, Wolinella, Arcobacter, and Campylobacter sequences and more than 1000 sequences of other bacteria. Similarity matrices were constructed from the aligned sequences using only those base positions for which data were available for 90% of the strains and were corrected for multiple base changes by the method of Jukes and Cantor [33]. A phylogenetic tree from the distance matrix was created with growtree using the UPGMA method [34].
Restriction Fragment Length Polymorphism (RFLP) of the 16S rRNA Gene
For restriction fragment analysis of the 16S rRNA gene, we amplified by PCR a specific and conserved 1.2-kb subfragment using primers C97 (5′- GCT ATG ACG GGT ATC C - 3′) and CO5 (5′- ACT TCA CCC CAG TCG CTG - 3′) by the method of Fox et al. [28]. RFLP patterns of amplified respective PCR products were obtained with each of the following enzymes: AluI, HhaI, ApaLI [25,35] in the appropriate buffer as recommended by the manufacturer (New England Biolabs, Acton, MA, USA). The resulting DNA cleavage products were compared with the RFLP patterns of H. magdeburgensis and known Helicobacter spp. after separation on agarose gels.
Amplification and Sequencing of PCR Products of the 23S rRNA Gene
Using primers O68 (forward), M86 (reverse), M93 (forward), and P46 (reverse), a 2,258-bp segment of the 23S rRNA gene was amplified as described [36] and sequenced using an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA). The consensus sequence was deposited in GenBank (accession number HM222564).
Randomly Amplified Polymorphic DNA (RAPD) Fingerprinting PCR
The RAPD fingerprinting method established for H. pylori strains [37] was used to compare the diversity of the DNA sequences among the Helicobacter strains tested. This method uses arbitrarily chosen oligonucleotides to prime DNA synthesis from genomic sites to which they are fortuitously matched, or almost matched. We used 20 ng genomic DNA from each strain as template, 20 pmol of each primer (5′- GAG CGG CCA AAG GGA GCA GAC-3′ D8635, 5′-CCG GAT CCG TGA TGC GGT GCG-3′ D9355, 5′-GGT TGG GTG AGA ATT GCA CG-3′ D14307), 1U Taq DNA-polymerase (Qiagen, Hilden, Germany) and 250 µm from each dNTP, 1× buffer, and water for a total volume of 50 µL. A Perkin-Elmer thermal cycler model 9700 was used for amplification reactions. The cycling program was four cycles of 94 °C, 5 min; 40 °C, 5 min; 72 °C, 5 min; low stringency amplification, and a final incubation at 72 °C for 10 min.
Agarose Gel Electrophoresis
Five microliters aliquots of each PCR reaction or 20 µL restriction digests were electrophoretically analyzed in 0.8–2.0 g/mL agarose gels containing 0.5 µg/mL ethidium bromide in 1× Tris acetate running buffer. The electrophoresis was for 2 hours at 100V. The 1-kb or 100-bp DNA ladder (Fermentas GeneRuler) was used as the size marker (M) in all gels.
Scanning Electron Microscopy (SEM)
Bacterial cells were harvested and fixed in a sterile solution containing 5% formaldehyde, 2% glutaraldehyde in cacodylate buffer (0.1 m cacodylate, 0.01 m CaCl2, 0.01 m MgCl2, 0.09 m sucrose, pH 6.9) for 1 hour on ice. The solution was centrifuged and passed through a sterile filter. After several washes with cacodylate buffer and TE buffer (20 m m Tris, 1 m m EDTA, pH 6.9), samples were dehydrated in serial dilutions of acetone (10%, 30%, 50%, 70%, 90%, and 100%) on ice for 15 min each step. Samples were then allowed to reach room temperature before another change of 100% acetone, after which they were subjected to critical-point drying with liquid CO2 (CPD030; Bal-Tec, now Leica, Wetzlar, Germany). Samples were finally covered with a ca. 10.0–11.0 nm thick gold film by sputter coating (SCD040; Bal-Tec) and examined in a field emission scanning electron microscope (Zeiss DSM 982 Gemini) using an Everhart Thornley SE detector and in-lens detector in a 50:50 ratio at an acceleration voltage of 5.0 kV.
Electron Microscopic Analysis of Negative Staining
For negative staining, thin carbon support films were prepared by indirect sublimation of carbon on freshly cleaved mica. Samples were then absorbed to the carbon film and negatively stained with 1% (wt/vol) aqueous uranyl acetate (pH 4.5). After air drying, samples were examined by transmission electron microscopy (TEM) in a Zeiss TEM 910 at an acceleration voltage of 80 kV.
Gram-staining and Biochemical Characterization Using the API Campy kit
Gram-staining of the isolated bacteria (Crystal violet, Gram’s iodine solution, acetone/ethanol (50 : 50 v:v), 0.1% basic fuchsin solution) was applied. For the biochemical characterization, the API Campy kit was used according to the recommendations of the manufacturer (bioMerieux, Marcy I’Etoile, France). API Campy is a standardized system for the identification of enzymatic activities in Campylobacter-like bacteria, which uses miniaturized tests, as well as a specially adapted database. Briefly, the API Campy strip consists of 20 microtubes containing dehydrated substrates. It is made up of two parts. The first part of the strip (enzymatic and conventional tests) is inoculated with a dense suspension, which rehydrates the substrates. During incubation (in aerobic conditions), metabolism produces color changes that are either spontaneous or revealed by the addition of reagents. The second part of the strip (assimilation or inhibition tests) is inoculated with a minimal medium and incubated in microaerobic conditions. The bacteria grow if they are capable of utilizing the corresponding substrate or if they are resistant to the antibiotic tested. The reactions were read and evaluated in accordance with the manufacturer’s identification table (bioMerieux).
Preparation of Genomic DNA in Low Melting Point (LMP) Agarose Plugs
After 24–48 hours of growth, bacterial colonies were suspended in TE buffer (50 mm Tris, 5 mm EDTA, pH 8.0) and embedded in low melting point (LMP) agarose (Mo Bio Laboratories, Inc., Carlsbaad, CA, USA), which were subsequently placed in lysis solution containing 0.25 m EDTA (pH 9.0), 0.5% lauroyl sarcosyl, and 0.5 mg of proteinase K per ml, as described previously [38]. One millimeter slices of the LMP agarose blocks were washed with phenylmethylsulfonyl fluoride solution (PMSF, 0.175 mg/mL) for 15 min, at least three times, and then washed three times with EB buffer. The agarose plugs were stored in EB buffer at 4 °C until further analyses.
Restriction Digests and Pulsed-field Gel Electrophoresis (PFGE)
The LMP agarose plugs containing genomic DNA were preincubated with 100 µL of the appropriate 1× restriction enzyme buffer before digestion was carried out with 50 U of enzyme in fresh 1× buffer. All restriction digests were incubated overnight at the temperature recommended by the manufacturer. Genomic DNA was digested with the following enzymes (Roche, Indianapolis, IN, USA) ApaI, AscI, BamHI, BglII, ClaI, HindIII, KpnI, KspI (SacII), MluI, NotI, NruI, PacI, SacI, SalI, SmaI, SpeI, XbaI, and XmaI. Restricted DNA fragments were separated by the contour-clamped homogeneous electric field method (CHEF Mapper, Bio-Rad, Hercules, CA, USA) in 1% SeaKem® Gold Agarose (Lonza, Basel, Switzerland) gels. Three different switch times were used, ranging from 3 to 35.38 seconds. Electrophoresis times varied from 12 to 18 hours at 6V/cm to visualize fragments of differing sizes. Two DNA markers were used to determine the sizes of the fragments. A Low Range PFGE Marker (New England Biolabs, Ipswich, MA, USA) was used as the DNA marker for 12 hour gels, and a digestion of Salmonella choleraesuis ss. Choleraesuis serotype Braenderup H9812 (ATCC BAA-664) genomic DNAs were used as the size markers for 18-hour gels. The markers were included in both sides of each PFGE gel with another one in the middle. Gels were stained with ethidium bromide, visualized with a UV transilluminator (Gel-Doc System, Bio-Rad), and pictures were recorded using GeneSnap (Syngene, Frederick, MD, USA). BioNumerics version 4.50 (Applied Maths, Austin, TX, USA) was used to perform a 10% background substraction of densitometric curves to identify bands and to determine band sizes and to perform genome calculations.
Cloning of Chromosomal DNA and Species-specific PCR Assay
To design species-specific PCR primers, we cloned fragments of chromosomal DNA. For this purpose, purified genomic DNA was prepared as described earlier and was digested at 37 °C for 3 hours using restriction endonuclease Sau3AI in 1× buffer provided by the manufacturer (New England BioLabs) and the resulting fragments were ligated into pBluescript II SK (+/−) vector. Ligated DNA was then transformed into E. coli Top 10 cells. Twenty five clones were sequenced as described earlier. Based on the sequence derived from one clone, specific primers with the following sequences 538F (5′-ATG CCG CCC TTG CAT CTG TC-3′) and 538R (5′- GGC GTA AAA ACT GAT GAA GCG AT-3′) were synthesized (Eurofins MWG GmbH, Ebersberg, Germany). The amplification conditions were 1U Taq DNA12 polymerase (Qiagen, Hilden), 1× 12.5 µm MgCl2, 1× PCR buffer, 20 ng template DNA, 200 µm dNTP, 20 pmol of each primer in a total volume of 50 µL at 95 °C, 5 min; 94 °C, 30 s; 68 °C, 30 s; 72 °C, 1 min (30–35 cycles); and finally 72 °C for 10 min. This PCR protocol specifically amplified a 750-bp PCR product of the cloned 800-bp Sau3AI fragment from the H. magdeburgensis genome.
Results
Isolation of Helicobacter from Mouse Intestines
Recently, the presence of EHS-DNA in 35 of 40 mouse strains harbored at our animal facility was detected. Direct sequencing of the PCR amplicons revealed that the mouse strains were infected with different known EHS including H. ganmani, H. hepaticus, H. typhlonicus as well as with novel EHS, which were not yet characterized [32]. Here we applied a more direct approach for the identification, isolation, culturing, and identification of putative novel EHS that were found to be present in the intestinal tract of some mice. For this purpose, we screened animals from mouse lines that were potentially harboring EHS. Altogether 13 mice (named HM001 to HM013) belonging to three mouse lines (BALB/c, C3H, and C57BL/6) were identified to be infected by EHS. Infected mice were then euthanized to prepare the organs and to obtain bacterial cultures. For this purpose, the ileum and the large intestine were cut into small pieces and incubated with prewarmed BHI medium. Suspensions were prepared and then cultured in different dilutions (100, 50, 25 or 5 µL) on multiple agar plates as described in Materials and Methods. We included Helicobacter pylori (HP) selective agar plates, GC agar plates with 10% horse serum, Campylobacter selective plates, Müller-Hinton agar plates, and Colombia agar plates containing 5% sheep blood. These plates were incubated from 2 to 7 days using Campygen (5% O2, 10% CO2, and 85% N2), Anaerogen (1.0% O2), Anaerocult (O2-deficient, CO2-enriched) or in an anaerobic chamber (5% N2, 4.5% CO2 and 3% H2), respectively. Of the complete microbiologic setting, EHS-like colonies were only identified from a subset of mice and only under anaerobic conditions using either GC agar plates with 10% horse serum or Columbia agar plates with 5% sheep blood. EHS-like bacterial colonies were obtained from HM003 (n = 1), HM004 (n = 1), HM006 (n = 2), HM007 (n = 2), HM009 (n = 4), HM010 (n = 1), and HM013 (n = 1). Gram-staining indicated that all these bacteria were Gram-negative (data not shown). In six other mice (HM001, HM002, HM005, HM008, HM011, and HM012), EHS-like colonies could not be cultured.
16S rRNA and 23S rRNA Phylogenetic Analysis
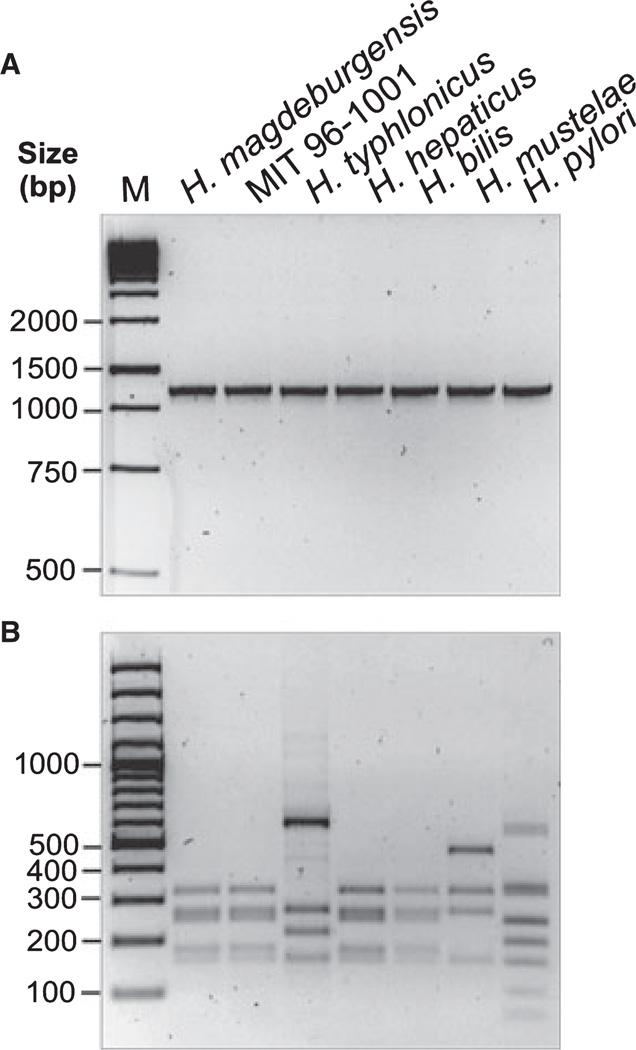
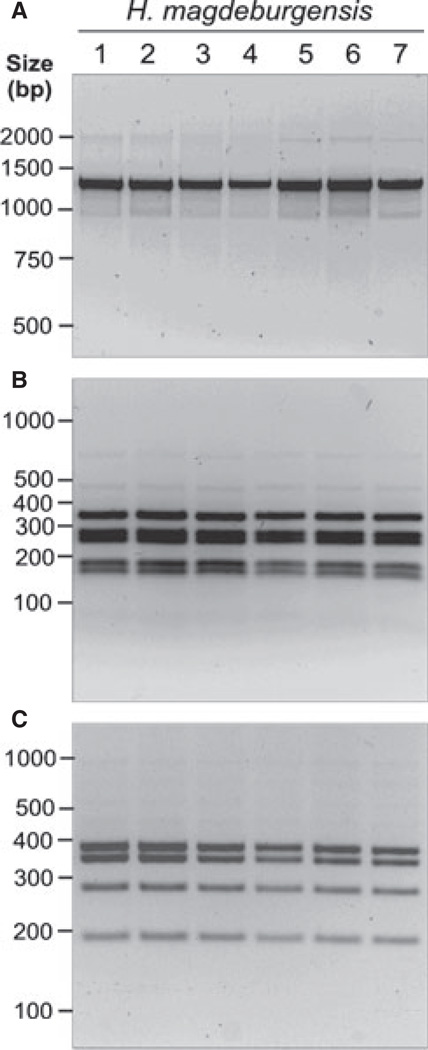
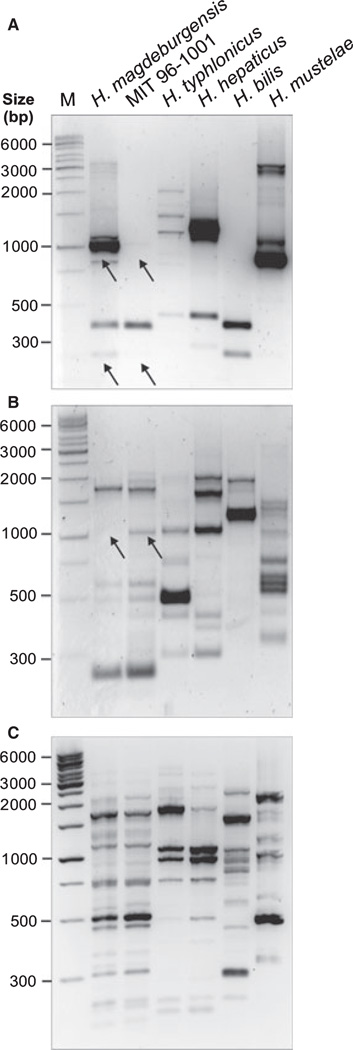
We suspected that the isolated bacteria belonged to the genus Helicobacter. To test this hypothesis, we isolated DNA for conventional PCR of the 16S rRNA gene. For this purpose, we amplified a 1.2-kb DNA subfragment of the 16S rRNA gene, which is highly conserved within the genus Helicobacter [28]. We also included controls of other known Helicobacter species such as H. typhlonicus, H. hepaticus, H. bilis, H. mustelae, H. pylori, and the MIT strain 96-1001. Amplification of the 1.2-kb PCR product was achieved for each of the species as expected (Fig. 1A). To confirm the specificity of these fragments, all PCR products were then digested with the restriction endonuclease AluI, which yields specific band patterns [25]. Indeed, the AluI pattern was identical between the new Helicobacter isolates and MIT strain 96-1001, H. hepaticus, and H. bilis but was different to that of H. typhlonicus, H. mustelae and H. pylori (Fig. 1B). Similar results were obtained using other recommended restriction enzymes such as HhaI and ApaLI (data not shown). Next, the 16S rRNA gene product from six other bacterial isolates from three mice (HM006, HM007, and HM009) was amplified and digested with AluI and HhaI, respectively. The results show that the PCR products and restriction fragment sites were identical suggesting the isolation of identical bacterial species from different mice (Fig. 2A–C).
Figure 1.
Investigation of 16S rRNA genes of different Helicobacter species by PCR and RFLP analyses. (A) DNA isolated from bacteria belonging to the genus Helicobacter (H. magdeburgensis, H. typhlonicus, H. hepaticus, H. bills, H. mustelae, H. pylori, and the MIT strain 96-1001) was applied for conventional PCR of the 16S rRNA gene. A conserved 1.2-kb DNA fragment in the genus Helicobacter was amplified [28]. (B) To confirm the specificity of these fragments, all PCR products were then digested with the restriction endonuclease AluI, which gives raise to a specific band pattern as described [25].
Figure 2.
Analysis of 16S rRNA of different Helicobacter magdeburgensis isolates by PCR and RFLPs. (A) DNA isolated from seven individual clones belonging to H. magdeburgensis was investigated by conventional PCR of the 16S rRNA gene. A conserved 1.2-kb DNA fragment in the genus Helicobacter was amplified [28]. To confirm the specificity of these fragments, all PCR products were then digested with the restriction endonuclease AluI (B) or HhaI (C) gives raise to a specific band pattern as described [25] and was identical among all investigated clones.
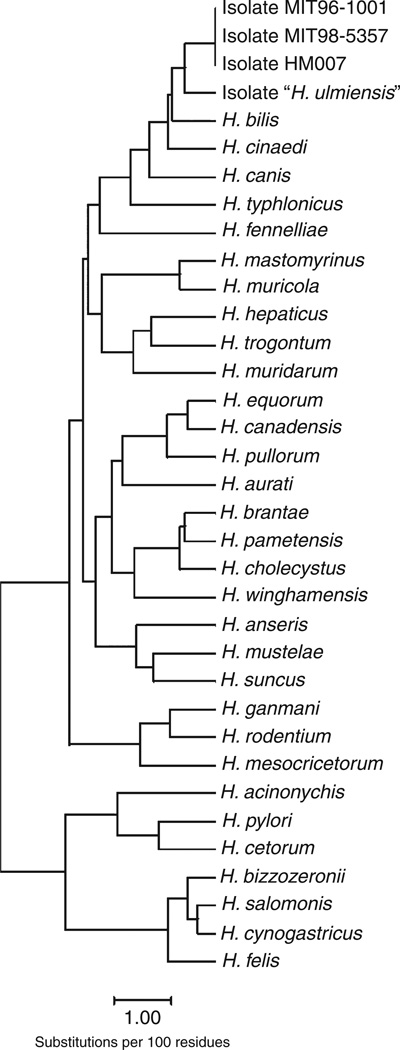
Next, the complete 16S rRNA gene sequence of the isolates HM006, HM007, and HM009 was determined by sequencing of a 1617-bp PCR product as described in Materials and Methods. All sequenced 16S rRNA genes from these mice gave rise to completely identical sequences. The 16S rRNA gene sequence of HM007-1, representative for these isolates, was deposited in the NCBI GenBank (accession number EF990624). Phylogenetically, the novel Helicobacter species isolated from HM006, HM007, and HM009 (hereafter named H. magdeburgensis) belong to a specific 16S rRNA gene cluster, which includes the species H. bilis, H. canis, H. cinaedi, H. typhlonicus, and the isolates MIT 96-1001 and MIT 98-5357, and H. ulmiensis. The 16S rRNA sequences of H. magdeburgensis and that of MIT 96-1001 and MIT 98-5357 were identical but varied clearly in comparison with other Helicobacter species as indicated in the phylogenetic tree (Fig. 3).
Figure 3.
Phylogenetic tree of 16S rRNA sequences of different Helicobacter species including H. magdeburgensis.
Interestingly, the 16S rRNA gene of H. magdeburgensis contained an intervening sequence (IVS) of 179 bp. Intervening sequences have also been described in H. bilis, H. typhlonicus, H. ulmiensis, and the EHS isolates MIT 96-1001 and MIT 98-5357. The IVS of H. magdeburgensis is identical to that of H. bilis and the Helicobacter isolates MIT 96-1001 and MIT 98-5357, while H. typhlonicus and H. ulmiensis have distinct IVS types.
The 23S rRNA from HM007-1 was also amplified and sequenced as described in Materials and Methods (accession number HM222564). Sequence analysis of this gene yielded a dendrogram, which was discordant with the dendrogram generated by the analysis of the 16S rRNA gene. But this discordance is not surprising in Helicobacter species because of the possible mosaic molecules in the 16S rRNA gene and the presence of intervening sequences in the 23S rRNA genes, which alter or may even produce a loss of phylogenetic information in these genes [36].
Morphologic Description of the Isolated Helicobacter ssp
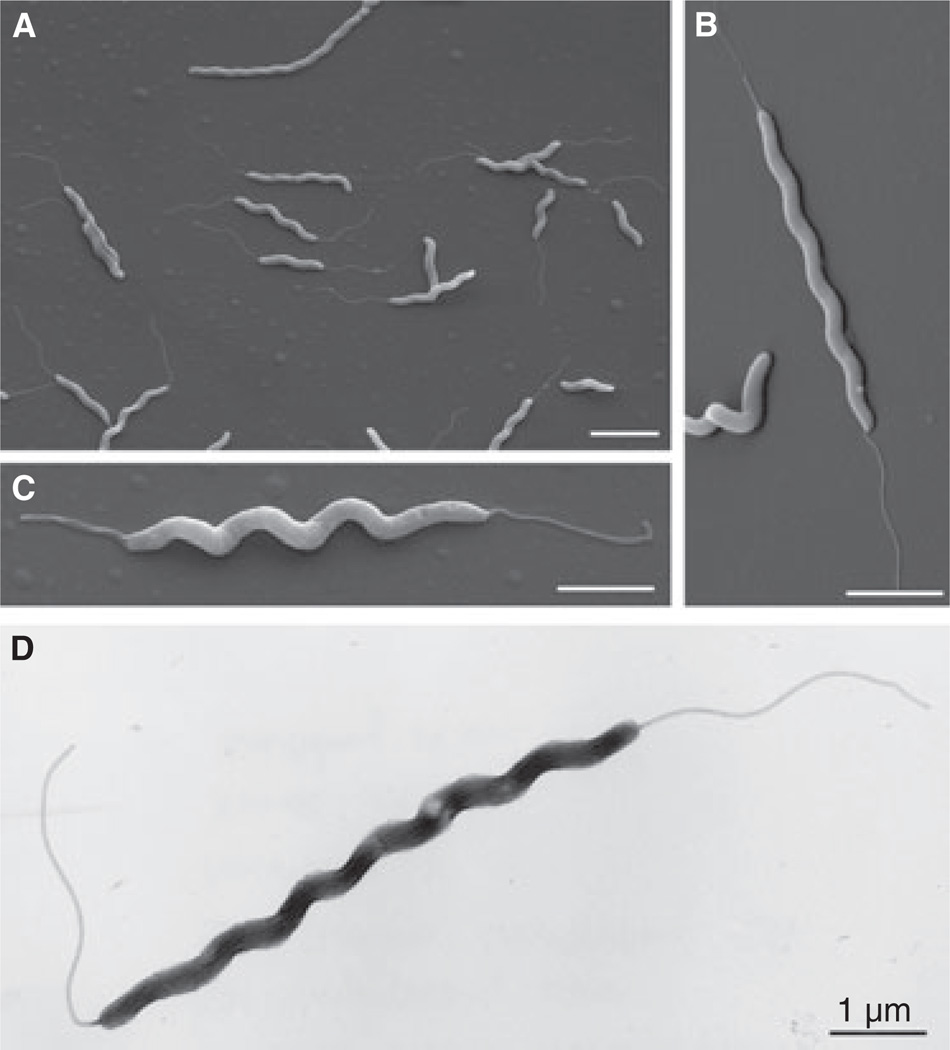
Next, we visualized the isolated H. magdeburgensis from HM006, HM007, and HM009. For this purpose, single bacterial colonies were grown for 2 days on Columbia agar plates containing 5% sheep blood and prepared as described. Scanning electronic microscopic investigation revealed in all cases spiral-shaped bacteria (Fig. 4A–C). These bacteria were about 0.18–0.22 µm in diameter and varied in length from 2.5 to 6 µm. The majority of these bacteria contained single monopolar or single bipolar flagella with lengths of about 1.5–2.5 µm. These flagella were commonly sheathed and about 28–32 nm in diameter. The nonsheathed flagella were about 16.5–17.5 nm in diameter. Preparation of H. magdeburgensis by another method (negative staining) revealed similar results, and thus confirmed our findings (example in Fig. 4D).
Figure 4.
Morphologic analyses of novel Helicobacter species by electron microscopy. (A–C) Scanning electron microscopy revealed spiral-shaped bacteria that were about 0.18–0.22 µm in diameter and varied in length from about 2.5–6 µm. The majority of bacteria contained single monopolar or single bipolar flagella. Representative pictures are shown from three preparations. (D) Investigation of the Helicobacter species by another method (negative staining) revealed similar results with respect to size and morphology. Each bar corresponds to 1 µm.
RAPD Fingerprinting of Helicobacter DNA
To further investigate the genetic relatedness between our strain and the closest known relative, MIT strain 96-1001 and other strains, we performed PCR-based randomly amplified polymorphic DNA (RAPD) fingerprinting analysis as described elsewhere [37]. This method uses a set of single primers (D14307, D9355 or D8635), which arbitrarily anneal and amplify genomic DNA resulting in strain-specific fingerprinting patterns [39]. Typical RAPD fingerprinting profiles with each of the three primers are shown in Fig. 5A–C. All control strains tested gave different RAPD profiles, indicating that they represent unrelated Helicobacter isolates. In agreement with the 16S rRNA analysis described earlier, we found that the RAPD pattern of our strain with two primers was very similar to that of MIT 96-1001 (Fig. 5B,C). However, using the primer D14307, we obtained a strong band at about 1 kb and a 1 weaker band at 200 bp, which were present in H. magdeburgensis and absent in MIT 96-1001 (Fig. 5A, arrows). In addition, a 1.1-kb band present in MIT 96-1001 is absent in our strain using primer D9355 (Fig. 5B, arrow) implying that H. magdeburgensis and the MIT 96-1001 represent different strains.
Figure 5.
PCR-based randomly amplified polymorphic DNA (RAPD) fingerprinting of Helicobacter species. To investigate the genetic relatedness between H. magdeburgensis and the closest known relative (MIT 96-1001) and other strains, we performed RAPD analysis as described [37]. (A–C) This method uses a set of single primers (D14307, D9355 or D8635), which arbitrarily anneal and amplify genomic DNA resulting in strain-specific fingerprinting patterns [39]. Typical RAPD fingerprinting profiles with each of the three primers are shown. Arrows indicate some bands either present or missing in H. magdeburgensis and MIT 96-1001, respectively.
Biochemical Characterization
To further characterize our H. magdeburgensis isolate, we determined the biochemical activity of specific enzymes using the conventional API Campy kit and compared the data of the representative HM007 isolate with that obtained for the MIT strain 96-1001 (Table 1). Helicobacter magdeburgensis was urease-negative as most of the EHS strains as assessed with a simple urease test, which is in concordance with our observation that it was found in the intestine of infected mice and does not require urease activity like H. pylori in the stomach. The major difference identified between the tested strains was that H. magdeburgensis was nitrate reductase-positive, while MIT 96-1001 was not. A minor difference was observed in the reductase of tetracoleum that showed weak activity for MIT 96-1001 but strong activity for H. magdeburgensis (Table 1). These results further demonstrate that H. magdeburgensis and MIT 96-1001 are closely related but not identical.
Table 1.
Enzymatic analysis of Helicobacter spp. by api® Campy test
| Tests | Reactions | Resultsa |
|
|---|---|---|---|
|
H. magdeburgensis |
MIT 96-1001 |
||
| URE | UREase | - | - |
| NIT | Reduction of NITrates | + | - |
| EST | ESTerase | - | - |
| HIP | HIPpurate | - | - |
| GGT | Gamma Glutamyl Transferase | - | - |
| TTC | Reduction of Triphenyl Tetrazolium Chloride | + | ± |
| PyrA | Pyrrolidonyl Arylamidase | - | - |
| ArgA | L-Arginine Arylamidase | - | - |
| Asp A | L-Aspartate Arylamidase | - | - |
| PAL | ALkaline Phosphatase | + | + |
| H2S | Production of H2S | - | - |
| GLU | Assimilation (GLUcose) | - | - |
| SUT | Assimilation (sodium SUccinaTe) | - | - |
| NAL | Growth inhibition (NALidixic acid) | - | - |
| CFZ | Growth inhibition (sodium CeFaZoline) | - | - |
| ACE | Assimilation (sodium ACEtate) | - | - |
| PROP | Assimilation (PROPionate) | - | - |
| MLT | Assimilation (MaLaTe) | - | - |
| CIT | Assimilation (trisodium CITrate) | - | - |
| ERO | Susceptibility - therapeutic prediction(ERythrOmycin) | - | - |
URE, urea; NIT, potassium nitrate; EST, 5-bromo-4-chloro-3-indoxyl acetate; HIP, sodium hippurate; GGT, γ-L-glutamic acid-β-naphthylamide; TTC, triphenyltetrazolium chloride; PyrA, pyroglutamic acid β–naphtilamide; ArgA, L-arginine-4-methoxy-β-naphthylamide; AspA, aspartic acid-β-naphthylamide; PAL, 2-naphthyl phosphate; H2S, sodium thiosulfate; GLU, D-glucose; SUT, sodium succinate; NAL, nalidixic acid; CFZ, sodium cefazoline; ACE, sodium acetate; PROP, propionic acid; MLT, malic acid; CIT, trisodium citrate; ERO, erythromycin.
Negative result, −; positive result, +; intermediate values, ±
Pulsed-Field Gel Electrophoresis (PFGE) Analysis of Chromosomal DNA and Calculation of Genome Size
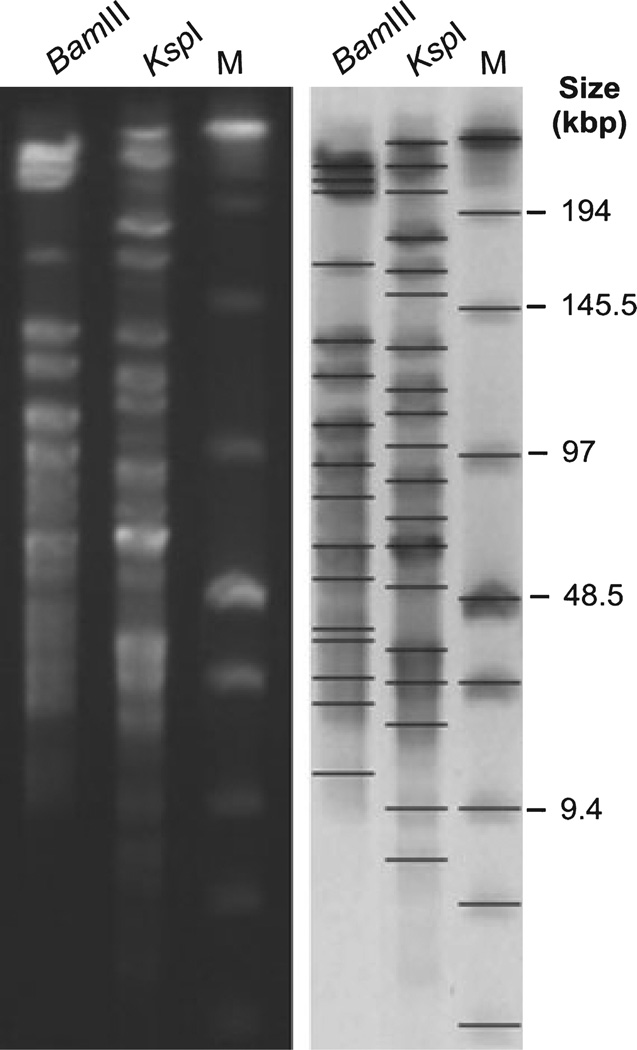
In the next set of experiments, we analyzed H. magdeburgensis HM007-1 in terms of restriction enzyme digests and estimation of its genome size. For this purpose, a series of commonly used restriction enzymes were tested to determine which ones were adequate for genome mapping using PFGE. Interestingly, ApaI, AscI, ClaI, KpnI, MluI, NotI, PacI SalI SmaI, and XmaI failed to digest the genome, while BglII, HindIII, NruI, SacI, SpeI, XbaI yielded a large number of short DNA fragments that were difficult to discriminate by PFGE (Fig. 6). However, the digestion with BamIII resulted in 16 DNA fragments with sizes ranging from 12.86 to 231.98 kbp, and the digestion with KspI resulted in 17 fragments with sizes between 7.99 and 243.23 kbp (Table 2). These two restriction enzymes proved to be the most suitable for PFGE analysis of the H. magdeburgensis genome. In addition, we used these two enzymes to calculate the approximate genome size of H. magdeburgensis (Table 2). Digests with BamIII revealed a total genome size of about 1695 kb and the restriction with KspI yielded a size of about 1793 kb, respectively.
Figure 6.
Pulsed-field gel electrophoresis (PFGE) analysis of Helicobacter magdeburgensis. Chromosomal DNA was digested with the restriction endonucleases BamIII and KspI, respectively. Low Range PFGE Marker was used as the DNA size marker (M). BioNumerics software was used to identify bands and to determine band sizes. The values from the genome calculations are summarized in Table 2.
Table 2.
Determination of Helicobacter magdeburgensis genome size by Restriction fragment length polymorphism (RFLP) and pulsed-field gel electrophoresis (PFGE) analysisa
| Restriction Enzyme |
||
|---|---|---|
| BomIII | KspI | |
| Detected band sizes (in kbp) | 230.15 | 243.23 |
| 220.12 | 227.48 | |
| 210.53 | 210.67 | |
| 169.21 | 183.99 | |
| 137.83 | 166.55 | |
| 126.16 | 134.49 | |
| 111.18 | 123.69 | |
| 97.72 | 114.39 | |
| 87.19 | 92.37 | |
| 78.72 | 77.90 | |
| 67.83 | 69.02 | |
| 57.29 | 55.38 | |
| 41.68 | 33.69 | |
| 26.40 | 24.86 | |
| 20.66 | 17.83 | |
| 12.60 | 9.90 | |
| - | 7.99 | |
| Total size | 1695.27 | 1793.43 |
A representative PFGE gel is shown in Fig. 6.
Development of a H. magdeburgensis-specific PCR Assay
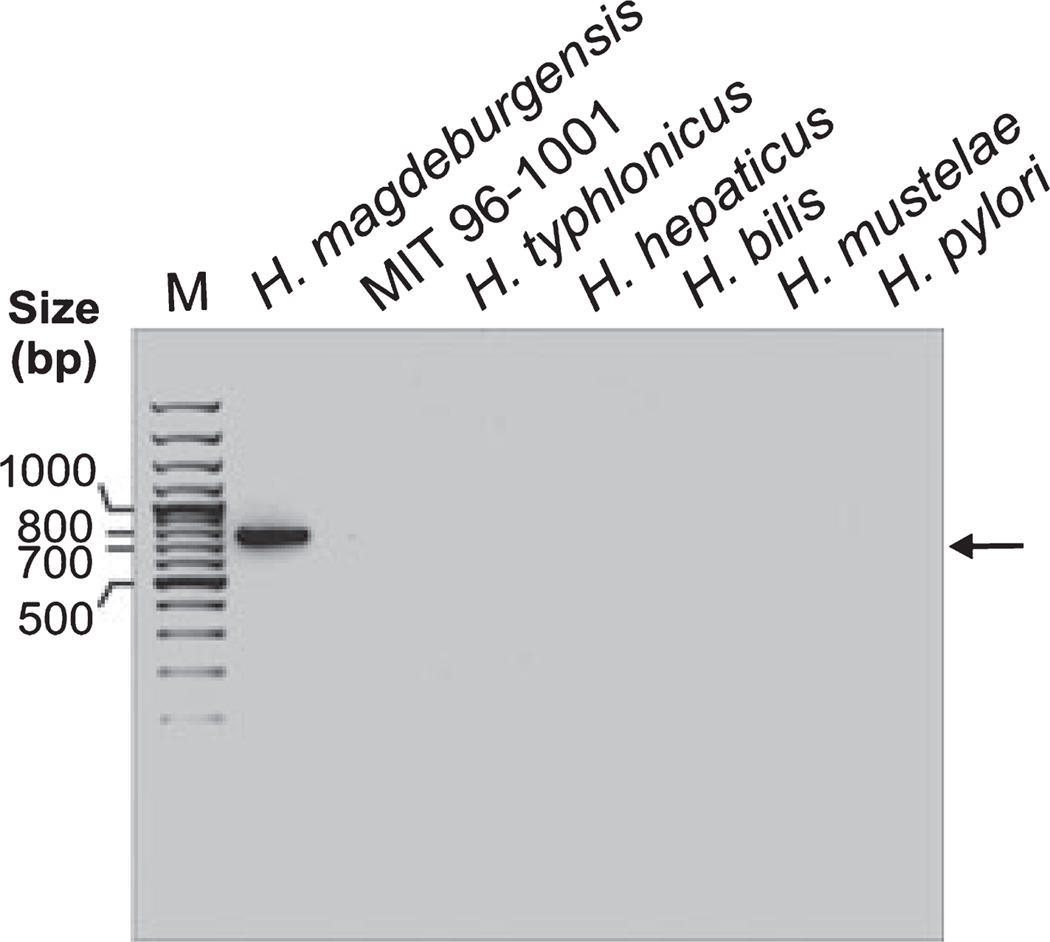
Finally, to discriminate our new isolate from other Helicobacter species, we developed a H. magdeburgensis-specific PCR assay. For this purpose, we digested isolated H. magdeburgensis DNA with Sau3AI, a frequent cutting restriction enzyme, which produced 0.05–10-kb DNA fragments on conventional agarose gels. These fragments were cloned into the pBluescript vector and 25 randomly selected single clones were sequenced. The results showed that 12 fragments had some weak homology to chromosomal DNA from H. hepaticus. The majority of the other cloned inserts exhibited weak homology to DNA from other bacteria but were mainly very small in size (< 100 bp) and therefore not useful for a PCR assay. However, one of the clones having the size of about 800 bp was of particular interest because it did not show any homology to known sequences in the NCBI database. Thus, the latter DNA fragment was used to design species-specific PCR primers as described in Materials and Methods. Using these primers, we developed a PCR assay for the detection of a single H. magdeburgensis-specific 750-bp DNA fragment. This DNA fragment is clearly absent in all other Helicobacter isolates tested, including the MIT strain 96- 1001, H. typhlonicus, H. hepaticus, H. bilis, H. mustelae, and H. pylori (Fig. 7).
Figure 7.
Development of a H. magdeburgensis-specific PCR detection assay. We designed specific PCR primers as described in the Materials and Methods section. Using these primers, we developed a PCR assay giving rise to a single H. magdeburgensis-specific 750-bp DNA fragment, which is clearly absent in all other Helicobacter isolates tested, even after 35 PCR cycles, including the MIT strain 96-1001, H. typhlonicus, H. hepaticus, H. bills, H. mustelae, and H. pylori.
Discussion
Helicobacter is a rapidly expanding bacterial genus with a wide host range but limited biologic niches. The respective ecologic division and taxa are often referred as gastric Helicobacter species (GHS) and enterohepatic Helicobacter species (EHS) [3,40]. EHS are emerging as important pathogens within this genus [7]. EHS can colonize the lower gastrointestinal tract, including the ileum, cecum, colon, and biliary tree. Similarly to GHS, EHS can cause persistent infections associated with chronic inflammation and epithelial cell hyperproliferation leading to neoplastic transformation [29]. EHS are also confounding factors in inflammatory bowel disease in the mouse animal model [32]. The interest in keeping healthy mice in research facilities, and the studies of the zoonotic potential of these mouse populations [41] has stimulated researchers to investigate EHS in more detail. Importantly, there is very little information concerning the Helicobacter status in noncommercial animal facilities. Numerous recent studies using culture and PCR methods indicated that the presence of EHS can become a very common problem in commercial mouse colonies [32,42–44]. These infections often remain nonrecognized but can cause severe health complications and, thus, can also change the results of animal experiments.
In recent studies using PCR assays from feces of our laboratory mice, we identified that the most frequently detected DNA from Helicobacter species corresponds to that of H. ganmani and MIT 98-5357 [32]. No species-specific PCR assays or other detection methods have been established for the analysis of these bacteria in animal health screens. Therefore, these rather uncommon Helicobacter species remain undetected by the routine screening procedures, which can explain the relatively high prevalence of these rare species in Helicobacter-infected mice. To avoid the spread of Helicobacter infections in any animal facility of research institutions, it is important to elucidate novel Helicobacter species, characterize them at the molecular level, and study the transmission route and possible disease outcome.
In the present report, we describe the direct isolation and molecular characterization of a novel urease-negative, straight spiral or curved rod-shaped Gram-negative Helicobacter species from laboratory mice in our animal facility. This bacterium is highly motile by means of single monopolar or bipolar sheathed flagella without helical periplasmic fibers. Using two different electron microscopic methods (SEM and negative staining), these bacteria were measured 0.3–0.6 µm in width and had lengths ranging from about 1.0 to 6.0 µm. Analysis of the 16S rRNA revealed that the bacterium is a novel member of the genus Helicobacter but distinct from known species; thus, we propose to name it Helicobacter magdeburgensis. Further analysis of biochemical traits and morphologic characteristics as well as genetic analysis revealed that this bacterium is closely related to a MIT 96-1001 Helicobacter strain [45,46]; however, we could differentiate these two strains by means of ApiCampy, RAPD fingerprinting, and other methods. Interestingly, Helicobacter magdeburgensis grows under anaerobic conditions, but its definition as a strict anaerobic bacterium will require further studies.
Our RFLP and subsequent PFGE analysis of chromosomal DNA revealed that this bacterium has an approximate genome size of 1.7 to 1.8 Mbp. Restriction enzymes including ApaI, AscI, ClaI, KpnI, MluI, NotI, PacI, SalI, SmaI, and XmaI failed to digest the genome, while other commonly used enzymes such as BglII, HindIII, NruI, SacI, SpeI, XbaI yielded a large number of short DNA fragments. The latter findings indicated incomplete digests; therefore, the resulting bands were found difficult to discriminate on PFGE gels. However, the digestion of chromosomal H. magdeburgensis DNA with two other enzymes, BamIII or KspI, resulted in suitable PFGE patterns to map the entire genome. Both enzymes were therefore used to calculate the approximate genome size of H. magdeburgensis, being in the size range of about 1.7 or 1.8 Mbp, respectively. These values are slightly higher than the genome of H. hepaticus but are in agreement with other Helicobacter spp. [47,48]. Finally, we cloned some genomic DNA fragments for sequencing and to develop a species-specific PCR assay that can be efficiently used for rapid and specific differentiation of H. magdeburgensis from other common EHS. Interestingly, this 750-bp genomic DNA fragment is obviously absent in the MIT 96-1001, further demonstrating that H. magdeburgensis and MIT 96-1001 are different EHS strains.
Taken together, we have identified and characterized morphologically, biochemically, and genetically, a novel EHS isolated from the intestine of certified specific-pathogen-free laboratory mice. As unrecognized infections with diverse microorganisms may change the results of animal experiments, our studies are very important for unraveling the presence/absence of these unknown bacteria in laboratory animals such as mice. The results of this study are also important for future studies of the pathophysiologic relevance of such infections. The isolate most closely related to H. magdeburgensis, strain MIT 96-1001, has been shown to exhibit pathogenic properties in both the liver and lower bowel of infected A/J and scid mice and to express a CDT ortholog [45,46]. Future studies will therefore define whether and how H. magdeburgensis may contribute to disease conditions in mice and other infected animals, and the interaction of H. magdeburgensis with other microbial species in the mouse intestine.
Acknowledgements
The authors thank Dana Zabler and Diana Schmidt (University of Magdeburg) and Robert Scott Miller (Auburn University) for excellent technical assistance. The work of SB was supported through Priority Program SPP1150 of the Deutsche Forschungsgemeinschaft (DFG) Ba1671/3-3 and DFG grant Ba1671/8-l. Francisco Rivas Traverso was supported by a scholarship from Secretaría Nacional de Ciencia y Tecnología (SENACYT) in Panama.
References
- 1.Dewhirst FE, Fox JG, On SL. Recommended minimal standards for describing new species of the genus Helicobacter. Int J Syst Evol Microbiol. 2000;50:2231–2237. doi: 10.1099/00207713-50-6-2231. [DOI] [PubMed] [Google Scholar]
- 2.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 3.Marshall BJ, Royce H, Annear DI, et al. Original isolation of Campylobacter pyloridis from human gastric mucosa. Microbios. 1984;25:83–88. [Google Scholar]
- 4.Romaniuk PJ, Zoltowska B, Trust TJ, et al. Campylobacter pylori, the spiral bacterium associated with human gastritis, is not a true Campylobacter sp. J Bacterial. 1987;169:2137–2141. doi: 10.1128/jb.169.5.2137-2141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paster BJ, Dewhirst FE. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 6.Goodwin CS, Armstrong JA, Chilvers T, et al. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Campylobacter pylori comb. nov. and Helicobacter mustelae comb. nov., respectively. Int J Syst Bacteriol. 1989;39:397–405. [Google Scholar]
- 7.Fox JG. The non-H pylori Helicobacters their expanding role in gastrointestinal and systemic diseases. Gut. 2002;50:273–283. doi: 10.1136/gut.50.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewhirst FE, Chien CC, Paster BJ, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jalava K, On SL, Vandamme PA, et al. Isolation and identification of Helicobacter spp. From canine and feline gastric mucosa. Appl Env Microbiol. 1998;64:3998–4006. doi: 10.1128/aem.64.10.3998-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Megraud F, Bonnet F, Garnier M, et al. Characterization of “Campylobacter pyloridis” by Culture, Enzymatic Profile, and Protein Content. J Clin Microbiol. 1985;22:1007–1010. doi: 10.1128/jcm.22.6.1007-1010.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.On SL, Holmes B. Effect of inoculum size on the phenotypic characterization of Campylobacter species. J Clin Microbiol. 1991;29:923–926. doi: 10.1128/jcm.29.5.923-926.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.On SL, Holmes B. Reproducibility of tolerance tests that are useful in the identification of campylobacteria. J Clin Microbiol. 1991;29:1785–1788. doi: 10.1128/jcm.29.9.1785-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.On SL, Holmes B. Assessment of enzyme detection tests useful in identification of campylobacteria. J Clin Microbiol. 1992;30:746–749. doi: 10.1128/jcm.30.3.746-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.On SL, Holmes B. Classification and identification of Campylobacters and Helicobacters and allied taxa numerical analysis of phenotypic characters. Syst Appl Microbiol. 1995;18:374–390. [Google Scholar]
- 15.Paster BJ, Lee A, Fox JG, et al. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991;41:31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- 16.Barrow GI, Feltham RKA. Cowan and Steel’s Manual for the Identification of Medical Bacteria. Cambridge: University Press; 1993. [Google Scholar]
- 17.Murray RG, Schleifer KH. Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int J Syst Bacteriol. 1994;44:174–176. doi: 10.1099/00207713-44-1-174. [DOI] [PubMed] [Google Scholar]
- 18.Murray RG, Stackebrandt E. Taxonomic note: implementation of the provisional status candidatus for incompletely described procaryotes. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 19.De Groote D, van Doorn LJ, Ducatelle R, et al. ‘Candidatus Helicobacter suis ‘, a gastric helicobacter from pigs, and its phylogenetic relatedness to other gastrospirilla. Int J Syst Bacteriol. 1999;49:1769–1777. doi: 10.1099/00207713-49-4-1769. [DOI] [PubMed] [Google Scholar]
- 20.De Groote D, van Doom LJ, Ducatelle R, et al. Phylogenetic characterization of ‘Candidatus Helicobacter bovis’, a new gastric helicobacter in cattle. Int J SystBacteriol. 1999;49:1707–1715. doi: 10.1099/00207713-49-4-1707. [DOI] [PubMed] [Google Scholar]
- 21.Solnick JV, O’Rourke J, Lee A, et al. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 22.Fox JG, Dewhirst FE, Tully JG, et al. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox JG, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 24.Bohr URM, Glasbrenner B, Primus A, et al. Identification of enterohepatic Helicobacter species in patients suffering from inflammatory bowel disease. J Clin Microbiol. 2004;42:2766–2768. doi: 10.1128/JCM.42.6.2766-2768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia A, Xu S, Dewhirst FE, et al. Enterohepatic Helicobacter species isolated from the ileum, liver and colon of a baboon with pancreatic islet amyloidosis. J Med Microbiol. 2006;55:1591–1595. doi: 10.1099/jmm.0.46707-0. [DOI] [PubMed] [Google Scholar]
- 26.Maurer KJ, Ihrig MM, Rogers AB, et al. Identification of cholelithogenic enterohepatic Helicobacter species and their role in murine cholesterol gallstone formation. Gastroenterology. 2005;128:1023–1033. doi: 10.1053/j.gastro.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Rogers AB, Fox JG. Inflammation and Cancer. I. Rodent models of infectious gastrointestinal and liver cancer. Am J Physiol Gastrointest Liver Physiol. 2004;286:G361–G366. doi: 10.1152/ajpgi.00499.2003. [DOI] [PubMed] [Google Scholar]
- 28.Fox JG, Dewhirst FE, Shen Z, et al. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 29.Taylor NS, Xu S, Nambiar P, et al. Enterohepatic Helicobacter species are prevalent in mice from commercial and academic institutions in Asia, Europe, and North America. J Clin Microbiol. 2007;45:2166–2172. doi: 10.1128/JCM.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WM, Lior H. A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb Pathog. 1988;4:115–126. doi: 10.1016/0882-4010(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 31.Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal distending toxin. Cell Microbiol. 2008;10:1599–1607. doi: 10.1111/j.1462-5822.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- 32.Bohr URM, Selgrad M, Ochmann C, et al. Prevalence and Spread of Enterohepatic Helicobacter Species in Mice Reared in a Specific-Pathogen-Free Animal Facility. J Clin Microbiol. 2006;44:738–742. doi: 10.1128/JCM.44.3.738-742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jukes TH, Cantor CR. Evolution of protein molecules. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 34.Sneath PEA, Sokal RR. Numerical Taxonomy: The Principles and Practice of Numerical Classification. San Francisco: Freeman; 1973. [Google Scholar]
- 35.Fox JG, Chien CC, Dewhirst FE, et al. Helicobacter canadensis sp. nov. isolated from humans with diarrhea as an example of an emerging pathogen. J Clin Microbiol. 2000;38:2546–2549. doi: 10.1128/jcm.38.7.2546-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewhirst FE, Shen Z, Scimeca M, et al. Discordant 16S and 23S rRNA phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. J Bacteriol. 2005;187:6106–6118. doi: 10.1128/JB.187.17.6106-6118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akopyanz N, Bukanov NO, Westblom TU, et al. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang N, Taylor DE. Use of pulsed-field agarose gel electrophoresis to size genomes of Campylobacter species and to construct a SalI map of Campylobacter jejuni UA580. J Bacteriol. 1990;172:5211–5217. doi: 10.1128/jb.172.9.5211-5217.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akada JK, Ogura K, Daiva D, et al. Helicobacter pylori tissue tropism: mouse colonizing strains can target different gastric niches. Microbiology. 2003;149:1901–1909. doi: 10.1099/mic.0.26129-0. [DOI] [PubMed] [Google Scholar]
- 40.On SL. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria:current status, future prospects and immediate concerns. J Appl Microbiol. 2001;90:1s–15s. doi: 10.1046/j.1365-2672.2001.01349.x. [DOI] [PubMed] [Google Scholar]
- 41.Azevedo NF, Almeida C, Fernandes I, et al. Survival of gastric and enterohepatic Helicobacter spp. in water: implications for transmission. Appl Eviron Microbiol. 2008;74:1805–1811. doi: 10.1128/AEM.02241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goto K, Ohashi H, Takakura A, et al. Current status of Helicobacter contamination of laboratory mice, rats, gerbils, and house musk shrews in Japan. Curr Microbiol. 2000;41:161–166. doi: 10.1007/s002840010111. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson HO, Ouis IS, Stenram U, et al. High prevalence of Helicobacter species detected in laboratory mouse strains by multiplex PCR-denaturing gradient gel electrophoresis and pyrosequencing. J Clin Microbiol. 2004;42:3781–3788. doi: 10.1128/JCM.42.8.3781-3788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seok S, Park J, Cho S, et al. Health surveillance of specific pathogen-free and conventionally-housed mice and rats in Korea. Exp Anim. 2005;54:85–92. doi: 10.1538/expanim.54.85. [DOI] [PubMed] [Google Scholar]
- 45.Chien CC, Taylor NS, Ge Z, et al. Identification of cdtB homologues and cytolethal distending toxin activity in enterohepatic Helicobacter spp. J Med Microbiol. 2000;49:525–534. doi: 10.1099/0022-1317-49-6-525. [DOI] [PubMed] [Google Scholar]
- 46.Shomer NH, Dangler CA, Schrenzel MD, et al. Cholangiohepatitis and inflammatory bowel disease induced by a novel urease-negative Helicobacter species in A/J and Tac:ICR:HascidfRF mice. Exp Biol Med (Maywood) 2001;226:420–428. doi: 10.1177/153537020122600505. [DOI] [PubMed] [Google Scholar]
- 47.Saunders KE, MCGovern KJ, Fox JG. Use of pulsed-field gel electrophoresis to determine genomic diversity in strains of Helicobacter hepaticus from geographically distant locations. J Clin Microbiol. 1997;35:2859–2863. doi: 10.1128/jcm.35.11.2859-2863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suerbaum S, Josenhans C, Sterzenbach T, et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. PNAS. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]