Abstract
In humans, the S100 protein family is composed of 21 members that exhibit a high degree of structural similarity, but are not functionally interchangeable. This family of proteins modulates cellular responses by functioning both as intracellular Ca2+ sensors and as extracellular factors. Dysregulated expression of multiple members of the S100 family is a common feature of human cancers, with each type of cancer showing a unique S100 protein profile or signature. Emerging in vivo evidence indicates that the biology of most S100 proteins is complex and multifactorial, and that these proteins actively contribute to tumorigenic processes such as cell proliferation, metastasis, angiogenesis and immune evasion. Drug discovery efforts have identified leads for inhibiting several S100 family members, and two of the identified inhibitors have progressed to clinical trials in patients with cancer. This Review highlights new findings regarding the role of S100 family members in cancer diagnosis and treatment, the contribution of S100 signalling to tumour biology, and the discovery and development of S100 inhibitors for treating cancer.
The term S100 was first used in 1965 to denote a mixture of the two founding family members, S100A1 and S100B1. This term alludes to the solubility of these approximately 10,000 Da proteins in 100% saturated ammonium sulphate. Although S100 family members exhibit a high degree of sequence and structural similarity, they are not functionally interchangeable and they participate in a wide range of biological processes such as proliferation, migration and/or invasion, inflammation and differentiation2–4. The structure and function of the S100 proteins are regulated by Ca2+ binding, which allows them to act as Ca2+ sensors that can translate fluctuations in intracellular Ca2+ levels into a cellular response5,6. Individual family members show unique affinities for divalent metal ions, oligomerization properties, post-translational modifications and spatiotemporal expression patterns. Intracellular S100 proteins bind to and regulate the activity of many targets; in some cases, multiple S100 family members may regulate one target2–4. Several S100 proteins are present in the extracellular space where they can participate in local intercellular communication (autocrine and paracrine), enter the systemic circulation and coordinate biological events over long distances. S100 proteins lack a signal peptide for secretion via the conventional Golgimediated pathway, and whether extracellular S100 proteins are actively secreted from living cells or passively released is still debated2,4. Extracellular S100 proteins interact with a variety of cell-surface receptors including receptor for advanced glycosylation end products (RAGE; also known as AGER), G protein-coupled receptors, Toll-like receptor 4 (TLR4), scavenger receptors, fibroblast growth factor receptor 1 (FGFR1), CD166 antigen (also known as ALCAM), interleukin-10 receptor (IL-10R), extracellular matrix metalloproteinase inducer (EMMPRIN; also known as basigin) and the bioactive sphingolipid ceramide 1-phosphate4,7–10. The functional diversity of S100 proteins and the unique repertoire of family members expressed in cells and tissues enable individual cells to generate unique and adaptive responses to changes in intracellular Ca2+ levels and the extracellular environment.
There are 21 S100 proteins — which are exclusively found in vertebrates — encoded in the human genome11. As new family members were discovered, the S100 nomenclature evolved with the consequence that numerous aliases exist for some S100 proteins4,12. Four family members are dispersed throughout the genome: S100B on chromosome 21, S100G on the X chromosome, S100P on chromosome 4 and S100Z on chromosome 5. The remaining 17 family members (S100A1–S100A14, the S100A7 genes and S100A16) are encoded in two tandem clusters within a 2 Mb region on chromosome 1q21 that is referred to as the epidermal differentiation complex (EDC). The EDC also contains genes encoding the S100-fused type proteins (SFTPs) trichohyalin (TCHH), TCHH-like 1 (TCHHL1), repetin (RPTN), hornerin (HRNR), filaggrin (FGL), FGL2 and cornulin (CRNN)13. SFTPs contain a full-length S100 protein domain fused in-frame to multiple tandem repeats composed of one or two sequences, for which the function is not well characterized. The five genomic loci that encode S100 proteins are highly conserved, but there are differences among species that affect the extrapolation of results from preclinical studies to human cancers. For example, the mouse genome lacks genes encoding S100A12 and S100P, and the protein encoded by the single mouse S100a7 locus (S100a7a) differs substantially from the proteins encoded by the three human S100A7 loci (S100A7, S100A7A and S100A7L2)11. Furthermore, although 1q21–25 is a hotspot for chromosomal alterations, mutations and/or translocations in S100 genes are rare. The only reported event involving chromosomal deletions of S100 family members is oral cancer (in which there is a deletion of S100A1–S100A16)14. Of the four S100A14 polymorphisms reported in oesophageal squamous cell carcinoma, only the mutation 461G>A is associated with increased cancer susceptibility due to diminished binding of S100A14 to p53 (REFS 14,15). S100A2 polymorphisms have been reported in non-small-cell lung cancer (NSCLC), but they are not associated with altered S100A2 expression or function16.
Nonetheless, dysregulation of S100 protein expression is a common occurrence in many human cancers. In vivo studies have shown that altered expression of ten family members contributes to the growth, metastasis, angiogenesis and immune evasion of numerous tumours (TABLE 1). Inhibitors directly targeting two family members, S100B and S100A9, are in clinical trials for melanoma and prostate cancer, respectively. This Review focuses on new advances regarding the role of S100 proteins in cancer diagnosis and treatment, the contribution of S100 signalling to cancer cell biology and the development of new S100 protein inhibitors for treating cancer.
Table 1.
In vivo cancer phenotypes of S100 family members
| Cancer | Family member | Model systems | Phenotype | Refs |
|---|---|---|---|---|
| Breast | S100A1 | Xenograft | ↑ Growth | 168 |
| S100A4 | Xenograft and GEMM | ↑ Metastasis | 77–79,87–89,169,170 | |
| S100A7 | Xenograft and GEMM | ↑ ↓ Growth | 54,69,71 | |
| S100A8 | Xenograft | ↑ Metastasis | 103 | |
| Osteosarcoma | S100A4 | Xenograft | ↑ Metastasis | 171–173 |
| Oral | S100A2 | Xenograft | ↓ Growth | 174 |
| S100A7 | Xenograft | ↓ Proliferation and ↓ metastasis | 38 | |
| Head and neck | S100A4 | Xenograft | ↑ ↓ Growth and ↑ metastasis | 175–177 |
| Lung | S100A2 | Xenograft | ↔ Growth and ↑ metastasis | 178 |
| S100A4 | Xenograft | ↑ Metastasis | 179 | |
| S100A9 | GEMM | ↓ Inflammation | 180 | |
| S100A10 | Xenograft | ↑ Growth and ↑ immune evasion | 181 | |
| S100A11 | Xenograft | ↑ Growth | 37 | |
| S100P | Xenograft | ↑ Angiogenesis and ↑ metastasis | 132 | |
| Prostate | S100A3 | Xenograft | ↑ Growth | 182 |
| S100A4 | Xenograft and GEMM | ↑ Growth, ↑ metastasis and ↑ angiogenesis | 183–185 | |
| S100A9 | GEMM | ↑ Growth, ↑ metastasis and ↑ angiogenesis | 19,148 | |
| S100A8 and S100A9 | Xenograft | ↔ ↓ Growth, ↔ metastasis and ↓ inflammation | 186 | |
| S100P | Xenograft | ↑ Growth | 187 | |
| Colorectal | S100A4 | Xenograft | ↑ Metastasis | 188 |
| S100A8 and S100A9 | Xenograft | ↑ Growth and ↑ metastasis | 18 | |
| S100P | Xenograft | ↑ Growth and ↑ metastasis | 133,134 | |
| Brain | S100A4 | Xenograft | ↑ Metastasis | 189 |
| S100A9 | Xenograft | ↑ Growth | 190 | |
| S100B | Xenograft | ↑ Growth, ↑ angiogenesis and ↑ inflammation | 30 | |
| Gastric | S100A4 | Xenograft | ↑ Growth | 191 |
| S100A6 | Xenograft | ↑ Growth and ↓ metastasis | 192 | |
| Bladder | S100A4 | Xenograft | ↑ Metastasis | 193 |
| Lymphoma | S100A9 | Xenograft | ↑ Growth and ↑ immune evasion | 19,100 |
| Pancreas | S100A4 | Xenograft | ↑ Growth and ↑ angiogenesis | 94 |
| S100P | Xenograft | ↑ Growth and ↑ metastasis | 143,194 | |
| Melanoma | S100A4 | Xenograft | ↑ Growth and ↑ angiogenesis | 94 |
| S100A9 | Xenograft | ↑ Metastasis | 9 | |
| S100B | Xenograft and GEMM | ↑ Growth | 123,145 | |
| Renal | S100A4 | Xenograft | ↑ Growth and ↑ metastasis | 195 |
| Liver | S100A14 | Xenograft | ↑ Growth and ↑ metastasis | 196 |
| Thyroid | S100A4 | Xenograft | ↑ Growth and ↑ metastasis | 197 |
| S100A8 | Xenograft | ↑ Growth and ↑ metastasis | 198 | |
| S100A11 | Xenograft | ↑ Growth | 199 | |
| Thymus | S100A8 and S100A9 | Xenograft | ↑ Growth and ↑ immune evasion | 144 |
↓, decreased; ↑, increased; ↔, no change; GEMM, genetically engineered mouse model.
Conformation and structure
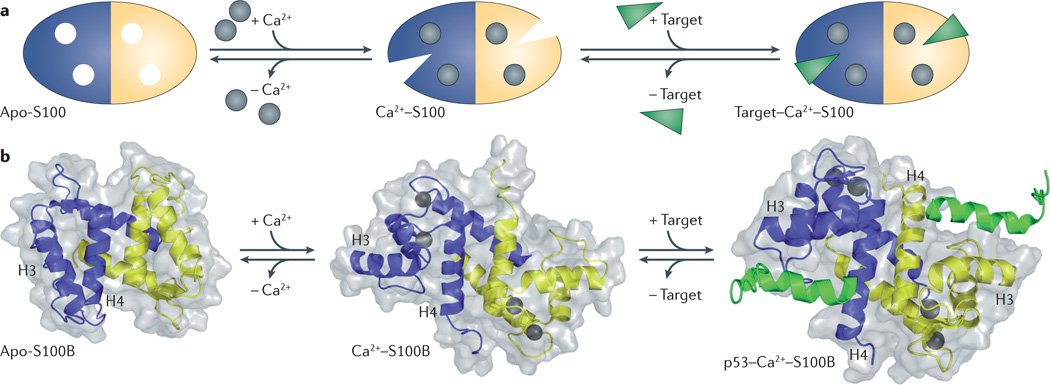
The S100 proteins are typically symmetric dimers with each S100 subunit containing four α-helices4. Although there are several reports of in vitro heterodimerization among family members, and mixtures of S100A1 homodimers, S100B homodimers, and S100A1–S100B heterodimers can be isolated from brain17, only the S100A8–S100A9 heterodimer has been documented to have physiologically relevant functions in vivo18–20. Each of the two S100 subunits contains two Ca2+-binding domains: a carboxyterminal canonical EF-hand (a helix–loop–helix domain) composed of 12 amino acids, and an amino-terminal ‘pseudo’ or ‘S100’ EF-hand that is unique to S100 proteins and is composed of 14 amino acids4. These motifs are connected by a ‘hinge’ region (loop 2), which consists of 10–12 residues and is crucial for target interactions. In the absence of target, the Ca2+-binding affinity of most S100 proteins is low, but when bound to target, the Ca2+-binding affinity increases by 5–300-fold21–23. This biochemical coupling can be understood in terms of structural transitions, as upon binding Ca2+, S100 proteins undergo a substantial conformational rearrangement that reorients helix 3 to expose a hydrophobic cleft that is required for target binding (FIG. 1). Recent studies have suggested that in the absence of a target, Ca2+-bound S100 proteins exist as an equilibrium population of dynamic conformers with an overall weak Ca2+-binding affinity, and that target binding narrows the distribution to favour S100 sub-states with high affinities for Ca2+ ions24. As a consequence, target binding is typically Ca2+-dependent. Importantly, the three-dimensional structures of S100–target complexes have revealed that individual family members exhibit distinct modes of target recognition owing, in part, to differences in surface geometries, hydrophobic residue distribution and charge density25. In addition, some family members undergo a variety of post-translational modifications, such as oxidative modification and sumoylation, which can modulate S100–target complex formation and/or intracellular localization26–29. Structural and biochemical considerations are particularly relevant to the development of therapies targeting S100 family members. Importantly, the 3D structures of S100 proteins permit an extensive analysis of target selectivity, which can be exploited for drug discovery, as described later in this Review.
Figure 1. S100 protein structural organization.
a | Apo-S100 protein shown with one blue subunit and one yellow subunit. S100 proteins are regulated by Ca2+ binding (grey circles), which allows them to act as Ca2+ sensors that can translate alterations in intracellular Ca2+ levels into a cellular response. Ca2+ binding induces a conformational rearrangement that exposes a hydrophobic cleft, allowing the S100 protein to bind its cellular targets (green) and elicit a physiological response. b | Ribbon and surface diagrams of apo–S100B, Ca2+–S100B and the Ca2+–S100B–p53 peptide complex. Individual subunits are shown in blue and yellow, the Ca2+ ions are shown as dark grey spheres, and the TP53 peptide is shown in green. The conformational rearrangement that occurs upon Ca2+ binding is referred to as the ‘Ca2+ switch’, and involves the reorientation of helix 3 (H3) and subsequent exposure of hydrophobic residues that participate in target protein binding.
Expression in cancer
Cancers exhibit a distinctive S100 protein profile that can be both stage-specific and subtype-specific. In gliomas, S100B expression positively correlates with proneuronal, neuronal and classic — but not mesenchymal — subtypes, whereas S100A8 and S100A9 expression positively correlate with mesenchymal subtypes30. Despite the availability of protein signatures from breast31, head and neck32, prostate33, melanoma34 and colorectal35,36 cancers, comprehensive analyses of S100 protein expression have not been carried out. Nonetheless, discernible trends and notable exceptions emerge when the S100 protein expression profiles in human cancers are compared (see Supplementary information S1 (table)). Dysregulation of multiple S100 family members occurs in most cancers and typically involves upregulation. One family member, S100P, is upregulated in all cancers that have been examined and the remaining S100 family members are upregulated in most but not all cancers. Two exceptions are head and neck, and ocular cancers, in which 11 out of 13, and 6 out of 12 dysregulated S100 proteins are downregulated, respectively (see Supplementary information S1 (table) and references therein). The contradictory expression profiles reported for some family members may be attributable to cancer subtype, disease stage, cellular distribution, or issues associated with S100 protein and/or mRNA detection. For example, S100A11 expression is increased in NSCLC, but is decreased in small-cell lung cancer37. S100A7 is expressed in pre-invasive, well-differentiated and early-stage oral squamous cell carcinomas, but not in non-invasive, poorly differentiated, late-stage tumours38. For S100A7, it is important to note that studies carried out before the development of paralogue-specific PCR primers and antibodies probably report on multiple loci39. As the S100A7 paralogues are differentially expressed and regulated, and exhibit diverse functions, it is important to discriminate between them in order to understand their distinct functional roles in tumorigenesis. In addition, only a limited number of commercial S100 antibodies have been tested for cross-reactivity against multiple family members40, and the generic term anti-S100 indicates potential cross-reactivity with multiple family members.
S100 protein expression profiles can be used to facilitate diagnosis and/or prognosis, inform on treatment options and monitor patient response to therapy (see Supplementary information S2, S3 (tables) and references therein). S100B expression in the primary tumour has been used as a diagnostic marker for malignant melanoma in human and veterinary medicine since the 1980s. S100P levels have diagnostic utility in a variety of human cancers as S100P can be detected in primary tumours (breast, colorectal, pancreatic and ovarian cancer), metastatic lesions (lung cancer), serum (breast and colorectal cancer), saliva (head and neck cancer), bile (cancer of the bile duct) and faeces (colorectal cancer) (see Supplementary information S2 (table) and references therein). The combined expression levels of S100A2 and S100A10 are used for prognosis in recurrent colon cancer after adjuvant 5-fluorouracil (5-FU) therapy and radical surgery5 (also see Supplementary information S3 (table) and references therein). In addition, autoantibodies directed against S100A7 have been reported in ovarian cancer and may facilitate diagnosis24(also see Supplementary information S2 (table) and references therein). It should be noted that the utility of S100 proteins as cancer biomarkers may be limited by their elevated expression in other pathologies, including cardiovascular, neurological and inflammatory diseases.
The expression of S100 family members in human cancers is controlled by a complex regulatory network, which includes epigenetic mechanisms and signal transduction pathways, including pathways that are activated in response to chemotherapeutic agents. Epigenetic changes regulate S100P expression in prostate, pancreatic and cervical cancer; S100A4 expression in pancreatic, endometrial, gastric, breast, ovarian, renal and brain tumours; S100A2 in prostate and breast cancer; S100A6 in prostate and gastric cancer; and S100A10 expression in pituitary cancer41–45. In colon cancer, seven S100 genes are direct targets of histone-lysine methyltransferase MLL2 (also known as KMT2B and KMT2D)46; however, coordinated regulation of S100 proteins within a given cancer is atypical. For example, in colon cancer, S100A4 expression is controlled by WNT–β-catenin signalling47, whereas S100P expression is regulated by activation of both the prostaglandin E2 (PGE2) receptor EP4 subtype (PTGER4) and the MEK–ERK–cAMP-responsive element-binding protein (CREB) pathway48. In addition, the regulatory mechanisms modulating S100 expression can be cancer type-specific. For instance, the expression of S100A8 and S100A9 is regulated by hypoxia-inducible factor 1 (HIF1) and PGE2–protein kinase A catalytic subunit (PKA-C)– CCAAT/enhancer-binding protein-β (CEBPβ) signalling in prostate cancer49,50; by nuclear factor-κB (NF-κB) signalling in liver cancer51; and by ultraviolet radiation, intrinsic ageing and photo-ageing in skin cancer52. Finally, the modulation of S100 expression is a common downstream event in S100 signalling cascades, resulting in feedback loops that can sustain and exacerbate tumour progression. This is exemplified by the expression of S100A8 and S100A9 in skin carcinogenesis, in which RAGE activation by extracellular S100A8 and S100A9 upregulates the expression of these ligands, resulting in a feedforward loop that promotes tumorigenesis53.
S100 signalling in cancer biology
Ten S100 family members actively contribute to in vivo tumour growth, metastasis, angiogenesis and immune evasion (TABLE 1). Although S100 proteins can also act as tumour suppressors, examples are rare and cancer type-specific. S100A2 functions as a tumour suppressor in oral cancer and as a tumour promoter in lung cancer174,178. S100A7 acts as a tumour suppressor in oestrogen receptor-α (ERα)-positive breast cancer but promotes ERα-negative breast tumour growth54. The roles of S100 proteins have been most widely examined in breast cancer and melanoma. As cell lines do not mimic the complex pathology or the S100 protein signatures that are observed in tumours in vivo55,56, our discussion of S100 signalling in melanoma and breast cancer focuses on in vivo studies.
S100 signalling in breast cancer
Overexpression of several S100 family members (such as S100A1, S100A4, S100A6, S100A7, S100A8, S100A9, S100A11, S100A14 and S100P) has been reported in breast cancer57–65. Although alterations in S100 protein expression levels have been correlated with aggressive disease, the mechanistic contribution of individual family members to disease progression has only been evaluated for S100A4, S100A7 and the heterodimer S100A8–S100A9.
S100A7 expression is not detected in epithelial cells in the normal breast. However, high levels of S100A7 are observed in ductal carcinoma in situ, as well as in a subset of invasive breast cancers66. With respect to invasive breast carcinomas, S100A7 overexpression is associated with aggressive, high-grade, ERα-negative lesions with lymphocytic infiltration, and is an independent prognostic indicator of poor outcome in patients with these tumours67. In vitro studies indicate that S100A7 promotes the survival of ERα-negative breast tumour cells under conditions of anchorage-independent growth68. In vivo, S100A7 induces ductal hyperplasia in the mammary glands of transgenic mice69 and enhances tumour growth in orthotopic breast cancer models68,70,71.
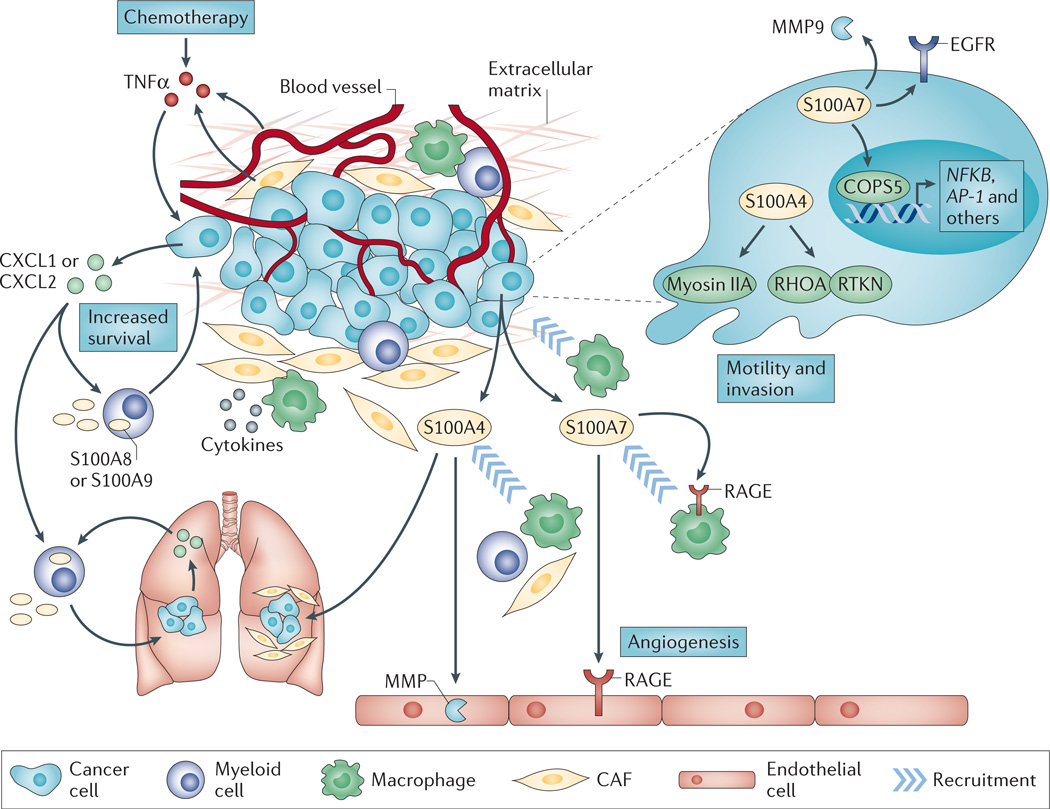
In ERα-positive breast cancer cells, S100A7 inhibits proliferative capacity by mediating the degradation of β-catenin through a mechanism that involves glycogen synthase kinase 3β (GSK3β) and E-cadherin signalling54. However, in ERα-negative breast cancer cells, S100A7 activates several pro-survival pathways by interacting with the transcription cofactor COPS5 (also known as JAB1), including upregulation of the activator protein 1 (AP-1) and NF-κB pathways, increased phospho-AKT and downregulation of cyclin-dependent kinase inhibitor CDKN1B (also known as p27 or Kip1)68,70. In addition to mediating pro-survival effects, S100A7 enhances the invasive capabilities of ERα-negative breast cancer cells by augmenting epidermal growth factor receptor (EGFR) signalling and matrix metalloproteinase 9 (MMP9) secretion72,73. S100A7 is also secreted by tumour cells69,74, and extracellular S100A7 may facilitate tumour angiogenesis75 and the recruitment of tumour-associated macrophages69 through interactions with RAGE on endothelial cells and macrophages, respectively (FIG. 2).
Figure 2. S100 signalling in breast cancer.
Intracellular and extracellular S100A4 and S100A7, and extracellular S100A8 and S100A9 mediate breast tumour progression and metastasis. In cells that do not express oestrogen receptor-α(ERα), intracellular S100A7 enhances cell survival and invasion by upregulating nuclear factor-κB (NF-κB; which is encoded by NFKB) and epidermal growth factor receptor (EGFR) signalling. Tumour cell-derived extracellular S100A7 binds receptor for advanced glycosylation end products (RAGE) on macrophages to mediate their recruitment to the tumour microenvironment. In addition, extracellular S100A7 also binds RAGE on endothelial cells to promote angiogenesis. S100A4 expression in tumour cells enhances cell migration and invasion through interactions with cytoskeletal effectors such as myosin IIA and Rhotekin (RTKN). Stromal cell-derived S100A4 mediates the recruitment of myeloid cells to the tumour microenvironment and is required for metastatic colonization in the lung. Whether these responses are elicited by extracellular or intracellular S100A4 is unknown; however, extracellular S100A4 stimulates matrix metalloproteinase (MMP) production in endothelial cells. Within the tumour microenvironment, S100A8 and S100A9 have both autocrine and paracrine functions that sustain myeloid recruitment and/or immune suppression and NF-κB signalling, respectively. The release of cytokines by the primary tumour also promotes the recruitment of myeloid cells expressing S100A8 or S100A9 to the pre-metastatic lung to maintain a pro-inflammatory milieu that promotes tumour metastasis. Lastly, chemotherapy induces the release of tumour necrosis factor-α (TNFα) from endothelial and other stromal cells, which results in a CXC-chemokine ligand 1 (CXCL1)- or CXCL2-induced S100A8 or S100A9 signalling axis between tumour cells and myeloid cells that facilitates the survival of chemoresistant tumour cells both within the primary tumour and at metastatic sites. AP-1, activator protein 1; CAF, cancer-associated fibroblast.
Several studies have suggested that in early-stage breast cancer, expression of S100A4 in combination with expression of either hepatocyte growth factor receptor (HGFR; also known as MET) or osteopontin is a predictive indicator of metastatic disease and poor survival58,59,76. Consistent with these observations, engineered overexpression of S100A4 in non-metastatic rat mammary Rama 37 tumour cells induces a metastatic phenotype in orthotopic mammary tumours77. Although S100A4 overexpression in the mammary epithelium is not tumorigenic78, it significantly enhances tumour metastasis in pre-existing tumorigenic backgrounds, such as those in the mouse mammary tumour virus (MMTV)-Neu transgenic79 and GRS/A78 mouse models of breast cancer. In both transgenic and orthotopic models, S100A4 expression has a negligible effect on tumour latency, suggesting that S100A4 specifically modulates tumour metastasis rather than tumour growth.
In breast cancer cells, S100A4 overexpression is also associated with increased migratory capacity. Accordingly, S100A4 is found in the pseudopodia of migrating cells80,81, and Ca2+-bound activated S100A4 localizes to the leading edge of breast cancer cells that are undergoing polarized migration82. Furthermore, the Ca2+-dependent interaction of S100A4 with non-muscle myosin IIA regulates the formation and stability of lamellipodia to enhance chemotactic migration83–85. S100A4 also interacts with the Rhotekin–RHOA complex to promote membrane ruffling and invasion in EGF-stimulated breast cancer cells86. In addition to the interactions with myosin IIA and Rhotekin, S100A4 can bind several other cytoskeletal and adhesion proteins, including F-actin, non-muscle tropomyosin and liprin β1. However, the regulation of these putative S100A4 targets is not well characterized.
Not only does S100A4 drive metastasis when expressed in the tumour, but S100A4 expression in the host stroma also contributes to metastatic dissemination. The metastatic potential of orthotopic mammary tumours is significantly reduced in S100a4−/− mice compared with in S100a4+/+ mice87, and metastasis to the lungs is inhibited in MMTV-polyoma middle T antigen (MMTV-PyMT)–S100a4−/− transgenic mice88,89. Despite these observations, the specific stromal cell types that contribute to S100A4-mediated tumour metastasis have not been well characterized. In the breast tumour microenvironment, the majority of S100A4+ stromal cells originate from the bone marrow90 and in MMTV-PyMT–S100A4−/− transgenic mice, the recruitment of CD45+ leukocytes and CD3+ T lymphocytes to tumours is significantly reduced89, suggesting that stromal cell-derived S100A4 modulates the tumour immune response. In addition, recent studies indicate that non-bone-marrow-derived S100A4+ cells, such as fibroblasts, are required for meta-static colonization of breast tumour cells to the lungs87. Currently, a molecular understanding of how stromal cell-derived S100A4 promotes metastasis is lacking and it remains to be determined whether this is mediated by intracellular and/or extracellular S100A4. Both macro-phages and fibroblasts can secrete S100A4 (REFS 91,92). S100A4 monoclonal antibodies significantly limit breast tumour invasion and metastasis in vivo, consistent with an extracellular function for S100A4 (REFS 93,94). Extracellular S100A4 has been shown to stimulate production of MMP13 in endothelial cells and may contribute to tumour angiogenesis95,96, and recent studies suggest that extracellular S100A4 induces the expression and secretion of pro-inflammatory cytokines in tumour cells to elicit a pro-tumoural response in the microenvironment97,98. Although the cell-surface receptors responsible for S100A4 binding and its associated signal transduction pathways remain largely unknown, the identification of these molecules will provide the foundation for a mechanistic understanding of the signalling cascades that are crucial to the metastatic process.
Expression of both S100A8 and S100A9 is upregulated in invasive ductal carcinoma of the breast63. Notably, S100A9 upregulation is associated with basal type tumours, high-grade lesions, ERα- and progesterone receptor (PR)-negative status, and HER2- and EGFR-positive tumours99. The increased levels of S100A8 and S100A9 observed in breast tumour samples are due, in part, to the recruitment of S100A8- and S100A9-expressing myeloid-derived suppressor cells (MDSCs) to the tumour stroma100,101. S100A9 expression is down-regulated during the normal differentiation of myeloid precursors to macrophages and dendritic cells. However, in cancer, tumour-derived factors upregulate S100A9 expression in myeloid precursors, which inhibits macrophage and dendritic cell differentiation, and promotes MDSC accumulation100. The expression of S100A9 is strictly required for MDSC recruitment, as MDSC accumulation in the tumour is ablated in S100A9-null mice100. In addition, MDSC-secreted S100A8–S100A9 heterodimer binds to carboxylated N-glycans on RAGE on the MDSC cell surface102. Thus, S100A8 and S100A9 maintain an autocrine feedback loop that sustains MDSC recruitment and the maintenance of immune suppression within the tumour microenvironment.
S100A8 and S100A9 heterodimers and homodimers also have paracrine functions through interactions with RAGE and TLR4 on tumour cells18,19,103. In tumour cells, RAGE binding activates MAPK and NF-κB signalling pathways, and upregulates the expression of genes associated with tumour growth and invasion18,103. With respect to intracellular functions, in phagocytes, S100A8 and/or S100A9 bind arachidonic acid and activate NADPH oxidase through the direct interaction of S100A8 with cytochrome b558, resulting in the formation of reactive oxygen species (ROS) and the activation of NF-κB signalling20,104. In addition, the S100A8–S100A9 complex is prone to oxidative modifications (such as nitrosylation, glutathionylation and oxidation) by various forms of ROS27,105,106, but how these modifications modulate the tumour-promoting functions of the complex has not been determined.
Extracellular S100A8 and/or S100A9 also contribute to the formation of the pre-metastatic niche. The release of tumour necrosis factor-α (TNFα), transforming growth factor-β (TGFβ) and vascular endothelial growth factor A (VEGFA) from the primary tumour promotes expression of S100A8 and/or S100A9 in pre-metastatic lung endothelium and lung-associated myeloid cells107. Furthermore, extracellular expression of S100A8 and S100A9 induces the expression of serum amyloid A3 (SAA3), which potentiates its own secretion via a TLR4-mediated NF-κB signalling cascade that recruits CD11b+ myeloid cells to the pre-metastatic lung108. This produces a pro-inflammatory milieu that mobilizes circulating tumour cells and promotes pulmonary metastasis108. Signalling via extracellular S100A8 and S100A9 also supports the establishment of a pre-metastatic niche in the brain. The expansion of bone marrow-derived CD11b+GR1+ cells, which express high levels of S100A8 and S100A9, creates a local inflammatory environment that mediates the further recruitment of CD11b+GR1+ myeloid cells and tumour cells through TLR4 signalling to promote brain metastasis109. These observations indicate that for breast cancer, S100A8 and S100A9 are crucial factors for establishing the pre-metastatic niche at multiple organ sites. However, it is unknown whether the upregulation of S100A8 and S100A9 within each of these target organs occurs through similar signalling pathways or whether organ-selective factors mediate S100A8 and/or S100A9 expression, as well as the recruitment of bone marrow-derived cells. Lastly, the requirement for S100A8 and/or S100A9 in creating a permissive environment at other organ sites that are relevant to breast cancer metastasis (such as bone) requires further investigation.
In addition to functions in the tumour microenvironment and the pre-metastatic niche, S100A8 and/or S100A9 also mediate chemoresistance and subsequent metastasis of breast cancer cells101. Chemotherapy induces the release of cytokines and chemokines, including TNFα, by the tumour stroma. Stromal cell-derived TNFα can then boost the expression and secretion of CXC-chemokine ligand 1 (CXCL1) and CXCL2 by breast tumour cells, resulting in the recruitment of CD11b+GR1+ myeloid cells and the release of S100A8 and S100A9 within the tumour microenvironment101. Activation of the pro-survival ERK1, ERK2 and ribosomal protein S6 kinase β1 (S6K1; also known as P70S6K) pathways by S100A8 and/or S100A9 facilitates the expansion of chemoresistant breast cancer cells both within the primary tumour and at distant metastatic sites101, thus providing a survival advantage to breast cancer cells that are under chemotherapeutic stress. Clinical targeting of multiple aspects of the TNFα–CXCL1/CXCL2– S100A8/S100A9 signalling axis may provide a mechanism for limiting drug resistance and metastatic dissemination.
S100 signalling in melanoma
Malignant melanoma is a highly proliferative and heterogeneous cancer that is resistant to conventional chemotherapy34. In melanoma, unlike most cancers, mutations in TP53 are rare. Instead, driver mutations that activate oncogenes (such as BRAF and NRAS) and inactivate cell cycle regulators (such as CDKN2A (which encodes p16) and PTEN) prevent wild-type TP53 from activating downstream target genes and inducing cell cycle arrest and/or apoptosis. Twelve S100 family members are expressed in melanoma: four exhibit no change in expression (S100A8, S100A9, S100A10 and S100A11); one is downregulated (S100A2); and seven are upregulated (S100A1, S100A4, S100A6, S100A13, S100B and S100P; see Supplementary information S1 (table)). The considerable number of S100 family members that are expressed in melanoma is consistent with the localization of S100A1–S100A16 genes to the EDC on human chromosome 1, and with previous observations that the skin expresses the largest number of S100 family members110. However, there is little information regarding the cellular distribution of S100 family members in melanoma. S100A4 is primarily expressed in stromal cells, whereas S100B, S100A6 (REFS 111, 112) and S100A10 (REF. 113) are expressed in tumour cells. Tumour and serum levels of S100B have been used as a diagnostic marker for melanoma for many years and more recently, urinary S100A7 levels have been reported as a potential diagnostic tool114. Decreased levels of serum S100B are associated with the dramatic initial clinical responses to targeted therapies for melanoma. Nonetheless, durable responses to these and other agents are rare owing to the rapid development of multifactorial resistance within individual tumours and patients. To date, increased expression of only one S100 family member, S100A13, has been associated with melanoma resistance to chemotherapy (specifically, resistance to the DNA-modifying agents dacarbazine and temozolomide; see Supplementary information S3 (table)). In vivo studies have confirmed that S100A4, S100A9 and S100B contribute to melanoma progression and may be therapeutic targets.
In melanoma, signalling via extracellular S100A4, S100A8 and S100A9 culminates in the expression of cytokines, chemokines, MMPs, and angiogenic and anti-apoptotic factors (FIG. 3). However, their molecular mechanisms of action are different. Extracellular S100A9, but not S100A8, binds the EMMPRIN receptor and requires the adaptor protein TNF receptor-associated factor 2 (TRAF2) to upregulate the expression of TNFα, IL-1, IL-6 and other factors9. Whether extracellular S100A8 homodimers and S100A8–S100A9 heterodimers also mediate cytokine expression has not been examined. Moreover, upregulation of S100A8 and S100A9 expression by these cytokines generates a feedforward mechanism that can drive tumour progression9. The interaction of stromal cell-derived S100A4 with RAGE on tumour cells also activates NF-κB-dependent gene expression97. The subsequent release of pro-inflammatory cytokines and paracrine factors by tumour cells stimulates endothelial cells and monocytes to promote angiogenesis and protumour immune responses, respectively97. On the basis of the well-established link between chronic inflammation and cancer115, the role of S100B in chronic inflammation116 and the presence of S100B in the systemic circulation of patients with melanoma, it is likely that signalling mediated by extracellular S100B also contributes to melanoma progression. Finally, melanoma-derived exosomes induce the expression of S100A8–S100A9 at pre-metastatic sites117.
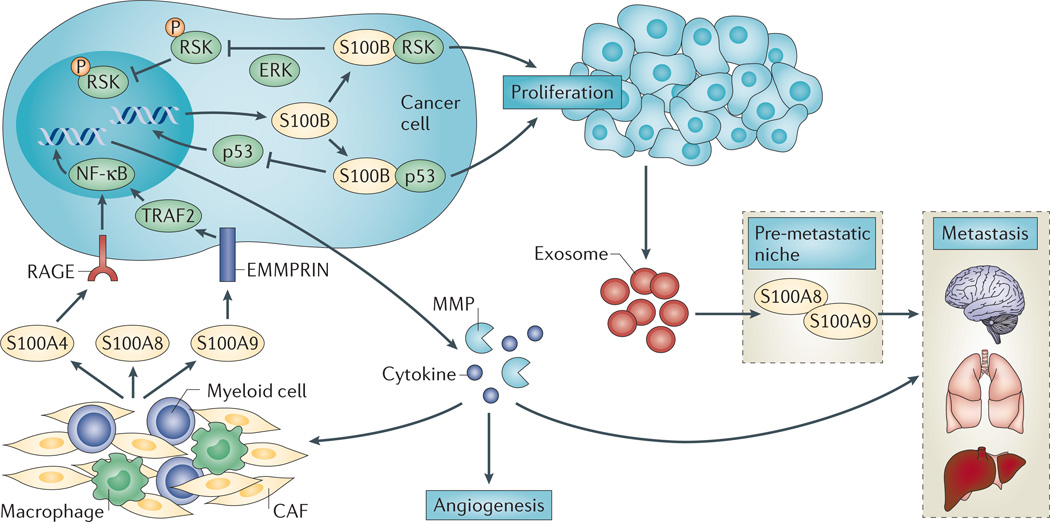
Figure 3. S100 signalling in melanoma.
Intracellular and extracellular S100A4, S100A8 and S100A9 contribute to melanoma proliferation and metastasis. Stroma-derived extracellular S100A4 and S100A9 — by signalling through receptor for advanced glycosylation end products (RAGE) and extracellular matrix metalloproteinase inducer (EMMPRIN), respectively — activate nuclear factor-κB (NF-κB)-mediated tumour cell expression of cytokines and matrix metalloproteinases (MMPs) that promote tumour cell invasion and metastasis. Extracellular S100A8 and S100A8–S100A9 heterodimers are also released from stromal cells, but their contribution to melanoma biology and their molecular mechanisms of action have not been elucidated. Melanoma-derived exosomes promote S100A8 and S100A9 expression at metastatic niches. In tumour cells, wild-type TP53 upregulates S100B expression, which promotes proliferation via p90 ribosomal S6 kinase (RSK) and a negative feedback loop involving TP53. S100B binding to TP53 blocks oligomerization and stimulates TP53 polyubiquitylation and degradation, which decreases TP53 transcriptional activity and downregulates the expression of pro-apoptotic genes. S100B binding to RSK blocks ERK-dependent phosphorylation and cytoplasmic sequestration of phospho-RSK. CAF, cancer-associated fibroblast; TRAF2, tumour necrosis factor receptor-associated factor 2.
There is interest in identifying the mechanisms in melanoma that prevent wild-type TP53 from activating cell cycle arrest and/or apoptosis in response to DNA-damaging agents or UV radiation118–120. In melanoma cell lines, the regulation of growth and survival by intracellular S100B involves a feedback loop that inactivates TP53 (REFS 121–125). Wild-type TP53 upregulates S100B expression by directly binding to the S100B promoter121. However, S100B downregulates TP53 levels and activity by directly binding to and dissociating the TP53 tetramer126, and stimulating TP53 polyubiquitylation and degradation122,126. Additionally, S100B blocks covalent modification of TP53 (such as phosphory lation and acetylation)127. Notably, a reduction in S100B levels or activity by as little as twofold elevates TP53 and phospho-TP53 levels, reduces survival and restores UV sensitivity125,128. There are a number of additional TP53-binding proteins that directly compete with S100B and modulate the TP53-S100B interaction. For example, TP53 and S100B both bind MDM2 and MDM4, which may allow for synergy in TP53 downregulation127. As is typical for S100 proteins, the effects of S100B expression on melanoma cell growth are not limited to a single pathway. S100B binding to the p90 ribosomal S6 kinase (RSK) blocks ERK-dependent phosphorylation and results in cytoplasmic sequestration of RSK124. However, it is not known how the shift in the subcellular distribution of RSK affects cell growth. Ascertaining the pathways that regulate both anti-tumorigenic and protumorigenic melanoma responses will require a careful assessment of the number, relative abundance and cellular distribution of S100 family members and their target proteins. Nonetheless, the beneficial effects of S100B inhibition on multiple pathways that contribute to the melanoma cell phenotype make it an excellent target for drug discovery.
Other S100 family members
Although there is a considerable amount of literature on the contributions of S100A4, S100A7, S100A8, S100A9 and S100B to tumour growth and metastasis in a number of cancers (see TABLE 1 and references therein), the role of the other S100 family members in modulating tumorigenesis is less well defined. In murine models of cancer, S100A1, S100A2, S100A3, S100A6, S100A10, S100A11, S100A14 and S100P are all reported to affect tumour growth. However, with the exception of S100P, the contribution of these S100 family members to promoting a cancerous phenotype has only been examined in one or two model systems (TABLE 1) and the mechanistic basis for the observed effects on tumour progression has not been delineated. By contrast, S100P has been shown to enhance tumour growth in several cancers, including lung, prostate, colorectal and pancreatic cancer (TABLE 1). Although S100P enhances proliferation in all models that have been examined, transcriptional regulation of S100P expression is highly dependent on the type of cancer. In colon cancer, expression is mediated by PGE2–PTGER4 signalling, which activates CREB via the ERK–MEK pathway48; in prostate cancer by IL-6 (REF. 129); and in breast and cervical cancer by glucocorticoids130. Consistent with the observation that S100P enhances the migratory capabilities of tumour cells in vitro131 and metastasis in vivo132–134, the majority of S100P targets are cytoskeletal regulators. S100P–mediated activation of ezrin135 (a membrane–cytoskeleton linker protein) and Ras GTPase-activating-like protein 1 (IQGAP1; a juxtamembrane scaffolding protein that links plasma membrane receptors with downstream signalling pathways)136provides a direct mechanism for the regulation of tumour cell migration by S100P. In addition, the binding of extracellular S100P to RAGE upregulates NF-κB activity to enhance cell survival137. Currently, extracellular S100P is thought to stimulate tumour cell proliferation through an autocrine signalling mechanism. However, a complete examination of potential paracrine functions and the cell types involved has been limited by the absence of the S100P gene in the mouse genome.
Targeting S100 proteins
S100 family members are excellent targets for cancer treatment as mouse models suggest that genetic deletion has minimal effects on normal physiology. In addition to cancer, some members of the S100 family represent attractive targets for the treatment of other diseases. For example, S100B and S100A1 inhibitors may delay the progression of Alzheimer’s disease116,138. However, S100A1 inhibitors may be contraindicated in patients with heart disease, as S100A1 delays the development of cardiomyopathy139.
A number of pharmacological approaches have been used to modulate S100 signalling in models of and in patients with cancer. Calcimycin (a calcium ionophore), niclosamide (an antihelminth drug) and sulindac (a non-steroidal anti-inflammatory drug) have all been identified as inhibitors of S100A4 transcription140,141. However, the efficacy of this approach may be limited owing to the long half-life of S100 proteins, which could prevent the achievement of sufficiently low steady-state protein levels to elicit a therapeutic effect. Transcriptional modulators may also have clinically significant toxic off-target effects owing to their ability to regulate the expression of numerous proteins under the control of the same or related transcriptional regulatory assemblies. Although gene therapy has not been used to modulate the expression of S100 family members in patients with cancer, it has been used in preclinical animal models, in which it beneficially upregulates S100A1 expression in heart disease142. Other approaches to modulate S100 protein activity include S100A4- and S100P–neutralizing antibodies93,94,143, and peptibodies (peptide–Fc fusion proteins) directed against S100A8 and S100A9 (REF. 144); both approaches reduce tumour growth in murine cancer models. Although the specificity of antibody-based therapies may reduce toxicity and off-target effects, their efficacy may be limited by their ability to target only extracellular S100 proteins. However, conformationally constrained inhibitory peptides directed against S100B, which are capable of penetrating cells, have been shown to reduce tumour growth in a melanoma xenograft model145.
The most common strategies for inhibiting S100 proteins exploit small molecules that block the hydrophobic cleft required for the recognition of S100 targets, and for eliciting biological effects5. Examples include paquinimod (also known as ABR-215757) and tasquinimod (also known as ABR-215050), which are quinoline-3-carboxamide derivatives that block the interaction of S100A8 and S100A9 with RAGE and TLR4, respectively (REFS 19,146). Paquinimod exerts anti-inflammatory effects in a number of in vivo disease models, but has not been specifically tested in cancer models. However, tasquinimod improves progression-free survival in patients with metastatic castration-resistant prostate cancer, possibly by reducing the recruitment of MDSCs and inhibiting metastasis147,148. Cromolyn, an anti-histaminic drug, disrupts the S100P– RAGE interaction, reducing pancreatic tumour growth and increasing the effectiveness of the chemotherapeutic drug gemcitabine149,150. Cromolyn binds several other S100 family members (S100A1, S10012 and S100A13), but the biochemical and biological consequences of these interactions have not been examined. Amlexanox, another anti-inflammatory anti-allergic immunomodulatory drug, interacts with several S100 proteins (S100A1, S100A4 and S100A13)151–153. Amlexanox inhibits S100A13 secretion, disrupts the interaction of S100A13 with fibroblast growth factor 1 (FGF1), and antagonizes the mitogenic and angiogenic effects of FGF1 (REF. 152). Phenothiazines also interact with multiple S100 family members. In the case of S100A4, phenothiazine-mediated S100A4 oligomerization results in the sequestration of S100A4 away from its protein targets154,155. It should be noted that all of these drugs were developed for other indications and their lack of selectivity within the S100 family is not surprising given the high degree of structural similarity between S100 proteins. Ongoing efforts to target S100 proteins are focused on improving selectivity and other pharmacological properties of S100 inhibitors such as affinity and biological half-life.
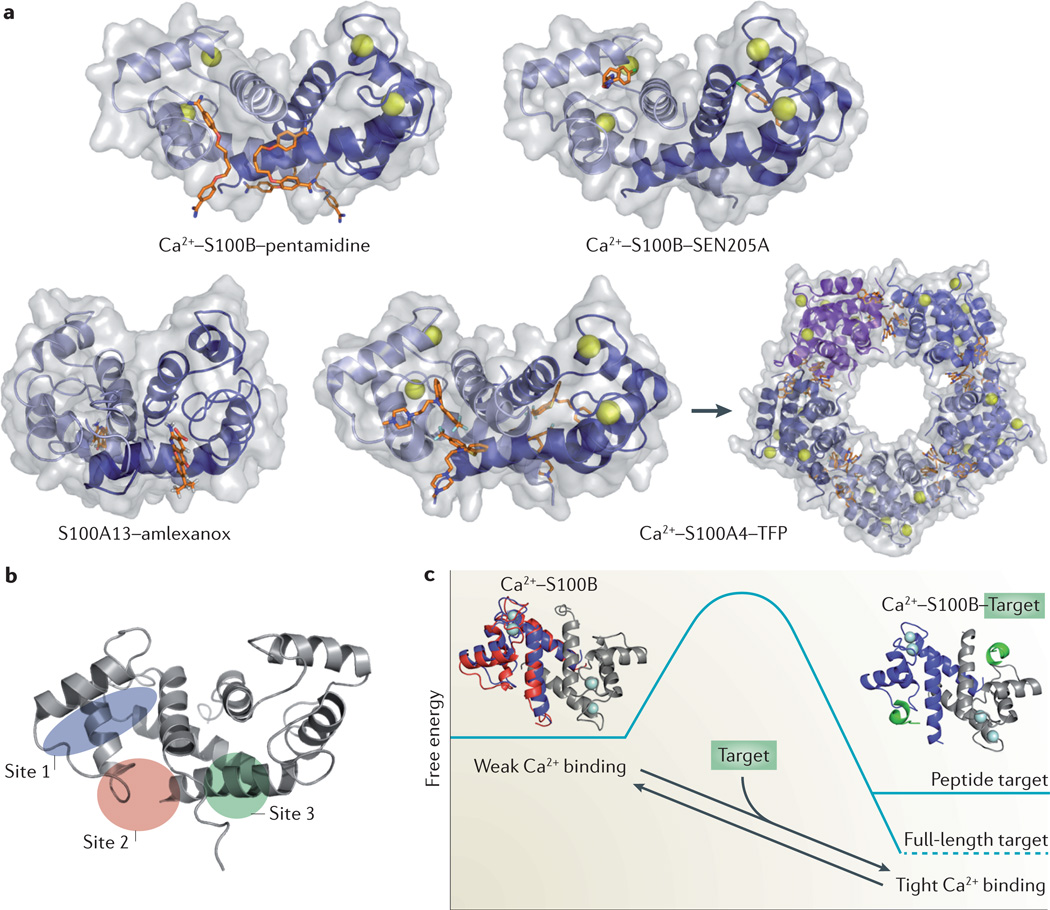
The large, shallow and relatively ‘featureless’ interfaces typified by most protein–protein interactions (PPIs) pose unique challenges for the development of high-affinity PPI inhibitors. In contrast to typical protein–protein interfaces, the relatively deep target-binding clefts of S100 proteins can readily accommodate small molecules that have been discovered through traditional high-throughput experimental screening and computer-aided drug design. These approaches have successfully identified new compounds that inhibit the interactions of S100P149, S100A4 (REFS 156,157), S100A9 (REF. 156), S100A10 (REF 158) and S100B123,159 with their respective targets. An examination of these S100–small-molecule complexes has revealed that most small molecules target one of three distinct pockets within S100 proteins160 (FIG. 4). Site 1 is exposed by Ca2+-mediated conformational rearrangements and involves residues from helices 3 and 4, and loop 2 (the hinge region), as occurs in the S100B– SEN205A interaction, for example161 (FIG. 4a). Interactions at sites 2 and 3 involve residues from loop 2 and helix 4, and the C-terminal loop and helix 1, respectively. In some instances, multiple copies of the same compound occupy both sites 2 and 3 (as occurs in the S100B–pentamidine162and S100A4–trifluoperazine interactions154). However, compounds have also been identified that target only one of these sites (such as S100B–SBiX-inhibitor complexes160or the S100A13–amlexanox interaction152). A comparison of S100 protein–small-molecule structures reveals that the location and orientation of ligands bound in sites 2 and 3 are less well conserved than those bound in site 1. The diversity of binding positions observed at sites 2 and 3 probably reflects differences in the highly specific interactions that mediate the binding of various targets (such as H-bonds, hydrophobic interactions and other types of interactions) within the S100 protein family and thus provides opportunities for specific S100–small-molecule interactions.
Figure 4. S100 protein–inhibitor complexes.
a | Ribbon and surface diagrams of the Ca2+–S100B–pentamidine complex (RCSB Protein Data bank (PDB) identifier: 3CR4), the Ca2+–S100B–SEN205A complex (PDB identifier: 3HCM), the Ca2+–S100A13–amlexanox complex (PDB identifier: 2KOT) and the Ca2+–S100A4–trifluoperazine (TFP) complex (PDB identifier: 3KO0). Individual S100 subunits are shown in light and dark blue, the Ca2+ ions are shown as grey spheres and the inhibitors as orange sticks. Small-molecule inhibitors can bind S100 proteins in distinct orientations. TFP binding induces the assembly of five Ca2+–S100A4–TFP dimers into a pentameric ring. b | Ribbon diagram of Ca2+–S100B showing three small-molecule binding sites. Site 1 involves residues from helices 3 and 4, and loop 2 (the ‘hinge’ region); site 2 involves residues from loop 2 and helix 4; and site 3 involves residues from the carboxy-terminal loop and helix 1. c | The ‘binding and functional folding’ (BFF) model for S100 protein–protein interactions (PPIs). In the absence of a molecular target, Ca2+-bound S100 proteins (that is, Ca2+–S100B) sample a large ensemble of dynamic sub-states with a range of Ca2+-binding affinities (red and blue ribbon diagrams) that result in a low net apparent affinity for Ca2+. Target binding induces a mini-folding event that stabilizes these dynamic features and biases the ensemble towards those sub-states (blue ribbon diagram), with high affinity for Ca2+ and a lower (that is, more favourable) global free energy (a complete description of the model can be found in Liriano et al24 and Markowitz et al167). Typically, complexes with full-length targets exhibit lower free energies than complexes involving target-derived peptides. This property allows for high intracellular S100 protein concentrations (>1 µM) without substantial sequestration of free Ca2+ or disruption of Ca2+ oscillations, but provides a highly responsive system that is poised to regulate cellular processes upon target binding. Drugs that also enhance S100 protein Ca2+ occupancy can be identified and/or better engineered by monitoring changes in the structure and dynamic properties of the S100 protein upon drug and/or target binding. Part c reprinted from J. Mol. Biol423, Liriano, M. A. et al. Target binding to S100B reduces dynamic properties and increases Ca2+-binding affinity for wild type and EF-hand mutant proteins, 365–385 © (2012), with permission from Elsevier.
Given that S100 proteins can accommodate small molecules at several sites, it is not unexpected that multiple mechanisms of inhibition are observed, including classic competitive binding between an inhibitor and physiological target to the same or overlapping sites on a S100 protein, such as amlexanox binding to S100A13 or SEN205A binding to S100B152,161 (FIG. 4a). However, some inhibitors of S100 proteins may exhibit subtle allosteric effects as exemplified by pentamidine binding to S100B. Each S100B subunit binds two pentamidines, with one binding site adjacent to the target binding site and the second at the dimer interface162. Although the pentamidine binding sites exhibit minimal overlap with the S100B target-binding cleft, pentamidine disrupts the S100B–TP53 interaction and potently inhibits the growth of malignant melanoma cells163, suggesting that inhibition occurs due to allosteric effects. More recently, structural, biophysical, and protein dynamics studies have shown that the binding of small-molecule inhibitors and peptides can increase Ca2+-binding affinity upon complex formation in a manner analogous to that associated with the binding of bona fide targets24.
When considering how small molecules and peptides that bind at some distance from the S100 EF-hands may affect Ca2+-binding affinities, it is important to take into account the structural features as well as dynamic properties of S100 proteins24,164–166. In the absence of a protein target, S100 proteins sample a large ensemble of conformational sub-states that exhibit a wide range of Ca2+ affinities but, overall, their apparent affinity is low. Target binding biases the distribution of sub-states towards those with high Ca2+ affinity. This model, termed the ‘binding and functional folding’ (BFF) model (FIG. 4c), provides a foundation for understanding how the biochemical activities of S100 proteins fit into overall cell physiology. The coupling of Ca2+ and target binding allows for high intracellular S100 protein concentrations (>1 µM) without substantial sequestration of free Ca2+ or disruption of Ca2+ oscillations, and thus cells remain highly responsive to changes in calcium and/or target availability. S100 inhibitor screens are now incorporating an examination of Ca2+ affinity and protein dynamics24, which should allow for the identification of inhibitors with increased binding affinities and specificities.
Future directions
In the past decade, important advances regarding the expression, structure and signalling of S100 proteins have improved our understanding of normal cell physiology as well as the pathophysiology of cancer. The development of molecular probes — such as antibodies and small-molecule inhibitors — will be instrumental for deciphering the in vivo functions of specific S100 proteins, as well as distinguishing the contribution of intracellular and extracellular S100 proteins. Moreover, these probes may also have therapeutic potential for cancer or other diseases. Despite considerable progress in S100 protein biology, we currently have little information on how post-translational modifications or heterodimer formation affect S100 signalling. A mechanistic examination of both S100 protein biology and biochemistry will be required to define how each family member contributes to the proliferation, metastasis, angiogenesis and immune evasion of cancers and other diseases.
Supplementary Material
Acknowledgements
The authors apologize to the numerous colleagues whose important contributions could not be included in this Review owing to space limitations. The authors thank S. C. Almo and J. M. Backer for critical reading of this Review. The authors’ research is funded by grants from the New York State Department of Health, Health Research Science Board (H11R-040; to A.R.B.), the National Cancer Institute (US National Institutes of Health; CA100324 to A.R.B. and CA107331 to D.J.W.), the Albert Einstein Cancer Center (NCI, NIH; CA013330), the Marlene and Stewart Greenebaum Cancer Center (NCI, NIH; CA134274) and the Center for Biomolecular Therapeutics, University of Maryland School of Medicine, USA (to D.J.W. and D.B.Z.).
Glossary
- Sub-states
Closely related interconverting conformational states that can be sampled by a protein under a given set of conditions
- GRS/A
An inbred mouse strain carrying the Mtv2a allele, which controls the expression of endogenous mouse mammary tumour virus and the early development of hormone-induced mammary tumours
- Lamellipodia
Transient cellular protrusions that form during cell migration
- Chemotactic migration
Directional cell migration in response to soluble extracellular ligands
- MMTV-polyoma middle T antigen
(MMTV-PyMT). A murine breast cancer model with expression of PyMT under the control of the mouse mammary tumour virus (MMTV) promoter
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
RCSB Protein Data Bank: http://www.rcsb.org/pdb
S100 gene family: http://www.genenames.org/genefamilies/S100
SUPPLEMENTARY INFORMATION
See online article: S1 (table) | S2 (table) | S3 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Anne R. Bresnick, Email: anne.bresnick@einstein.yu.edu.
David J. Weber, Email: dweber@som.umaryland.edu.
Danna B. Zimmer, Email: dzimmer@som.umaryland.edu.
References
- 1.Moore BW. A soluble protein characteristic of the nervous system. Biochem. Biophys. Res. Commun. 1965;19:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 2.Leclerc E, Heizmann CW. The importance of Ca2+/ Zn2+ signaling S100 proteins and RAGE in translational medicine. Front. Biosci. (Schol. Ed.) 2011;3:1232–1262. doi: 10.2741/223. [DOI] [PubMed] [Google Scholar]
- 3.Hermann A, Donato R, Weiger TM, Chazin WJ. S100 calcium binding proteins and ion channels. Front. Pharmacol. 2012;3:67. doi: 10.3389/fphar.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donato R, et al. Functions of S100 proteins. Curr. Mol. Med. 2013;13:24–57. [PMC free article] [PubMed] [Google Scholar]
- 5.Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins. 1999;37:499–507. doi: 10.1002/(sici)1097-0134(19991115)37:3<499::aid-prot17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer DB, Weber DJ. The calcium-dependent interaction of S100B with its protein targets. Cardiovasc. Psychiatry Neurol. 2010;2010:728052. doi: 10.1155/2010/728052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Bauer R, et al. CD166/ALCAM mediates proinflammatory effects of S100B in delayed type hypersensitivity. J. Immunol. 2013;191:369–377. doi: 10.4049/jimmunol.1201864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmytriyeva O, et al. The metastasis-promoting S100A4 protein confers neuroprotection in brain injury. Nature Commun. 2012;3:1197. doi: 10.1038/ncomms2202. [DOI] [PubMed] [Google Scholar]
- 9.Hibino T, et al. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. 2013;73:172–183. doi: 10.1158/0008-5472.CAN-11-3843. [DOI] [PubMed] [Google Scholar]
- 10.Hankins JL, et al. Ceramide-1-phosphate mediates endothelial cell invasion via the annexin a2/p11 heterotetrameric protein complex. J. Biol. Chem. 2013;288:19726–19738. doi: 10.1074/jbc.M113.481622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zimmer DB, Eubanks JO, Ramakrishnan D, Criscitiello MF. Evolution of the S100 family of calcium sensor proteins. Cell Calcium. 2012;53:170–179. doi: 10.1016/j.ceca.2012.11.006. This reference clarifies inconsistencies regarding the S100 nomenclature and addresses species-specific differences in S100 protein expression that affect the translation of mouse studies to human disease.
- 12.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res. Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 13.Henry J, et al. Update on the epidermal differentiation complex. Front. Biosci. 2012;17:1517–1532. doi: 10.2741/4001. [DOI] [PubMed] [Google Scholar]
- 14.Lunde ML, et al. Profiling of chromosomal changes in potentially malignant and malignant oral mucosal lesions from south and south-east Asia using array-comparative genomic hybridization. Cancer Genomics Proteomics. 2014;11:127–140. [PubMed] [Google Scholar]
- 15.Chen H, et al. Functional role of S100A14 genetic variants and their association with esophageal squamous cell carcinoma. Cancer Res. 2009;69:3451–3457. doi: 10.1158/0008-5472.CAN-08-4231. [DOI] [PubMed] [Google Scholar]
- 16.Strazisar M, Rott T, Glavac D. Frequent polymorphic variations but rare tumour specific mutations of the S100A2 on 1q21 in non-small cell lung cancer. Lung Cancer. 2009;63:354–359. doi: 10.1016/j.lungcan.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Isobe T, et al. A rapid separation of S100 subunits by high performance liquid chromatography: the subunit compositions of S100 proteins. Biochem. Int. 1983;6:419–426. [PubMed] [Google Scholar]
- 18.Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 2011;9:133–148. doi: 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallberg E, et al. S100A9 interaction with TLR4 promotes tumor growth. PLoS ONE. 2012;7:e34207. doi: 10.1371/journal.pone.0034207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthier S, et al. Molecular interface of S100A8 with cytochrome b558 and NADPH oxidase activation. PLoS ONE. 2012;7:e40277. doi: 10.1371/journal.pone.0040277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright NT, et al. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) J. Mol. Biol. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markowitz J, et al. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 23.Malashkevich VN, et al. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liriano MA, et al. Target binding to S100B reduces dynamic properties and increases Ca2+-binding affinity for wild type and EF-hand mutant proteins. J. Mol. Biol. 2012;423:365–385. doi: 10.1016/j.jmb.2012.07.011. This reference discusses three models for the observed coupling between the binding of targets and calcium to S100 proteins, with a careful consideration of both the structural features and the dynamic properties of S100 proteins.
- 25.Ramagopal UA, et al. Structure of the S100A4/ myosin-IIA complex. BMC Struct. Biol. 2013;13:31–46. doi: 10.1186/1472-6807-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orre LM, Pernemalm M, Lengqvist J, Lewensohn R, Lehtio J. Up-regulation, modification, and translocation of S100A6 induced by exposure to ionizing radiation revealed by proteomics profiling. Mol. Cell Proteomics. 2007;6:2122–2131. doi: 10.1074/mcp.M700202-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Lim SY, Raftery MJ, Geczy CL. Oxidative modifications of DAMPs suppress inflammation: the case for S100A8 and S100A9. Antioxid. Redox Signal. 2011;15:2235–2248. doi: 10.1089/ars.2010.3641. [DOI] [PubMed] [Google Scholar]
- 28.Bowers RR, Manevich Y, Townsend DM, Tew KD. Sulfiredoxin redox-sensitive interaction with S100A4 and non-muscle myosin IIA regulates cancer cell motility. Biochemistry. 2012;51:7740–7754. doi: 10.1021/bi301006w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miranda KJ, Loeser RF, Yammani RR. Sumoylation and nuclear translocation of S100A4 regulates IL-11β mediated production of matrix metalloprotinase-13. J. Biol. Chem. 2010;285:31517–31524. doi: 10.1074/jbc.M110.125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin. Cancer Res. 2013;19:3764–3775. doi: 10.1158/1078-0432.CCR-12-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter KD, Parkinson EK, Harrison PR. Profiling early head and neck cancer. Nature Rev. Cancer. 2005;5:127–135. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- 33.Barbieri CE, Demichelis F, Rubin MA. Molecular genetics of prostate cancer: emerging appreciation of genetic complexity. Histopathology. 2012;60:187–198. doi: 10.1111/j.1365-2559.2011.04041.x. [DOI] [PubMed] [Google Scholar]
- 34.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nature Rev. Cancer. 2012;12:349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 35.Sadanandam A, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nature Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legolvan MP, Taliano RJ, Resnick MB. Application of molecular techniques in the diagnosis, prognosis and management of patients with colorectal cancer: a practical approach. Hum. Pathol. 2012;43:1157–1168. doi: 10.1016/j.humpath.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Hao J, et al. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Mol. Cell Biochem. 2012;359:323–332. doi: 10.1007/s11010-011-1026-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhou G, et al. Reciprocal negative regulation between S100A7/psoriasin and β-catenin signaling plays an important role in tumor progression of squamous cell carcinoma of oral cavity. Oncogene. 2008;27:3527–3538. doi: 10.1038/sj.onc.1211015. [DOI] [PubMed] [Google Scholar]
- 39.Wolf R, Ruzicka T, Yuspa SH. Novel S100A7 (psoriasin)/S100A15 (koebnerisin) subfamily: highly homologous but distinct in regulation and function. Amino Acids. 2011;41:789–796. doi: 10.1007/s00726-010-0666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heizmann CW. S100B protein in clinical diagnostics: assay specificity. Clin. Chem. 2004;50:249–251. doi: 10.1373/clinchem.2003.027367. [DOI] [PubMed] [Google Scholar]
- 41.Horiuchi A, et al. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1α to a hypoxia-induced, methylation free hypoxia response element (HRE) of S100A4 gene. Int. J. Cancer. 2012;131:1755–1767. doi: 10.1002/ijc.27448. [DOI] [PubMed] [Google Scholar]
- 42.Lesniak W. Epigenetic regulation of S100 protein expression. Clin. Epigenetics. 2011;2:77–83. doi: 10.1007/s13148-011-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibadulinova A, Tothova V, Pastorek J, Pastorekova S. Transcriptional regulation and functional implication of S100P in cancer. Amino Acids. 2011;41:885–892. doi: 10.1007/s00726-010-0495-5. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, et al. S100P, a potential novel prognostic marker in colorectal cancer. Oncol. Rep. 2012;28:303–310. doi: 10.3892/or.2012.1794. [DOI] [PubMed] [Google Scholar]
- 45.Day T, Bianco-Miotto T. Common gene pathways and families altered by DNA methylation in breast and prostate cancer. Endocr. Relat. Cancer. 2013;20:R215–R232. doi: 10.1530/ERC-13-0204. [DOI] [PubMed] [Google Scholar]
- 46.Guo C, et al. Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways. Proc. Natl Acad. Sci. USA. 2012;109:17603–17608. doi: 10.1073/pnas.1208807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sack U, Stein U. Wnt up your mind — intervention strategies for S100A4-induced metastasis in colon cancer. Gen. Physiol. Biophys. 2009;28:F55–F64. [PubMed] [Google Scholar]
- 48.Chandramouli A, et al. The induction of S100P expression by the prostaglandin E (PGE)/EP4 receptor signaling pathway in colon cancer cells. Cancer Biol. Ther. 2010;10:1056–1066. doi: 10.4161/cbt.10.10.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grebhardt S, Veltkamp C, Strobel P, Mayer D. Hypoxia and HIF-1 increase S100A8 and S100A9 expression in prostate cancer. Int. J. Cancer. 2012;131:2785–2794. doi: 10.1002/ijc.27591. [DOI] [PubMed] [Google Scholar]
- 50.Miao L, et al. Prostaglandin E2 stimulates S100A8 expression by activating protein kinase A and CCAAT/ enhancer-binding-protein-beta in prostate cancer cells. Int. J. Biochem. Cell Biol. 2012;44:1919–1928. doi: 10.1016/j.biocel.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 51.Nemeth J, et al. S100A8 and S100A9 are novel nuclear factor kappa B target genes during malignant progression of murine and human liver carcinogenesis. Hepatology. 2009;50:1251–1262. doi: 10.1002/hep.23099. [DOI] [PubMed] [Google Scholar]
- 52.Lee YM, Kim YK, Eun HC, Chung JH. Changes in S100A8 expression in UV-irradiated and aged human skin in vivo. Arch. Dermatol. Res. 2009;301:523–529. doi: 10.1007/s00403-009-0960-8. [DOI] [PubMed] [Google Scholar]
- 53.Gebhardt C, et al. RAGE signaling sustains inflammation and promotes tumor development. J. Exp. Med. 2008;205:275–285. doi: 10.1084/jem.20070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deol YS, Nasser MW, Yu L, Zou X, Ganju RK. Tumor-suppressive effects of psoriasin (S100A7) are mediated through the β-catenin/T cell factor 4 protein pathway in estrogen receptor-positive breast cancer cells. J. Biol. Chem. 2011;286:44845–44854. doi: 10.1074/jbc.M111.225466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross SR, Sin CG, Barraclough R, Rudland PS. Joining S100 proteins and migration: for better or for worse, in sickness and in health. Cell. Mol. Life Sci. 2013;71:1551–1579. doi: 10.1007/s00018-013-1400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leclerc E, Heizmann CW, Vetter SW. RAGE and S100 protein transcription levels are highly variable in human melanoma tumors and cells. Gen. Physiol. Biophys. 2009;28:F65–F75. [PubMed] [Google Scholar]
- 57.Nikitenko LL, Lloyd BH, Rudland PS, Fear S, Barraclough R. Localisation by in situ hybridisation of S100A4 (p9Ka) mRNA in primary human breast tumour specimens. Int. J. Cancer. 2000;86:219–228. doi: 10.1002/(sici)1097-0215(20000415)86:2<219::aid-ijc11>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 58.Lee WY, et al. Expression of S100A4 and Met: potential predictors for metastasis and survival in early-stage breast cancer. Oncology. 2004;66:429–438. doi: 10.1159/000079496. [DOI] [PubMed] [Google Scholar]
- 59.de Silva Rudland S, et al. Association of S100A4 and osteopontin with specific prognostic factors and survival of patients with minimally invasive breast cancer. Clin. Cancer Res. 2006;12:1192–1200. doi: 10.1158/1078-0432.CCR-05-1580. [DOI] [PubMed] [Google Scholar]
- 60.Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 61.Al-Haddad S, et al. Psoriasin (S100A7) expression and invasive breast cancer. Am. J. Pathol. 1999;155:2057–2066. doi: 10.1016/S0002-9440(10)65524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emberley ED, Alowami S, Snell L, Murphy LC, Watson PH. S100A7 (psoriasin) expression is associated with aggressive features and alteration of Jab1 in ductal carcinoma in situ of the breast. Breast Cancer Res. 2004;6:R308–R315. doi: 10.1186/bcr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arai K, et al. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr. Cancer Drug Targets. 2008;8:243–252. doi: 10.2174/156800908784533445. [DOI] [PubMed] [Google Scholar]
- 64.McKiernan E, McDermott EW, Evoy D, Crown J, Duffy MJ. The role of S100 genes in breast cancer progression. Tumour Biol. 2011;32:441–450. doi: 10.1007/s13277-010-0137-2. [DOI] [PubMed] [Google Scholar]
- 65.Schor AP, Carvalho FM, Kemp C, Silva ID, Russo J. S100P calcium-binding protein expression is associated with high-risk proliferative lesions of the breast. Oncol. Rep. 2006;15:3–6. [PubMed] [Google Scholar]
- 66.Emberley ED, Murphy LC, Watson PH. S100A7 and the progression of breast cancer. Breast Cancer Res. 2004;6:153–159. doi: 10.1186/bcr816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emberley ED, et al. Psoriasin (S100A7) expression is associated with poor outcome in estrogen receptor-negative invasive breast cancer. Clin. Cancer Res. 2003;9:2627–2631. [PubMed] [Google Scholar]
- 68.Emberley ED, et al. The S100A7-c-Jun activation domain binding protein 1 pathway enhances prosurvival pathways in breast cancer. Cancer Res. 2005;65:5696–5702. doi: 10.1158/0008-5472.CAN-04-3927. [DOI] [PubMed] [Google Scholar]
- 69.Nasser MW, et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res. 2012;72:604–615. doi: 10.1158/0008-5472.CAN-11-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emberley ED, et al. Psoriasin interacts with Jab1 and influences breast cancer progression. Cancer Res. 2003;63:1954–1961. [PubMed] [Google Scholar]
- 71.Krop I, et al. A putative role for psoriasin in breast tumor progression. Cancer Res. 2005;65:11326–11334. doi: 10.1158/0008-5472.CAN-05-1523. [DOI] [PubMed] [Google Scholar]
- 72.Paruchuri V, et al. S100A7-downregulation inhibits epidermal growth factor-induced signaling in breast cancer cells and blocks osteoclast formation. PLoS ONE. 2008;3:e1741. doi: 10.1371/journal.pone.0001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sneh A, et al. Differential role of psoriasin (S100A7) in estrogen receptor α positive and negative breast cancer cells occur through actin remodeling. Breast Cancer Res. Treat. 2013;138:727–739. doi: 10.1007/s10549-013-2491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enerback C, et al. Psoriasin expression in mammary epithelial cells in vitro and in vivo. Cancer Res. 2002;62:43–47. [PubMed] [Google Scholar]
- 75.Shubbar E, Vegfors J, Carlstrom M, Petersson S, Enerback C. Psoriasin (S100A7) increases the expression of ROS and VEGF and acts through RAGE to promote endothelial cell proliferation. Breast Cancer Res. Treat. 2012;134:71–80. doi: 10.1007/s10549-011-1920-5. [DOI] [PubMed] [Google Scholar]
- 76.Rudland PS, et al. Prognostic significance of the metastasis-inducing protein S100A4 (p9Ka) in human breast cancer. Cancer Res. 2000;60:1595–1603. [PubMed] [Google Scholar]
- 77.Davies BR, Davies MP, Gibbs FE, Barraclough R, Rudland PS. Induction of the metastatic phenotype by transfection of a benign rat mammary epithelial cell line with the gene for p9Ka, a rat calcium-binding protein, but not with the oncogene EJ-ras-1. Oncogene. 1993;8:999–1008. [PubMed] [Google Scholar]
- 78.Ambartsumian NS, et al. Metastasis of mammary carcinomas in GRS/A hybrid mice transgenic for the mts1 gene. Oncogene. 1996;13:1621–1630. [PubMed] [Google Scholar]
- 79. Davies MP, et al. Expression of the calcium-binding protein S100A4 (p9Ka) in MMTV-neu transgenic mice induces metastasis of mammary tumours. Oncogene. 1996;13:1631–1637. Using murine models of breast cancer, references 77, 78 and 79 established S100A4 as a mediator of tumour metastasis.
- 80.Kim EJ, Helfman DM. Characterization of the metastasis-associated protein, S100A4. Roles of calcium binding and dimerization in cellular localization and interaction with myosin. J. Biol. Chem. 2003;278:30063–30073. doi: 10.1074/jbc.M304909200. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, et al. Profiling signaling polarity in chemotactic cells. Proc. Natl Acad. Sci. USA. 2007;104:8328–8333. doi: 10.1073/pnas.0701103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.House RP, Garrett SC, Bresnick AR. In: Signaling Pathways and Molecular Mediators in Metastasis. Fatatis A, editor. Netherlands: Springer; 2012. pp. 91–113. [Google Scholar]
- 83.Kriajevska MV, et al. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J. Biol. Chem. 1994;269:19679–19682. [PubMed] [Google Scholar]
- 84.Ford HL, Silver DL, Kachar B, Sellers JR, Zain SB. Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry. 1997;36:16321–16327. doi: 10.1021/bi971182l. [DOI] [PubMed] [Google Scholar]
- 85.Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage chemotaxis. Mol. Biol. Cell. 2010;21:2598–2610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen M, Bresnick AR, O’Connor KL. Coupling S100A4 to rhotekin alters Rho signaling output in breast cancer cells. Oncogene. 2012;32:3754–3764. doi: 10.1038/onc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O’Connell JT, et al. VEGF-A and Tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc. Natl Acad. Sci. USA. 2011;108:16002–16007. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xue C, Plieth D, Venkov C, Xu C, Neilson EG. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–3394. [PubMed] [Google Scholar]
- 89.Grum-Schwensen B, et al. Lung metastasis fails in MMTV-PyMT oncomice lacking S100A4 due to a T-cell deficiency in primary tumors. Cancer Res. 2010;70:936–947. doi: 10.1158/0008-5472.CAN-09-3220. [DOI] [PubMed] [Google Scholar]
- 90.Kidd S, et al. Origins of the tumor microenvironment: quantitative assessment of adipose-derived and bone marrow-derived stroma. PLoS ONE. 2012;7:e30563. doi: 10.1371/journal.pone.0030563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cabezon T, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int. J. Cancer. 2007;121:1433–1444. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 92.Forst B, et al. Metastasis-inducing S100A4 and RANTES cooperate in promoting tumor progression in mice. PLoS ONE. 2010;5:e10374. doi: 10.1371/journal.pone.0010374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klingelhofer J, et al. Anti-S100A4 antibody suppresses metastasis formation by blocking stroma cell invasion. Neoplasia. 2012;14:1260–1268. doi: 10.1593/neo.121554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hernandez JL, et al. Therapeutic targeting of tumor growth and angiogenesis with a novel anti-S100A4 monoclonal antibody. PLoS ONE. 2013;8:e72480. doi: 10.1371/journal.pone.0072480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ambartsumian N, et al. The metastasis-associated Mts1(S100A4) protein could act as an angiogenic factor. Oncogene. 2001;20:4685–4695. doi: 10.1038/sj.onc.1204636. [DOI] [PubMed] [Google Scholar]
- 96.Schmidt-Hansen B, et al. Extracellular S100A4(mts1) stimulates invasive growth of mouse endothelial cells and modulates MMP-13 matrix metalloproteinase activity. Oncogene. 2004;23:5487–5495. doi: 10.1038/sj.onc.1207720. [DOI] [PubMed] [Google Scholar]
- 97.Bettum IJ, et al. Metastasis-associated protein S100A4 induces a network of inflammatory cytokines that activate stromal cells to acquire pro-tumorigenic properties. Cancer Lett. 2014;344:28–39. doi: 10.1016/j.canlet.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 98.Hansen MT, et al. A link between inflammation and metastasis: serum amyloid A1 and A3 induce metastasis, and are targets of metastasis-inducing S100A4. Oncogene. 2014 doi: 10.1038/onc.2013.568. http://dx.doi.org/10.1038/onc.2013.568. [DOI] [PubMed] [Google Scholar]
- 99.Goncalves A, et al. Protein profiling of human breast tumor cells identifies novel biomarkers associated with molecular subtypes. Mol. Cell Proteomics. 2008;7:1420–1433. doi: 10.1074/mcp.M700487-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sinha P, et al. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yin C, et al. RAGE-binding S100A8/A9 promotes the migration and invasion of human breast cancer cells through actin polymerization and epithelial- mesenchymal transition. Breast Cancer Res. Treat. 2013;142:297–309. doi: 10.1007/s10549-013-2737-1. [DOI] [PubMed] [Google Scholar]
- 104.Siegenthaler G, et al. A heterocomplex formed by the calcium-binding proteins MRP8 (S100A8) and MRP14 (S100A9) binds unsaturated fatty acids with high affinity. J. Biol. Chem. 1997;272:9371–9377. doi: 10.1074/jbc.272.14.9371. [DOI] [PubMed] [Google Scholar]
- 105.Markowitz J, Carson WE3rd. Review of S100A9 biology and its role in cancer. Biochim. Biophys. Acta. 2012;1835:100–109. doi: 10.1016/j.bbcan.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gomes LH, et al. S100A8 and S100A9—oxidant scavengers in inflammation. Free Radic. Biol. Med. 2012;58:170–186. doi: 10.1016/j.freeradbiomed.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 107.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nature Cell Biol. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 108. Hiratsuka S, et al. The S100A8–serum amyloid A3–TLR4 paracrine cascade establishes a pre-metastatic phase. Nature Cell Biol. 2008;10:1349–1355. doi: 10.1038/ncb1794. References 107 and 108 demonstrate that tumour-derived factors induce S100A8 and/or S100A9 expression in the lung to establish a pre-metastatic niche and create a permissive environment for pulmonary metastasis.
- 109.Liu Y, et al. Premetastatic soil and prevention of breast cancer brain metastasis. Neuro Oncol. 2013;15:891–903. doi: 10.1093/neuonc/not031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zimmer DB, Chaplin J, Baldwin A, Rast M. S100-mediated signal transduction in the nervous system and neurological diseases. Cell. Mol. Biol. (Noisy-le-Grand) 2005;51:201–214. [PubMed] [Google Scholar]
- 111.Nonaka D, Chiriboga L, Rubin BP. Differential expression of S100 protein subtypes in malignant melanoma, and benign and malignant peripheral nerve sheath tumors. J. Cutan. Pathol. 2008;35:1014–1019. doi: 10.1111/j.1600-0560.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 112.Maelandsmo GM, et al. Differential expression patterns of S100A2, S100A4 and S100A6 during progression of human malignant melanoma. Int. J. Cancer. 1997;74:464–469. doi: 10.1002/(sici)1097-0215(19970822)74:4<464::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 113.Petersson S, Shubbar E, Enerback L, Enerback C. Expression patterns of S100 proteins in melanocytes and melanocytic lesions. Melanoma Res. 2009;19:215–225. doi: 10.1097/CMR.0b013e32832c6358. [DOI] [PubMed] [Google Scholar]
- 114.Brouard MC, Saurat JH, Ghanem G, Siegenthaler G. Urinary excretion of epidermal-type fatty acid-binding protein and S100A7 protein in patients with cutaneous melanoma. Melanoma Res. 2002;12:627–631. doi: 10.1097/00008390-200212000-00013. [DOI] [PubMed] [Google Scholar]
- 115.Hensler S, Mueller MM. Inflammation and skin cancer: old pals telling new stories. Cancer J. 2013;19:517–524. doi: 10.1097/PPO.0000000000000010. [DOI] [PubMed] [Google Scholar]
- 116.Roltsch E, Holcomb L, Young KA, Marks A, Zimmer DB. PSAPP mice exhibit regionally selective reductions in gliosis and plaque deposition in response to S100B ablation. J. Neuroinflammation. 2010;7:78. doi: 10.1186/1742-2094-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Geara FB, Ang KK. Radiation therapy for malignant melanoma. Surg. Clin. North Am. 1996;76:1383–1398. doi: 10.1016/s0039-6109(05)70521-1. [DOI] [PubMed] [Google Scholar]
- 119.Satyamoorthy K, et al. Aberrant regulation and function of wild-type p53 in radioresistant melanoma cells. Cell Growth Differ. 2000;11:467–474. [PubMed] [Google Scholar]
- 120.Sawa H, et al. Histone deacetylase inhibitors such as sodium butyrate and trichostatin A induce apoptosis through an increase of the bcl-2-related protein Bad. Brain Tumor Pathol. 2001;18:109–114. doi: 10.1007/BF02479423. [DOI] [PubMed] [Google Scholar]
- 121.Lin J, et al. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J. Biol. Chem. 2004;279:34071–34077. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 122.Lin J, et al. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J. Biol. Chem. 2001;276:35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 123.Zimmer DB, Lapidus RG, Weber D. J., In vivo screening of S100B inhibitors for melanoma therapy. Methods Mol. Biol. 2013;963:303–317. doi: 10.1007/978-1-62703-230-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hartman KG, et al. Complex formation between S100B and the p90 ribosomal S6 kinase (RSK) in malignant melanoma is Ca2+-dependent and inhibits ERK-mediated phosphorylation of RSK. J. Biol. Chem. 2014;289:12886–12895. doi: 10.1074/jbc.M114.561613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J. Biol. Chem. 2010;285:27487–27498. doi: 10.1074/jbc.M110.155382. References 121, 122 and 125 establish the molecular basis of S100B-mediated regulation of p53 in melanoma, thus informing the development of S100B inhibitors.
- 126.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc. Natl Acad. Sci. USA. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wilder PT, et al. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim. Biophys. Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 128.Smith J, et al. The effect of pentamidine on melanoma ex vivo. Anticancer Drugs. 2010;21:181–185. doi: 10.1097/CAD.0b013e3283340cee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hammacher A, Thompson EW, Williams ED. Interleukin-6 is a potent inducer of S100P, which is up-regulated in androgen-refractory and metastatic prostate cancer. Int. J. Biochem. Cell Biol. 2005;37:442–450. doi: 10.1016/j.biocel.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 130.Tothova V, et al. Glucocorticoid receptor-mediated transcriptional activation of S100P gene coding for cancer-related calcium-binding protein. J. Cell Biochem. 2011;112:3373–3384. doi: 10.1002/jcb.23268. [DOI] [PubMed] [Google Scholar]
- 131.Barry S, et al. S100P is a metastasis-associated gene that facilitates transendothelial migration of pancreatic cancer cells. Clin. Exp. Metastasis. 2013;30:251–264. doi: 10.1007/s10585-012-9532-y. [DOI] [PubMed] [Google Scholar]
- 132.Bulk E, et al. Adjuvant therapy with small hairpin RNA interference prevents non-small cell lung cancer metastasis development in mice. Cancer Res. 2008;68:1896–1904. doi: 10.1158/0008-5472.CAN-07-2390. [DOI] [PubMed] [Google Scholar]
- 133.Ding Q, et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J. Clin. Invest. 2011;121:4526–4536. doi: 10.1172/JCI45008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jiang L, et al. Targeting S100P inhibits colon cancer growth and metastasis by lentivirus-mediated RNA interference and proteomic analysis. Mol. Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Austermann J, Nazmi AR, Muller-Tidow C, Gerke V. Characterization of the Ca2+ -regulated ezrin-S100P interaction and its role in tumor cell migration. J. Biol. Chem. 2008;283:29331–29340. doi: 10.1074/jbc.M806145200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heil A, et al. S100P is a novel interaction partner and regulator of IQGAP1. J. Biol. Chem. 2011;286:7227–7238. doi: 10.1074/jbc.M110.135095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J. Biol. Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 138.Afanador L, et al. The Ca2+ sensor S100A1 modulates neuroinflammation, histopathology and Akt activity in the PSAPP Alzheimer’s disease mouse model. Cell Calcium. 2014;56:68–80. doi: 10.1016/j.ceca.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 139.Rohde D, et al. S100A1: a multifaceted therapeutic target in cardiovascular disease. J. Cardiovasc. Transl. Res. 2010;3:525–537. doi: 10.1007/s12265-010-9211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sack U, et al. S100A4-induced cell motility and metastasis is restricted by the Wnt/β-catenin pathway inhibitor calcimycin in colon cancer cells. Mol. Biol. Cell. 2011;22:3344–3354. doi: 10.1091/mbc.E10-09-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stein U, et al. Intervening in β-catenin signaling by sulindac inhibits S100A4-dependent colon cancer metastasis. Neoplasia. 2011;13:131–144. doi: 10.1593/neo.101172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weber C, et al. Therapeutic safety of high myocardial expression levels of the molecular inotrope S100A1 in a preclinical heart failure model. Gene Ther. 2014;21:131–138. doi: 10.1038/gt.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dakhel S, et al. S100P antibody-mediated therapy as a new promising strategy for the treatment of pancreatic cancer. Oncogenesis. 2014;3:e92. doi: 10.1038/oncsis.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Qin H, et al. Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice. Nature Med. 2014;20:676–681. doi: 10.1038/nm.3560. This reference uses an innovative and unbiased approach to identify novel diagnostic and therapeutic targets on the surface of MDSCs, which were subsequently determined to be S100 proteins.
- 145.Dhar A, et al. Simultaneous inhibition of key growth pathways in melanoma cells and tumor regression by a designed bidentate constrained helical peptide. Biopolymers. 2014;101:344–358. doi: 10.1002/bip.22505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bjork P, et al. Identification of human S100A9 as a novel target for treatment of autoimmune disease via binding to quinoline-3-carboxamides. PLoS Biol. 2009;7:e97. doi: 10.1371/journal.pbio.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pili R, et al. Phase II randomized, double-blind, placebo-controlled study of tasquinimod in men with minimally symptomatic metastatic castrate-resistant prostate cancer. J. Clin. Oncol. 2011;29:4022–4028. doi: 10.1200/JCO.2011.35.6295. [DOI] [PubMed] [Google Scholar]
- 148.Jennbacken K, et al. Inhibition of metastasis in a castration resistant prostate cancer model by the quinoline-3-carboxamide tasquinimod (ABR-215050) Prostate. 2011;72:913–924. doi: 10.1002/pros.21495. [DOI] [PubMed] [Google Scholar]
- 149.Arumugam T, Ramachandran V, Maxwell D, Bornmann WG, Logsdon CD. Designing and developing S100P inhibitor 5-methyl cromolyn (C5OH) for pancreatic cancer therapy. Mol. Cancer Ther. 2013;12:654–662. doi: 10.1158/1535-7163.MCT-12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim CE, Lim SK, Kim J. S., In vivo antitumor effect of cromolyn in PEGylated liposomes for pancreatic cancer. J. Control. Release. 2012;157:190–195. doi: 10.1016/j.jconrel.2011.09.066. [DOI] [PubMed] [Google Scholar]
- 151.Mack GS, Marshall A. Lost in migration. Nature Biotech. 2010;28:214–229. doi: 10.1038/nbt0310-214. [DOI] [PubMed] [Google Scholar]
- 152.Rani SG, Mohan SK, Yu C. Molecular level interactions of S100A13 with amlexanox: inhibitor for formation of the multiprotein complex in the nonclassical pathway of acidic fibroblast growth factor. Biochemistry. 2010;49:2585–2592. doi: 10.1021/bi9019077. [DOI] [PubMed] [Google Scholar]
- 153.Okada M, Tokumitsu H, Kubota Y, Kobayashi R. Interaction of S100 proteins with the antiallergic drugs, olopatadine, amlexanox, and cromolyn: identification of putative drug binding sites on S100A1 protein. Biochem. Biophys. Res. Commun. 2002;292:1023–1030. doi: 10.1006/bbrc.2002.6761. [DOI] [PubMed] [Google Scholar]
- 154.Malashkevich VN, et al. Phenothiazines inhibit S100A4 function by inducing protein oligomerization. Proc. Natl Acad. Sci. USA. 2010;107:8605–8610. doi: 10.1073/pnas.0913660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garrett SC, et al. A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bjork P, et al. Common interactions between S100A4 and S100A9 defined by a novel chemical probe. PLoS ONE. 2013;8:e63012. doi: 10.1371/journal.pone.0063012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dulyaninova NG, et al. Cysteine 81 is critical for the interaction of S100A4 and myosin-IIA. Biochemistry. 2011;50:7218–7227. doi: 10.1021/bi200853y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Reddy TR, Li C, Fischer PM, Dekker LV. Three-dimensional pharmacophore design and biochemical screening identifies substituted 1,2,4-triazoles as inhibitors of the annexin A2-S100A10 protein interaction. ChemMedChem. 2012;7:1435–1446. doi: 10.1002/cmdc.201200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yoshimura C, Miyafusa T, Tsumoto K. Identification of small-molecule inhibitors of the human S100B–p53 interaction and evaluation of their activity in human melanoma cells. Bioorg. Med. Chem. 2013;21:1109–1115. doi: 10.1016/j.bmc.2012.12.042. [DOI] [PubMed] [Google Scholar]
- 160. Cavalier MC, et al. Covalent small molecule inhibitors of Ca2+-bound S100B. Biochemistry. 2014;53:6628–6640. doi: 10.1021/bi5005552. Using different approaches, references 152, 154 and 160 identify small-molecule inhibitors of S100 proteins and demonstrate the multiple mechanisms for disrupting S100-target interactions.
- 161.Agamennone M, et al. Fragmenting the S100B–p53 interaction: combined virtual/biophysical screening approaches to identify ligands. ChemMedChem. 2010;5:428–435. doi: 10.1002/cmdc.200900393. [DOI] [PubMed] [Google Scholar]
- 162.Charpentier TH, et al. Divalent metal ion complexes of S100B in the absence and presence of pentamidine. J. Mol. Biol. 2008;382:56–73. doi: 10.1016/j.jmb.2008.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Markowitz J, et al. Identification and characterization of small molecule inhibitors of the calcium-dependent S100B–p53 tumor suppressor interaction. J. Med. Chem. 2004;47:5085–5093. doi: 10.1021/jm0497038. [DOI] [PubMed] [Google Scholar]
- 164.Wright PE, Dyson HJ. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nature Chem. Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proc. Natl Acad. Sci. USA. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Markowitz J, et al. Design of inhibitors for S100B. Curr. Top. Med. Chem. 2005;5:1093–1108. doi: 10.2174/156802605774370865. [DOI] [PubMed] [Google Scholar]
- 168.Wang G, et al. Mutually antagonistic actions of S100A4 and S100A1 on normal and metastatic phenotypes. Oncogene. 2005;24:1445–1454. doi: 10.1038/sj.onc.1208291. [DOI] [PubMed] [Google Scholar]
- 169.Grigorian M, et al. Effect of mts1 (S100A4) expression on the progression of human breast cancer cells. Int. J. Cancer. 1996;67:831–841. doi: 10.1002/(SICI)1097-0215(19960917)67:6<831::AID-IJC13>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 170.Lloyd BH, Platt-Higgins A, Rudland PS, Barraclough R. Human S100A4 (p9Ka) induces the metastatic phenotype upon benign tumour cells. Oncogene. 1998;17:465–473. doi: 10.1038/sj.onc.1201948. [DOI] [PubMed] [Google Scholar]
- 171.Fujiwara M, et al. Stable knockdown of S100A4 suppresses cell migration and metastasis of osteosarcoma. Tumour Biol. 2011;32:611–622. doi: 10.1007/s13277-011-0160-y. [DOI] [PubMed] [Google Scholar]
- 172.Maelandsmo GM, et al. Reversal of the in vivo metastatic phenotype of human tumor cells by an anti-CAPL (mts1) ribozyme. Cancer Res. 1996;56:5490–5498. [PubMed] [Google Scholar]
- 173.Zhang G, Li M, Jin J, Bai Y, Yang C. Knockdown of S100A4 decreases tumorigenesis and metastasis in osteosarcoma cells by repression of matrix metalloproteinase-9. Asian Pac. J. Cancer Prev. 2011;12:2075–2080. [PubMed] [Google Scholar]
- 174.Tsai WC, Tsai ST, Jin YT, Wu LW. Cyclooxygenase-2 is involved in S100A2-mediated tumor suppression in squamous cell carcinoma. Mol. Cancer Res. 2006;4:539–547. doi: 10.1158/1541-7786.MCR-05-0266. [DOI] [PubMed] [Google Scholar]
- 175.Lo JF, et al. The epithelial-mesenchymal transition mediator S100A4 maintains cancer-initiating cells in head and neck cancers. Cancer Res. 2011;71:1912–1923. doi: 10.1158/0008-5472.CAN-10-2350. [DOI] [PubMed] [Google Scholar]
- 176.Rasanen K, et al. Comparative secretome analysis of epithelial and mesenchymal subpopulations of head and neck squamous cell carcinoma identifies S100A4 as a potential therapeutic target. Mol. Cell Proteomics. 2013;12:3778–3792. doi: 10.1074/mcp.M113.029587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen D, et al. S100A4 silencing blocks invasive ability of esophageal squamous cell carcinoma cells. World J. Gastroenterol. 2012;18:915–922. doi: 10.3748/wjg.v18.i9.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bulk E, et al. S100A2 induces metastasis in non-small cell lung cancer. Clin. Cancer Res. 2009;15:22–29. doi: 10.1158/1078-0432.CCR-08-0953. [DOI] [PubMed] [Google Scholar]
- 179.Takenaga K, Nakamura Y, Sakiyama S. Expression of antisense RNA to S100A4 gene encoding an S100-related calcium-binding protein suppresses metastatic potential of high-metastatic Lewis lung carcinoma cells. Oncogene. 1997;14:331–337. doi: 10.1038/sj.onc.1200820. [DOI] [PubMed] [Google Scholar]
- 180.Ortiz ML, Lu L, Ramachandran I, Gabrilovich DI. Myeloid-derived suppressor cells in the development of lung cancer. Cancer Immunol. Res. 2014;2:50–58. doi: 10.1158/2326-6066.CIR-13-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Phipps KD, Surette AP, O’Connell PA, Waisman DM. Plasminogen receptor S100A10 is essential for the migration of tumor-promoting macrophages into tumor sites. Cancer Res. 2011;71:6676–6683. doi: 10.1158/0008-5472.CAN-11-1748. [DOI] [PubMed] [Google Scholar]
- 182.Kang M, et al. S100A3 suppression inhibits in vitro and in vivo tumor growth and invasion of human castration-resistant prostate cancer cells. Urology. 2014 doi: 10.1016/j.urology.2014.09.018. http://dx.doi.org/10.1016/j.urology.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 183.Siddique HR, et al. The S100A4 oncoprotein promotes prostate tumorigenesis in a transgenic mouse model: regulating NFκB through the RAGE receptor. Genes Cancer. 2013;4:224–234. doi: 10.1177/1947601913492420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Saleem M, et al. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc. Natl Acad. Sci. USA. 2006;103:14825–14830. doi: 10.1073/pnas.0606747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ochiya T, Takenaga K, Endo H. Silencing of S100A4, a metastasis-associated protein, in endothelial cells inhibits tumor angiogenesis and growth. Angiogenesis. 2013;17:17–26. doi: 10.1007/s10456-013-9372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Grebhardt S, Muller-Decker K, Bestvater F, Hershfinkel M, Mayer D. Impact of S100A8/A9 expression on prostate cancer progression in vitro and in vivo. J. Cell. Physiol. 2013;229:661–671. doi: 10.1002/jcp.24489. [DOI] [PubMed] [Google Scholar]
- 187.Basu GD, et al. Functional evidence implicating S100P in prostate cancer progression. Int. J. Cancer. 2008;123:330–339. doi: 10.1002/ijc.23447. [DOI] [PubMed] [Google Scholar]
- 188.Dahlmann M, et al. Systemic shRNA mediated knockdown of S100A4 in colorectal cancer xenografted mice reduces metastasis formation. Oncotarget. 2012;3:783–797. doi: 10.18632/oncotarget.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Takenaga K, et al. Modified expression of Mts1/ S100A4 protein in C6 glioma cells or surrounding astrocytes affects migration of tumor cells in vitro and in vivo. Neurobiol. Dis. 2007;25:455–463. doi: 10.1016/j.nbd.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 190.Chen S, et al. Comparative proteomics of glioma stem cells and differentiated tumor cells identifies S100A9 as a potential therapeutic target. J. Cell Biochem. 2013;114:2795–2808. doi: 10.1002/jcb.24626. [DOI] [PubMed] [Google Scholar]
- 191.Hua J, et al. Short hairpin RNA-mediated inhibition of S100A4 promotes apoptosis and suppresses proliferation of BGC823 gastric cancer cells in vitro and in vivo. Cancer Lett. 2010;292:41–47. doi: 10.1016/j.canlet.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 192.Zhang J, Zhang K, Jiang X. S100A6 as a potential serum prognostic biomarker and therapeutic target in gastric cancer. Dig. Dis. Sci. 2014;59:2136–2144. doi: 10.1007/s10620-014-3137-z. [DOI] [PubMed] [Google Scholar]
- 193.Levett D, et al. Transfection of S100A4 produces metastatic variants of an orthotopic model of bladder cancer. Am. J. Pathol. 2002;160:693–700. doi: 10.1016/S0002-9440(10)64889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Arumugam T, Ramachandran V, Logsdon CD. Effect of cromolyn on S100P interactions with RAGE and pancreatic cancer growth and invasion in mouse models. J. Natl Cancer Inst. 2006;98:1806–1818. doi: 10.1093/jnci/djj498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yang XC, et al. RNA interference suppression of A100A4 reduces the growth and metastatic phenotype of human renal cancer cells via NF-KB-dependent MMP-2 and bcl-2 pathway. Eur. Rev. Med. Pharmacol. Sci. 2013;17:1669–1680. [PubMed] [Google Scholar]
- 196.Zhao FT, Jia ZS, Yang Q, Song L, Jiang XJ. S100A14 promotes the growth and metastasis of hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2013;14:3831–3836. doi: 10.7314/apjcp.2013.14.6.3831. [DOI] [PubMed] [Google Scholar]
- 197.Shi Y, et al. Ribonucleic acid interference targeting S100A4 (Mts1) suppresses tumor growth and metastasis of anaplastic thyroid carcinoma in a mouse model. J. Clin. Endocrinol. Metab. 2006;91:2373–2379. doi: 10.1210/jc.2006-0155. [DOI] [PubMed] [Google Scholar]
- 198.Reeb AN, et al. S100A8 is a novel therapeutic target for anaplastic thyroid carcinoma. J. Clin. Endocrinol. Metab. 2014 doi: 10.1210/jc.2014-2988. http://dx.doi.org/10.1210/jc.2014-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Anania MC, et al. S100A11 overexpression contributes to the malignant phenotype of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013;98:E1591–E1600. doi: 10.1210/jc.2013-1652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.