Abstract
Inherently disparate cell growth and division, which are intimately coupled through a delicate network of intracellular and extracellular signaling, require ribosomal biogenesis. A number of events imparting instability to ribosomal biogenesis can cause nucleolar stress. In response to this stress, several ribosomal proteins bind to MDM2 and block MDM2-mediated p53 ubiquitination and degradation, resulting in p53-dependent cell cycle arrest. By doing so, the ribosomal proteins play a crucial role in connecting deregulated cell growth with inhibition of cell division. The ribosomal protein-MDM2-p53 signaling pathway provides a molecular switch that may constitute a surveillance network monitoring the integrity of ribosomal biogenesis.
Keywords: Ribosome biogenesis, RPL11, RPL5, RPL23, MDM2, p53, zinc finger, cancer, nucleolus
Introduction
Cell growth (increase in cell size) and cell division (increase in cell number) are two separable yet interconnected aspects of cell behavior in all organisms (Conlon and Raff, 1999). Cell growth is dominant and rate limiting for cell division (Hartwell, 1971). Blocking cell growth of cultured mammalian cells by reagents that inhibit ribosomal biogenesis leads to arrest of the cell cycle, usually in the G1 phase (Pardee, 1989). Deregulation of the molecular mechanisms controlling cell growth results in cells of altered size and leads to developmental errors and a variety of pathological conditions, including cancer (Ruggero and Pandolfi, 2003).
It is clear that mutations in genes involved in cell division control often contribute to cancer development, and one prominent example is the p53 tumor suppressor. In response to cellular stress, p53 is activated to induce cell cycle arrest, senescence or apoptosis. Mutations in p53 inactivating p53’s function are frequently found in both familial and sporadic human cancers. More than 50% of human cancers harbor mutations in TP53; in most of the remaining cancers, the p53 pathway is otherwise inactivated, such as through overproduction of the p53 inhibitor MDM2 (murine double minute 2, and also HDM2 for its human ortholog) (Toledo and Wahl, 2006). While disruption of normal cell cycle control is an established route to cancer, it is more controversial whether an increase in cell growth on its own can initiate or promote cancer development (Ruggero and Pandolfi, 2003). Overall, the mechanisms that link cell growth with cell division are poorly understood (Sulic et al., 2005b). However, recent studies began to offer some clues for associating the MDM2-p53 feedback loop with the sophisticated mechanisms involving ribosomal biogenesis.
The MDM2-p53 feedback loop is regulated in response to a multitude of cytotoxic and genotoxic stressors. One of these stressors is called nucleolar stress (a.k.a. ribosomal stress) (Rubbi and Milner, 2003). In principle, Ribosome biogenesis is an essential cellular process that involves three fundamental steps: coordinated expression of ribosomal RNA (rRNA) and ribosomal proteins (RPs), processing of rRNA, and assembly of the 40S and 60S ribosome subunits in the nucleolus (Perry, 2007). The 40S and 60S subunits are then exported from the nucleolus, through the nucleoplasm, into the cytoplasm for 80S Ribosome assembly and protein synthesis (Perry, 2007). Perturbation of any step in this process is thought to lead to nucleolar stress, triggering specific binding of several RPs to MDM2, which inhibits MDM2’s E3 ubiquitin ligase function toward p53, leading to p53 stabilization and activation (Lindstrom et al., 2007a). For instance, expression of a dominant negative mutant of Bop1 involved in rRNA processing can inhibit ribosomal biogenesis and elicit p53 activation (Pestov et al., 2001). Likewise, reduced production of individual RPs can also prompt p53 activation. In vivo inactivation of RPS6 activates a p53-dependent checkpoint response in thymocytes (Sulic et al., 2005a) or in embryonic fibroblast cells during gastrulation (Panic et al., 2006). Similar to the mammalian system, deficiency of several RPs in zebrafish also initiates p53-dependent cell growth arrest and apoptosis (Chakraborty et al., 2009; Danilova et al., 2008). Hence, these findings suggest an important role for the nucleolus as a cellular stress sensor in addition to as the workshop for ribosomal biogenesis. They also place p53 as a key molecule in the center of the signaling pathway sensing nucleolar stress and ribosomal malfunction. This review aims to illustrate recent progress toward understanding this newly acknowledged, yet under-studied, RP-MDM2-p53 signaling pathway [Readers are referred to recent reviews for other extraribosomal functions of RPs (Warner and McIntosh, 2009), and downstream functions (Vousden and Prives, 2009) and regulations (Kruse and Gu, 2009) of p53].
The ribosomal protein-MDM2-p53 pathway
In order to better illustrate the RP-MDM2-p53 pathway, it is necessary to briefly revisit what we have learned regarding the MDM2-p53 feedback loop and its regulation in response to cellular stresses. MDM2 regulates p53 primarily in two ways: (i) MDM2 binds directly to p53, thereby “masking” p53’s transactivation domain from access to the basal transcriptional machinery (Oliner et al., 1993), and (ii) MDM2 acts as an E3 ubiquitin ligase for p53, mediating the conjugation of ubiquitins to p53 and subsequent proteasomal degradation (Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997). Because MDM2 itself is a transcriptional target of p53 (Barak et al., 1993; Wu et al., 1993) and deletion of Tp53 completely rescues the lethality of Mdm2 knockout mice (Jones et al., 1995; Luna et al., 1995), the primary physiological function of MDM2 is thus to serve as a negative feedback ”knot” for p53, and therefore unhitching this MDM2 “knot” becomes critical for p53 activation by various stresses.
Increasing evidence has supported the notion that various stresses can activate distinct cellular signaling pathways that lead to the suppression of MDM2 activity and activation of p53 (Vogelstein et al., 2000; Vousden and Lu, 2002). For instance, DNA damage caused by genotoxic chemicals and ionizing or ultraviolet radiation triggers the activation of the ATM-Chk2 or ATR-Chk1 kinase cascades that leads to phosphorylation of both MDM2 and p53, blocking their functional or physical interactions (Appella and Anderson, 2001; Prives, 1998). Also, biological and oncogenic signals, such as viral infection or over expression of cellular oncogenes, induce the expression of the tumor suppressor ARF, which in turn binds to MDM2 and inhibits its activity (Sherr, 2006; Zhang and Xiong, 2001). Moreover, different types of stress have been shown to trigger a number of posttranslational modifications, including acetylation, sumoylation, methylation and neddylation, of p53 and/or MDM2 resulting in p53 activation [reviewed by (Brooks and Gu, 2003; Dai et al., 2006a; Huang and Berger, 2008; Melchior and Hengst, 2002; Prives and Manley, 2001)].
Recently, a new type of stress signal generated by disrupting ribosomal biogenesis and mediated by several RPs has been shown to inhibit MDM2 and activate p53. Ribosomal biogenesis can be disrupted by serum starvation, depletion of nucleotides, agents such as Actinomycin D, 5-Fluorouracil (5-FU), malfunction of nucleolar proteins involving in ribosome biogenesis (such as the dominant negative mutant Bop1) (Pestov et al., 2001), inhibition of B23 (a.k.a. nucleophosmin, NPM) activity by ARF (Itahana et al., 2003), and reduction of RPS6 (Fumagalli et al., 2009; Volarevic et al., 2000); all of these have been shown to generate nucleolar stress that signals to p53. This allows cells to halt proliferation under unhealthy and poor ribosomal biogenesis conditions. Although MDM2 was previously shown to interact with 5S rRNA and RPL5 (Marechal et al., 1994), the functional meaning of the interaction had not been realized until recently when several RPs, including RPL11 (Lohrum et al., 2003; Zhang et al., 2003), RPL23 (Dai et al., 2004; Jin et al., 2004), and RPL5 (Dai and Lu, 2004) were found to activate p53 through their interactions with MDM2.
Previously, RPL5 was shown to participate in MDM2 nuclear export (Roth et al., 1998). It was thought that the p53-MDM2 complex might “hitch a ride” on the Ribosome for cytoplasmic degradation of p53 (Sherr and Weber, 2000), and p53 accumulation after nucleolar stress was due to its failure to undergo nucleolus-dependent export and degradation (Rubbi and Milner, 2003). Later studies identified a direct interaction between MDM2 and RPs in response to Actinomycin D induced nucleolar stress, providing the first potential molecular mechanism for the nucleolar stress-p53 signaling pathway. Actinomycin D, a commonly used drug for human cancers (da Rocha et al., 2001), inhibits transcription catalyzed by RNA polymerases (RNA Pol) I, II and III at high concentrations (e.g. >30 nM), but it selectively inhibits RNA Pol-I-dependent transcription and hence rRNA production and ribosomal biogenesis at low concentrations (e.g. <10 nM) (Iapalucci-Espinoza and Franze-Fernandez, 1979; Perry and Kelley, 1970). Treating cells with either a lower dose of Actinomycin D or serum starvation inhibits ribosome assembly and consequently releases free RPs from the nucleolus to the nucleoplasm (Scheer and Hock, 1999). In response to nucleolar stress induced by a low dose of Actinomycin D (5 nM) (Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004) and 5-Fluorouracil (Gilkes et al., 2006; Sun et al., 2007), by serum depletion and contact inhibition (Bhat et al., 2004), by mycophenolic acid (MPA)-mediated depletion of GTP (Sun et al., 2008), or by interfering with nucleolar function via ectopic overexpression of nucleostemin (Dai et al., 2008), there is an increased binding of RPL5, RPL11, and RPL23 to MDM2. This binding inhibits MDM2’s E3 ligase function, resulting in p53 accumulation and activation. In addition to the three RPs, RPS7 (Chen et al., 2007; Zhu et al., 2009) and RPL26 (Ofir-Rosenfeld et al., 2008) have also been shown to interact with MDM2. The consequence of the RPS7-MDM2 interaction resembles that of the aforementioned three RPs in terms of their binding to MDM2 and activation of p53. However, surprising findings from a latest study show that MDMX can facilitate RPS7 suppression of MDM2 and that RPS7 itself is a substrate for MDM2 ubiquitination. Thus, it is proposed that RPS7 acts as both effector and affector of MDM2 (Zhu et al., 2009).
The interaction of RPL26 with MDM2 appears to perform a different function. RPL26 was found to increase the translational rate of p53 mRNA by binding to its 5′ untranslated region (UTR) (Takagi et al., 2005) and, in this case, MDM2 acts as a ubiquitin E3 ligase for ubiquitylation and degradation of RPL26, hence inhibiting p53 translation (Ofir-Rosenfeld et al., 2008). Recently, an RPS27-like protein (RPS27L) was identified as a direct p53 transcriptional target (He and Sun, 2007) and also thought to activate p53 by repressing MDM2 activity (personal communication with Y. Sun). Altogether, these findings indicate that RPs could play a pivotal role in p53 response to nucleolar stress (Figure. 1).
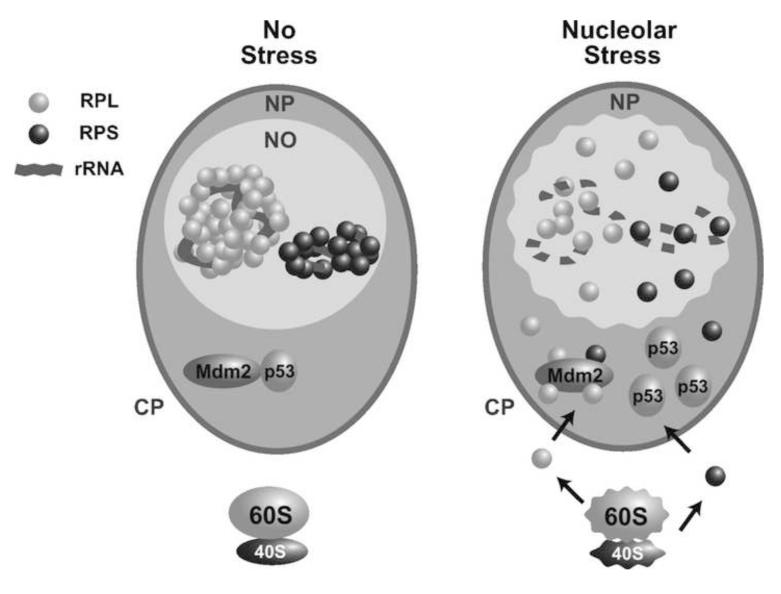
Figure 1. Schematic of RP-MDM2-p53 pathway regulation by nucleolar stress.
Under normal growth conditions (no stress), small (S, 40S) and large (L, 60S) RPs are assembled in the nucleolus (NO) and transported to the cytoplasm (CP) for protein synthesis. Under nucleolar stress, ribosomal biogenesis is inhibited and Ribosome-free forms of RPs (RPL and RPS) enter the nucleoplasm (NP) to interact with MDM2, resulting in p53 stabilization and activation. Similarly, RPs either released from breaking down (indicated by wavy edges) of cytoplasmic Ribosomes or overproduced in the cytoplasm can enter the nucleoplasm to interact with MDM2.
Multiple ribosomal players
Ribosomal biogenesis consumes a major part of cellular energy and resources and plays a key role in the life cycle of a cell (Conlon and Raff, 1999; Neufeld and Edgar, 1998; Warner, 1999) and thus is strictly monitored in the cell. Previous studies have been concentrated on how changes of proliferation lead to alteration of Ribosome production (Pyronnet and Sonenberg, 2001), but less attention has been paid to how alterations of ribosomal biogenesis may influence cell proliferation. As discussed above, recent studies using mammalian systems have begun to reveal the nucleolar stress-RP-MDM2-p53 signaling pathway and to unfold the role of this pathway in coordinating inhibition of cell growth with cell cycle arrest.
To monitor the status of cell growth and communicate it with cell division, both are very complex and sophisticated cell functions, it is not surprise for the cell to employ multiple RPs to regulate the MDM2-p53 feedback loop. Why are multiple RPs necessary? Before discussing this, it is necessary to briefly describe MDM2’s functional domains. MDM2 contains three conserved regions: an N-terminal p53 binding domain, a central acidic region encompassing a C4 zinc finger, and a C-terminal RING domain conferring the E3 ligase activity. The importance of the central acidic region and the C4 zinc finger for mediating MDM2 ubiquitination and degradation of p53 has been elegantly demonstrated by a domain-swapping experiment between MDM2 and its homolog MDMX (a.k.a. MDM4) (Kawai et al., 2003; Meulmeester et al., 2003). This notion is also underscored by the finding that ARF binds to this region to suppress MDM2 activity (Argentini et al., 2001; Kawai et al., 2003; Meulmeester et al., 2003). Importantly, these findings also provide a basis for how binding of the RPs to MDM2 may interfere with MDM2’s inhibition of p53 function.
Like ARF (Argentini et al., 2001; Kawai et al., 2003; Meulmeester et al., 2003), RPL5, RPL11, and RPL23 all bind to the central acidic region of MDM2 in an apparently similar but non-identical manner (Dai et al., 2004). Detailed analysis of these bindings reveals that these RPs require specific domains for efficient MDM2 binding. For example, the MDM2 C4 zinc finger is critical for its binding to RPL5 and RPL11, as the zinc finger mutant MDM2C305F cannot bind either one, but is not required for RPL23 binding (Dai et al., 2006b; Lindstrom et al., 2007b). Interestingly, a slightly different C4 zinc finger mutant, MDM2C305S, fails to bind RPL11, but was still capable of interacting with RPL5 and RPL23 (Gilkes et al., 2006). Whether RPL11 binds directly to the C4 zinc finger or whether the zinc finger only provides structural stability allowing MDM2 to bind to RPL11 remains unclear. Nevertheless, these studies suggest that the RPs, though interacting with the same MDM2 central acidic domain, have specific sequence requirements for binding. Surprisingly, these MDM2-interacting RPs have not been found to bind to MDMX (Gilkes et al., 2006; Jin et al., 2006), implying that fundamental structural differences exist between MDM2 and MDMX even though both contain a similar central acidic domain and C4 zinc finger.
It is still puzzling why RPL11’s ability to bind MDM2 relies so heavily on the MDM2 C4 zinc finger. Some clues may arise from the structural features of this domain, which belongs to a class of zinc fingers known as RanBP2/NZF with a defined consensus sequence X(4)-W-X-C-X(2- 4)-C-X(3)-N-X(6)-C-X(2)-C-X(5), where “X” is any amino acid. The NZF type of zinc finger is a compact zinc-binding module found in many proteins that function in ubiquitin-related processes, including direct interaction with ubiquitins (Alam et al., 2004). The solution structure of the MDM2 C4 zinc finger (Protein Data Bank accession 2C6B) (Yu et al., 2006) indeed discloses that MDM2 shares the zinc finger structure with a wide variety of proteins that have distinct functions and mostly involve binding other macromolecules. For instance, the C4 zinc finger of RanBP2 binds to the nuclear export receptor protein Crm-1 (Singh et al., 1999). Also, the C4 zinc finger domain of some other proteins mediates binding to RNA. For example, the zinc finger protein znf265 interacts with pre-mRNA and is able to alter splicing patterns (Adams et al., 2001), and the zinc finger domain of Npl4 binds to ubiquitin (Alam et al., 2004; Meyer et al., 2002). Although the MDM2 zinc finger may not bind to ubiquitin (Meyer et al., 2002), it could directly bind to rRNA, mRNA, or Crm-1, and if so, RPL11 (and other MDM2-binding RPs) might compete with MDM2 for binding to these molecules.
Consistent with the observation that RPL11, RPL5 and RPL23 bind to overlapping yet distinct domains within the central region of MDM2, these RPs appear to utilize similar yet non-identical mechanisms to regulate MDM2’s E3 ubiquitin ligase activity (Dai et al., 2006b). Like ARF, when RPL11 is over expressed in cells, it suppresses p53 polyubiquitination to a lesser degree than do RPL5 and RPL23 (Dai et al., 2006b). Also similar to ARF, RPL11 does not inhibit ubiquitination of Mdm2, instead it represses degradation of MDM2 via a postubiquitination mechanism, whereas RPL5 and RPL23 suppress MDM2 autoubiquitination when co-expressed with MDM2 (Dai et al., 2006b). However, it remains intangible how exactly these RPs negate MDM2’s E3 ligase activity (see more discussion in the following section).
It is intriguing that all the MDM2-interacting RPs are important for p53’s full response to nucleolar stress. Why, then, are multiple RPs needed to interact with MDM2 and suppress its activity, and why does knockdown of each of the RPs attenuate, at least partially, nucleolar stress-induced p53 activation? Although there has not been a definitive answer thus far, it is possible that multiple RPs might sense different growth inhibitory or ribosomal stress signals so that different steps of ribosomal biogenesis can be effectively monitored. Also, in addition to the aforementioned difference in their mechanisms, the amino acid sequence in MDM2 required for RPL23 binding is different from that for RPL11 binding, so that RPL23 and RPL11 can bind MDM2 simultaneously to form a ternary complex (Dai et al., 2006b; Lindstrom et al., 2007b). Consistent with this idea, endogenous RPL23 and RPL11 appear to react differently to Actinomycin D treatment, as the former decreased whereas the latter increased when cells were treated with Actinomycin D (Jin et al., 2004). Moreover, multiple MDM2-binding RPs may work together to synergize their inhibitory effects on MDM2 when an error in ribosomal biogenesis occurs, as exemplified by the cooperation of RPL11 with RPL5 to synergistically inhibit MDM2 and activate p53 (Horn and Vousden, 2008). This is consistent with the finding that RPL11 and RPL5 are more essential for p53 response to nucleolar stress than is RPL23, as knockdown of RPL11 or RPL5 substantially alleviates p53 activation induced by this stress, whereas knockdown of RPL23 actually induces p53 (Dai et al., 2004; Jin et al., 2004). Another explanation for why multiple RPs may perform similar functions to activate p53 could be that they may compensate for one another or serve as rescuers if any of them fails. Because ribosomal biogenesis is a highly coordinated process involving a number of vital cellular activities taking place in different subcellular compartments and involving numerous steps and co-factors, errors could occur at each of these steps and pose dire consequences for cells. To monitor these steps, each of the MDM2-interacting RPs may preferentially recognize specific signals of nucleolar stresses. In partial agreement with this hypothesis, knockdown of RPS6 impairs 40S ribosomal biogenesis, promotes the translation of RPL11 mRNA specifically, and subsequently initiates RPL11-dependent MDM2 inhibition and p53 activation (Fumagalli et al., 2009). In summary, for cells to grow and proliferate normally, multiple RPs appear to be needed to signal to the p53 pathway in response to a variety of nucleolar stresses (Figure. 1).
How do the ribosomal players work?
If multiple RPs are needed to sense and transduce nucleolar stress to the MDM2-p53 pathway, then how do they work? Where do the RPs find and bind to MDM2, as RPs mostly reside in the nucleolus and the cytoplasm, while MDM2 often stays in the nucleoplasm? One possibility is that the RPs may interact with MDM2 in the nucleoplasm on their way to the nucleolus, as they, once translated in the cytoplasm, must be transported through the nucleoplasm to the nucleolus for Ribosome assembly. An increase in RP translation would lead to increased traffic of the RPs through the nucleus, consequently an increased interaction with MDM2. In this regard, the RP-MDM2 interaction could function as a mechanism sensing elevated RP translation, such as the increase in global translation caused by overexpression of the proto-oncogene c-Myc (Schmidt, 2004) or the rise of RPL11 level by RPS6 knockdown (Fumagalli et al., 2009). It could also sense the breakdown of cytoplasmic polysomes and the resulting release of individual ribosomal RPs into the nucleoplasm (Dai et al., 2004; Gilkes et al., 2006; Jin et al., 2004). Alternatively, given that there is not a physical boundary between the nucleoplasm and the nucleolus, RPs and other nucleolar proteins could shuttle freely between the two cellular compartments (Chen and Huang, 2001), which would provide an opportunity for excess RPs to interact with MDM2 in the nucleoplasm. In a similar and reciprocal way, MDM2 may enter the nucleolus, as can be seen after cells treated by MG132 (Klibanov et al., 2001), presumably through its interaction with the nucleolar ARF (Tao and Levine, 1999; Weber et al., 1999), and thus interact with RPs in the nucleolus (Lohrum et al., 2003). However, the latter mechanism might be context specific, as MDM2 was not detected in the nucleolus under unstressed or nucleolar-stress conditions in recent studies (Dai et al., 2008; Sun et al., 2008).
Another quandary regarding how RPs regulate MDM2 concerns protein levels in a cell. Unlike DNA damage- or aberrant oncogene-induced p53 activation, which often requires either activation of a kinase (checkpoint kinases) or induction of a protein (the tumor suppressor ARF) that is usually maintained at basal levels under unstressed conditions, RPs are always abundant in cells. Then, how is the RP-MDM2 interaction prevented under normal and favorable growth conditions, yet promoted in response to nucleolar stress? It is conceivable that without stress, an appropriate amount of RPs is produced just for the need of Ribosome assembly, but any excess RPs beyond this demand undergo constant degradation (Lam et al., 2007). However, under nucleolar stress, rRNA synthesis is reduced or blocked and extra Ribosome-unbound RPL11 translocates to the nucleoplasm to interact with MDM2 and activate p53 (Bhat et al., 2004), even though the total amount of RPs is not altered.
As discussed above, like ARF, all MDM2-binding RPs inhibit the MDM2 ubiquitin ligase activity by binding to its central acidic domain. However, it is unclear how exactly the binding of these small inhibitory proteins in the middle of MDM2 leads to inhibition of the ubiquitin ligase activity of its C-terminal RING finger. One possible explanation is that the central acidic domain of MDM2 acts as a flexible arm to juxtapose the N-terminal-bound p53 within close proximity of the C-terminal RING domain in order to facilitate ubiquitin transfer. The binding of RPL11 or other small protein molecules to the MDM2 central domain may reduce its flexibility, and the rigid MDM2 is thus unable to bring its RING domain and p53 together. Although this prediction is tempting, direct evidence is lacking and a better mechanistic insight will come from a three-dimensional structure of the complex of MDM2 with RPL11 or other RPs.
Another possible mechanism by which RPs inhibit MDM2’s E3 ligase activity concerns the MDM2 homologue MDMX. Previous studies using protein overexpression and siRNA knockdown suggested that MDMX enhances the MDM2 E3 ligase activity (Linares et al., 2003). A later study showed that MDMX-deficient MEF cells (Mdm2+/−;MdmX−/−) had a moderately elevated level of endogenous p53 compared to that in MDMX-proficient cells (Mdm2+/−;MdmX+/+) (Francoz et al., 2006), suggesting that MDMX may contribute to p53 degradation in vivo. Although how MDMX contributes to MDM2’s E3 activity remains obscure, it is clear that MDMX is essential for negating p53 function as germline inactivation of Mdmx results in embryonic lethality that can be rescued by concomitant deletion of p53 (Migliorini et al., 2002; Parant et al., 2001). The RING finger domain of MDMX is crucial for MDM2-mediated p53 degradation (Poyurovsky et al., 2007; Uldrijan et al., 2007), probably by facilitating heterodimerization with MDM2 (Jackson and Berberich, 2000; Sharp et al., 1999; Stad et al., 2000; Tanimura et al., 1999). Hence, it is possible that RPs (probably ARF as well) might inhibit MDM2 function by disrupting the MDM2-MDMX interaction. Alternatively, RPL11 can promote MDMX degradation by binding to MDM2, consequently activating p53, as it fails to do so when the RPL11 binding-deficient MDM2C305S mutant is used (Gilkes et al., 2006).
In addition to mediating p53 ubiquitination, the C-terminal RING domain of MDM2 is shown to be the E3, at least in vitro, for its own ubiquitination and degradation (Fang et al., 2000; Honda and Yasuda, 2000). However, even though ectopic RPL11 can inhibit MDM2-mediated p53 ubiquitination, it does not inhibit MDM2 autoubiquitination (Dai et al., 2006b), just like ARF (Xirodimas et al., 2001). Thus, it appears that these two small proteins preferentially inhibit MDM2’s E3 activity toward p53 but not its autoubiquitination. This could be explained by the existence of a putative cellular E3 ligasese for MDM2, as in vivo MDM2 bearing a C462A mutation in its C-terminal RING domain, which abolishes its E3 ligase activity, is still ubiquitinated and degraded as quickly as wild type MDM2 under both unstressed and genotoxically stressed conditions (Itahana et al., 2007), which could also explain why RPL11 or ARF do not inhibit MDM2 “autoubiquitination” (Dai et al., 2006b).
Relevance to cancer and other genetic diseases
Identification of several RPs as crucial players in regulating p53 function not only reveals molecular insight into this under-appreciated signaling pathway but also raises new questions concerning the importance of RPs in cancer prevention and development. The role of RPs and ribosomal biogenesis in tumorigenesis has been regarded as a double-edged sword. On one hand, elevated levels of ribosomal biogenesis and protein translation, such as from overexpression of eIF-4E (Ruggero et al., 2004; Wendel et al., 2004) and tRNAs and 5S rRNA (Marshall et al., 2008), have been linked to tumor formation in multiple mouse tissues [reviewed by (Dai and Lu, 2008; Ruggero and Pandolfi, 2003)]. For example, overexpression of RPS3a can lead to transformation and tumorigenesis in nude mice (Naora et al., 1998). On the other hand, reduction of ribosomal biogenesis and translational capacity due to genetic hapoloinsufficiency, such as loss of one copy of an RP-encoding gene or mutation of a gene essential for ribosomal biogenesis, has been associated with a high incidence of cancer development in humans. For instance, mutation of the DKC1 gene, whose protein product dyskerin is a putative pseudouridine synthase important for pre-rRNA processing, is tightly linked to dyskeratosis congenital disease with characteristics of premature aging and an increased susceptibility to certain cancers (Ruggero et al., 2003). Also, RMRP, encoding an RNase vital for pre-rRNA processing, is mutated in the pleiotropic human disease Cartilage-Hair hypoplasia, which typically results in short stature, defective cellular immunity, and predisposition to cancer (Ridanpaa et al., 2001).
Mutations in several RPs have also been linked to cancer-prone genetic diseases. A prominent example is Diamond-Blackfan anemia (DBA), an inheritable disease initially found to be associated with mutation of RPS19. Heterozygous null mutations of RPS19 were found in 25% of DBA patients (Draptchinskaia et al., 1999), who often suffer chronic constitutional regenerative anemia, various degrees of congenital abnormalities, and an increased susceptibility to hematopoietic malignancies. Another RP-associated disease is 5q-syndrome, which is frequently linked with deletion of one allele of RPS14 and characterized by increased incidence of hematopoietic tumors and anemia (Ebert et al., 2008). Remarkably, recent studies have found various heterozygous mutations, including single point mutations, in RPL5, RPL11, and RPS7 in DBA patients (Cmejla et al., 2009; Gazda et al., 2008). Mutational screening in zebrafish has also revealed a number of RP genes are cancer related (Amsterdam et al., 2004) and mutations of these genes in zebrafish are linked with growth impairment and predispoition to tumorigenesis (Lai et al., 2009). Although it is still unknown how mutations in the RPs result in DBA and cancer, the fact that RPL5, RPL11 and RPS7, three RPs mutated in DBA (Gazda et al., 2008), are all MDM2-binding proteins cannot be regarded as coincidence.
In addition to the association of RP mutants with DBA and cancer, mutations in RPS19 or RPS20 are also tied with a p53-mediated dark skin effect in mice (McGowan et al., 2008). It is perplexing how the reduction of a specific RP can cause a tissue- and cell-specific pathological phenotype. The pathological changes may result from global decline of ribosomal biogenesis owing to loss of one copy of an rp gene and subsequent diminution of translation (Ellis and Lipton, 2008; Liu and Ellis, 2006). It is also speculated that some of these diseases can be explained by RPL11-mediated deregulation of c-Myc, an important player in ribosomal biogenesis (Dai et al., 2007). A testable hypothesis is that haploinsufficiency of certain RPs could reduce the production of tumor suppressors, such as p53 (MacInnes et al., 2008), and hence render DBA patients more susceptible to tumorigenesis. Finally, it is postulated that the high incidence of malignancies in the RP-pertinent diseases could be a result of a defect in the p53 pathway. As discussed above, several RPs including RPL11, RPL5, RPL23, and RPS7 can activate p53 by attenuating MDM2 function in response to stresses derived from ribosomal malfunction. Hence, it is possible that reduced expression of some RPs could impede stress-induced p53 activation due to their decreased MDM2 interaction.
Questions and prospects
Although accumulating evidence has begun to divulge a relatively new signaling pathway linking defects in ribosomal biogenesis with p53, suggesting another cellular surveillance mechanism for cancer prevention, more questions than answers are brought up by these studies. (1) Why are multiple RPs needed to signal to p53? So far, there are at least 4 confirmed RPs that bind to the central acidic domain of MDM2 and utilize apparently similar mechanisms to activate p53. Do multiple RPs have redundant roles, inhibiting MDM2 in response to the same type of stress? Or, do they have specific roles in sensing different types of stress? (2) What are the physiological stresses that activate the RP-p53 signaling pathway? Would cancer develop if this pathway were impaired? A mouse model that specifically targets this pathway is crucial for understanding the physiological role of the RP-MDM2 interaction. (3) Does Mdm2 have a general role in regulating RP turnover, like that shown for RPL26 and RPS7 (Ofir-Rosenfeld et al., 2008; Zhu et al., 2009)? If so, would this explain MDM2 overexpression-induced growth inhibition? (4) Do the MDM2-interacting RPs signal to p53 in response to rRNA damage? rRNAs are the most actively synthesized nucleic acid species in a cell, highly abundant and essential for cell growth and proliferation (Neufeld and Edgar, 1998; Warner, 1999). Thus, rRNA is vulnerable to both endogenous and exogenous genotoxic agents. A recent study showed that yeast utilizes similar mechanisms to repair genotoxic damage to both DNA and rRNA molecules (Fujii et al., 2009). How the integrity of rRNA is monitored during ribosomal biogenesis is not clear, nor do we know how p53 may sense rRNA damage. (5) Are any of RPs mutated in human cancers? As discussed above, mutations in RPL5 and RPL11 are associated with DBA (Cmejla et al., 2009; Gazda et al., 2008). Reports of direct connections between mutations in the p53-activating RPs and cancer, however, have been lacking. It is possible that these RPs may play a role in cancer in a manner similar to the so-called non-oncogene addiction (NOA) molecules (Luo et al., 2009; Solimini et al., 2007). Unlike oncoproteins, such as H-Ras or c-Myc that are often mutated in human cancers, NOA proteins are rarely mutated in human cancers. However, like oncoproteins, NOA proteins are required for cancer cell growth. Under normal growth conditions, the p53-activating RPs are essential components of Ribosome biogenesis. Under stress conditions, their intrinsic anti-tumor activities are awakened (Figure 1). Examples of other proteins that are crucial for p53 activation but are not normally mutated in cancers include Chk1, Chk2, p300, and CBP. The intrinsic anti-tumor activities of these molecules could be useful for anti-cancer drug discovery (Luo et al., 2009).
Indeed, identification of MDM2-RP interactions offers an opportunity for anti-cancer drug development. MDM2 is highly expressed in various human cancers, including breast cancer, sarcoma, glioma, and blood cancers [reviewed in (Onel and Cordon-Cardo, 2004)]. MDM2 is amplified in about 50% of leukemias and lymphomas, in which TP53 mutations are rare (Bueso-Ramos et al., 1996). Hence, MDM2 could be an ideal target for anti-cancer drugs. The previous focus of anti-MDM2 drug screening has been on either the N-terminal p53-binding domain (Issaeva et al., 2004; Shangary et al., 2008; Vassilev et al., 2004) or the C-terminal RING-finger E3 ubiquitin ligase domain (Yang et al., 2005) of MDM2. The MDM2 central acidic domain, including the zinc finger, is crucial for MDM2-induced p53 ubiquitination and degradation (Kawai et al., 2003). The interaction of ARF and RPs with this MDM2 domain suppresses MDM2’s E3 activity and activates p53. Thus, the central domain represents another targeting site in MDM2 for anti-cancer drug screening. Future efforts should be taken to identify small molecules that inhibit MDM2 activity by directly binding to its central domain. Gaining promising candidates from this attempt will not only shed light on the biological significance of RP-MDM2-p53 interplay, but also provide potential drugs for therapeutic intervention of cancers that harbor wild type p53 and high levels of MDM2.
Acknowledgements
We thank M. Oren, K. Vousden, Y. Sun, and H. Ke for sharing unpublished information, K. Itahana for figure art and H. Clegg for copyediting. We apologize for not being able to cite all of the relevant papers due to limited space. Y. Z. is support by grants from The Leukemia & Lymphoma Society, The American Cancer Society, and The National Institute of Health. H. L. is supported by The Efroymson Fund and grants from The National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams DJ, van der Weyden L, Mayeda A, Stamm S, Morris BJ, Rasko JE. ZNF265--a novel spliceosomal protein able to induce alternative splicing. J Cell Biol. 2001;154:25–32. doi: 10.1083/jcb.200010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. Ubiquitin interactions of NZF zinc fingers. Embo J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Argentini M, Barboule N, Wasylyk B. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene. 2001;20:1267–1275. doi: 10.1038/sj.onc.1204241. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. Mdm-2 expression is induced by wild-type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. Embo J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Bueso-Ramos C, Manshouri T, Haidar MA, Yang Y, McCown P, Ordonez N, Glassman A, Sneige N, Albitar M. Abnormal expression of MDM-2 in breast carcinoma. Breast Cancer Res& Treatment. 1996;37:179–188. doi: 10.1007/BF01806499. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS ONE. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–37. doi: 10.1038/sj.onc.1210327. 2. [DOI] [PubMed] [Google Scholar]
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Petrtylova K, Mihal V, Stary J, Cerna Z, Jabali Y, Pospisilova D. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat. 2009;30:321–327. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- da Rocha AB, Lopes RM, Schwartsmann G. Natural products in anticancer therapy. Curr Opin Pharmacol. 2001;1:364–369. doi: 10.1016/s1471-4892(01)00063-7. [DOI] [PubMed] [Google Scholar]
- Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 2007;26:3332–3345. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia. 2006a;8:630–644. doi: 10.1593/neo.06334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J Biol Chem. 2006b;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR, Lipton JM. Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol. 2008;82:217–241. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. JBiolChem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 2009;23:963–974. doi: 10.1101/gad.1775609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. Embo J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. Journal of Molecular Biology. 1971;59:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Letter. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Iapalucci-Espinoza S, Franze-Fernandez MT. Effect of protein synthesis inhibitors and low concentrations of actinomycin D on ribosomal RNA synthesis. FEBS Lett. 1979;107:281–284. doi: 10.1016/0014-5793(79)80390-7. [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein Involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y. Targeted Inactivation of Mdm2 RING Finger E3 Ubiquitin Ligase Activity in the Mouse Reveals Mechanistic Insights into p53 Regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. MolCell Biol. 2000;20:1001–1007. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207–1218. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23:4939–4947. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klibanov SA, O’Hagan HM, Ljungman M. Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J Cell Sci. 2001;114:1867–1873. doi: 10.1242/jcs.114.10.1867. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 Regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn. 2009;238:76–85. doi: 10.1002/dvdy.21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Deisenroth C, Zhang Y. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle. 2007a;6:434–437. doi: 10.4161/cc.6.4.3861. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-Associated Mutations in the MDM2 Zinc Finger Domain Disrupt Ribosomal Protein Interaction and Attenuate MDM2-Induced p53 Degradation. Mol Cell Biol. 2007b;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107:4583–4588. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- Luna RM, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci U S A. 2008;105:10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette J, Nicolas J-C, Levine AJ. The ribosomal protein L5 is associated with mdm-2 and mdm2-p53 complexes. MolCell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Hengst L. SUMO-1 and p53. Cell Cycle. 2002;1:245–249. [PubMed] [Google Scholar]
- Meulmeester E, Frenk R, Stad R, de Graaf P, Marine JC, Vousden KH, Jochemsen AG. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol. 2003;23:4929–4938. doi: 10.1128/MCB.23.14.4929-4938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. Embo J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naora H, Takai I, Adachi M. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP, Edgar BA. Connections between growth and the cell cycle. Curr Opin Cell Biol. 1998;10:784–790. doi: 10.1016/s0955-0674(98)80122-1. [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2:1–8. [PubMed] [Google Scholar]
- Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, Grabusic K, Volarevic S. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Perry RP. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. Embo J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107:815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Sonenberg N. Cell-cycle-dependent translational control. Curr Opin Genet Dev. 2001;11:13–18. doi: 10.1016/s0959-437x(00)00150-7. [DOI] [PubMed] [Google Scholar]
- Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. Embo J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. JBiolChem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF-p53 pathway. CurrOpinGenetDev. 2000;10:94–99. [Google Scholar]
- Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem. 1999;274:37370–37378. doi: 10.1074/jbc.274.52.37370. [DOI] [PubMed] [Google Scholar]
- Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Stad R, Ramos YFM, Litle N, Grivell S, Attema J, van der Eb AJ, Jochmsen AG. Hdmx stabilizes Mdm2 and p53. JBiolChem. 2000;275:28039–28044. doi: 10.1074/jbc.M003496200. [DOI] [PubMed] [Google Scholar]
- Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005a;19:3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulic S, Panic L, Dikic I, Volarevic S. Deregulation of cell growth and malignant transformation. Croat Med J. 2005b;46:622–638. [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem. 2008;283:12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. ProcNatlAcadSciUSA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. Embo J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nature Cell Biology. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Lain S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Yu GW, Allen MD, Andreeva A, Fersht AR, Bycroft M. Solution structure of the C4 zinc finger domain of HDM2. Protein Sci. 2006;15:384–389. doi: 10.1110/ps.051927306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell GrowthDiff. 2001;12:175. [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]