Summary
The gut microbiome is widely studied by fecal sampling, but the extent to which stool reflects the commensal composition at intestinal sites is poorly understood. We investigated this relationship in rhesus macaques by 16S sequencing feces and paired lumenal and mucosal samples from 10 sites distal to the jejunum. Stool composition correlated highly with the colonic lumen and mucosa, and moderately with the distal small intestine. The mucosal microbiota varied most based on location and was enriched in oxygen-tolerant taxa (e.g. Helicobacter, Treponema), while the lumenal microbiota showed inter-individual variation and obligate anaerobe enrichment (e.g. Firmicutes). This mucosal and lumenal community variability corresponded to functional differences, such as nutrient availability. Additionally, Helicobacter, Faecalibacterium, and Lactobacillus levels in stool were highly predictive of their abundance at most other gut sites. These results quantify the composition and biogeographic relationships between gut microbial communities in macaques and support fecal sampling for translational studies.
Introduction
Gut mucosal and lumenal microbial communities are distinct (Eckburg et al., 2005; Morgan et al., 2012; Stearns et al., 2011), and diseases such as colorectal cancer and inflammatory bowel disease induce site-specific epithelial inflammation at which the microbiota are disrupted relative to adjacent normal tissue (Darfeuille-Michaud et al., 2004; Kostic et al., 2013a; Sobhani et al., 2011). Understanding the relationship between stool and the mucosal microbiome is thus of great interest, but large-scale human health-related studies typically focus on the stool microbiota due to technical limitations (Human Microbiome Project, 2012; Qin et al., 2010; Qin et al., 2012; Yatsunenko et al., 2012).
Furthermore, human biopsy samples are near-universally collected after bowel preparation (Whitlock et al., 2008), which itself alters the mucosal community (Ahmed et al., 2007); paired stool data is rarely available. Previous studies of human gut biogeography have included only samples from different individuals and / or timepoints (Darfeuille-Michaud et al., 2004; Gevers et al., 2014; Huse et al., 2014; Morgan et al., 2012) or used a very small number of individuals (Eckburg et al., 2005; Stearns et al., 2011). While the mucosa and lumenal contents of mice are readily accessible for biogeographic studies, neither the pelleted, sparse nature of their colonic contents nor their native microbial composition are totally representative the human gut (Kostic et al., 2013b). The captive rhesus macaque (Macaca mulatta), widely used in biomedical research due to its genetic and physiological similarities to humans (Bauer et al., 2011; Handley et al., 2012; McKenna et al., 2008; Vallender and Miller, 2013), is an excellent model for detailed biogeographic study of the mucosal, lumenal, and stool microbiota. It further avoids confounding due to sample collection and manipulation methods (no colon preparation is required upon autopsy) or diet (synchronized meals).
In this study, we investigated i) the extent to which the stool microbiome reflects the composition of other intra-intestinal sites, ii) the biogeography of the composition of the rhesus macaque gut microbiome, and (iii) predictability of microbiota in the gut. Our results indicate that the stool microbiota community is a good proxy of the large intestinal (LI) lumen and mucosa and is surprisingly well-correlated with the small intestine (SI). The LI mucosa was highly enriched in Helicobacter, which is flagellated and facultatively anaerobic. In contrast, obligate anaerobic Firmicutes were primarily localized to the intestinal lumen. This study thus provides the quantitative relationship between mucosal and lumenal microbial communities as assessed using stool.
Results
The macaque intestinal mucosa is dominated by non-pathogenic Pasteurellaceae and Helicobacteriaceae
Similarly to humans (Eckburg et al., 2005; Human Microbiome Project, 2012; Qin et al., 2010), the macaque intestine was colonized primarily by Bacteroidetes, Firmicutes, and Proteobacteria (Fig. 1A). In contrast, the Actinobacteria and Verrucomicrobia were rare in macaques, and Spirochaetes and Helicobacter were much more abundant. To assess our data in the context of other human (Human Microbiome Project, 2012; Yatsunenko et al., 2012) and macaque (Handley et al., 2012; McKenna et al., 2008) microbiome studies, we combined these datasets, calculated Bray-Curtis dissimilarity and weighted UniFrac distance, and performed principal coordinate analysis (Fig. S1D–E; Supplemental Methods). Despite differences in sequencing technology, the three macaque studies were similar to one another, and more similar to the Malawian and Amerindian than to the US microbiomes.
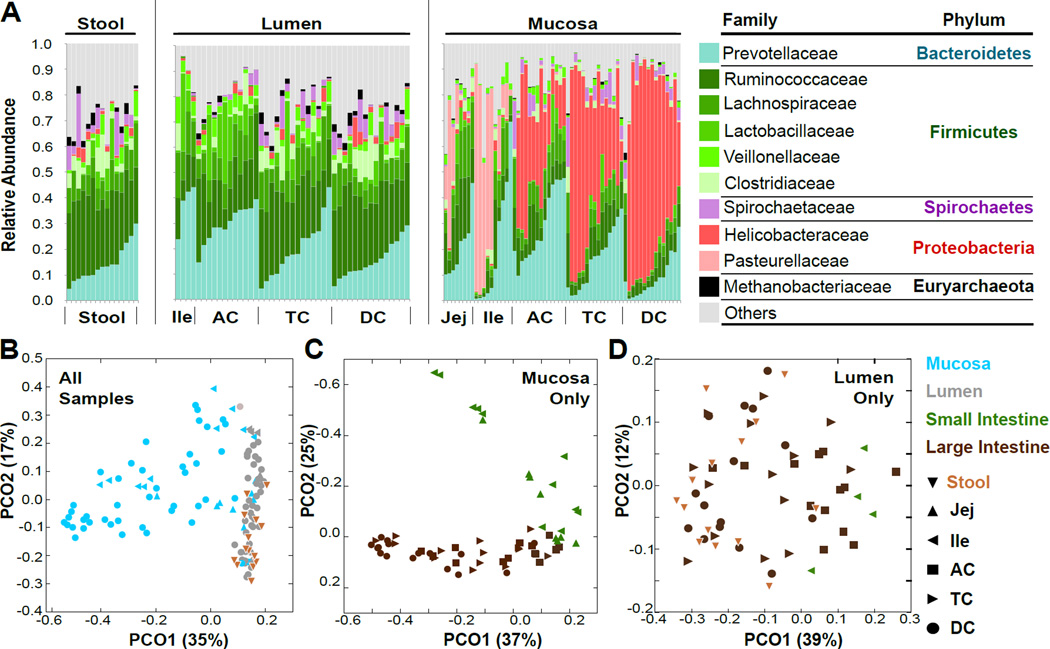
Figure 1. Biogeographic influences on macaque gut microbial composition.
A) Family-level relative abundance of intestinal microbiota in the stool (left), lumen (middle) and mucosa (right) of 15 healthy rhesus macaques. B) Principal coordinate analysis (PCoA) of all samples by weighted UniFrac distance. C) PCoA of mucosal-only samples. D) PCoA of lumen and stool-only samples. See also Fig. S1, Table S1.
We used univariate (Segata et al., 2011) and multivariate analyses (Morgan et al., 2012) to identify bacterial taxa significantly enriched (FDR q < 0.2) in the mucosa or lumen; multivariate analysis included location, sample type, weight, age, and primate center of origin as covariates (Table S1). Relative to mucosa, stool and lumen were enriched for obligately anaerobic, short chain fatty acid-producing clades such as the Lachnospiraceae, Clostridiaceae, and Prevotellaceae (Duncan et al., 2007). In the mucosa, facultatively anaerobic clades were more abundant; these were mostly Proteobacteria, such as Helicobacter in the LI and Pasteurella in the SI (Fig. 1A). This likely reflects the higher host-derived oxygen content in the mucous layer compared to the lumen. Helicobacteraceae in particular was strongly associated with mucosa (q < 10−21) and the ascending LI (q=0.0011). While H. macacae has been previously associated with chronic diarrhea and intestinal adenocarcinoma (Fox et al., 2007; Lertpiriyapong et al., 2014; Marini et al., 2010), our animals showed no evidence of tumorigenesis nor excess inflammation upon routine histopathologic examination of the ileal, cecal, and colonic tissues.
All the animals in our study were housed at the New England Primate Research Center (NEPRC) for 2 years prior to sample collection, but 11 animals came from Oregon National Primate Center, where they were housed outdoors. Research center was not associated with major systematic variation in microbial diversity, but was significantly associated with 23 Operational Taxonomic Units (OTUs) (effect size −0.05 – 0.04; q < 0.2 ; Table S1; Fig. S1A). Most of these OTUs were Ruminococcaceae and Lachnospiraceae, which are primarily lumenally-enriched taxa. However, several mucosally-enriched taxa, including Treponema, Desulfovibrio, and Corynebacterium, were enriched by primate center, suggesting that their presence in the colonic mucosa may be highly influenced by early exposure.
The mucosal microbiota is most influenced by location, while the lumenal microbiota is most influenced by individual
The largest covariation within microbial community structure (as assessed by weighted UniFrac dissimilarity (Lozupone and Knight, 2005) was explained by mucosal/lumenal sample origin (Fig. 1B). When mucosal and lumenal/stool samples were separated, the largest source of variation in mucosal samples corresponded to SI vs. LI sample origin (Fig. 1C), but no such pattern was observed for lumenal samples (Fig. 1D). As observed in previous human studies (Eckburg et al., 2005; Fox et al., 2000), both the stool and lumen showed high inter-individual variation; the latter was not substantially influenced by biogeographical location (Fig. S1B). The Bray-Curtis dissimilarity (based on species in common between sites) between stool and each of the other sites showed that stool was equally dissimilar to all mucosal sites regardless of anatomical proximity (Fig. S1C); in contrast, lumenal dissimilarity increased with colonic distance. This suggests that despite the close anatomical proximity of distal mucosa and stool, lumenal flux of microbiota occurs more readily than transfer of microbiota between mucosa and lumen.
Stool microbial composition accurately reflects the colonic lumen and mucosa
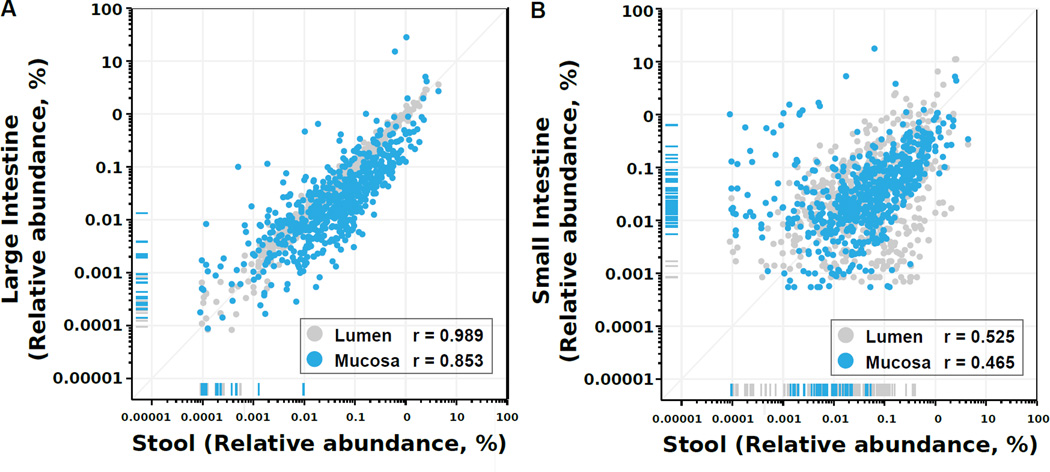
We assessed the extent to which the mucosal and lumenal community of each individual was reflected in the stool by measuring the Spearman correlation between stool and the four major subdivisions of the distal gut (SI mucosa, SI lumen, LI mucosa, and LI lumen), thus accounting for both OTU rank order and the magnitude of relative abundances between the two sites being compared. Stool composition was highly correlated with the LI lumen (Spearman’s r=0.98; p<0.001) and LI mucosa (r=0.85, p<0.001; Fig. 2A). Stool composition was also surprisingly correlated with the SI mucosa (r=0.465, p<0.001) and lumen (r=0.525, p<0.001; Fig. 2B). We examined these OTUs for a systematic taxonomic bias (Fig. S2) and found that most mucosal OTUs that do not appear in stool are primarily Proteobacteria.
Figure 2. Stool microbial composition mirrors that of the colonic lumen.
Each dot corresponds to the average relative abundance of an OTU across 15 animals for each of 4 intestinal regions (SI mucosa and lumen, LI mucosa and lumen). To measure correlation, Spearman’s r was calculated between stool and mean region OTU abundance. Marks on the x-axis (vertical lines) or y-axis (horizontal lines) margins represent OTUs with zero measured abundance at one site but non-zero abundance at the other. See also Fig. S2.
In the SI lumen and LI mucosa and lumen, over 97% of observed OTUs were also detected in stool, and stool-undetected OTUs had very low relative abundances (<10−3) in the mucosa and lumen, and thus may have been detected with deeper sequencing of stool. In contrast, 10% of SI mucosal OTUs were stool-undetected despite relative abundance typically >10−3; thus, increasing stool read depth may not improve the detection of these OTUs. Fusobacteria, β- and γ-proteobacteria are particularly likely to be stool-undetected (Fig S2; Table S2).
Nearly all (95%) OTUs detected within the LI mucosa lumen and in stool were detected in stool within within two orders of magnitude (10−1 – 101) of their lumenal and mucosal relative abundances; this was only true for 50% of SI content and 66% of SI mucosal OTUs. Stool is therefore an excellent proxy for the LI lumen and mucosa, as it contains nearly all OTUs at preserved proportions.
Most OTUs are shared between adjacent sites, but each site has a small site-specific community
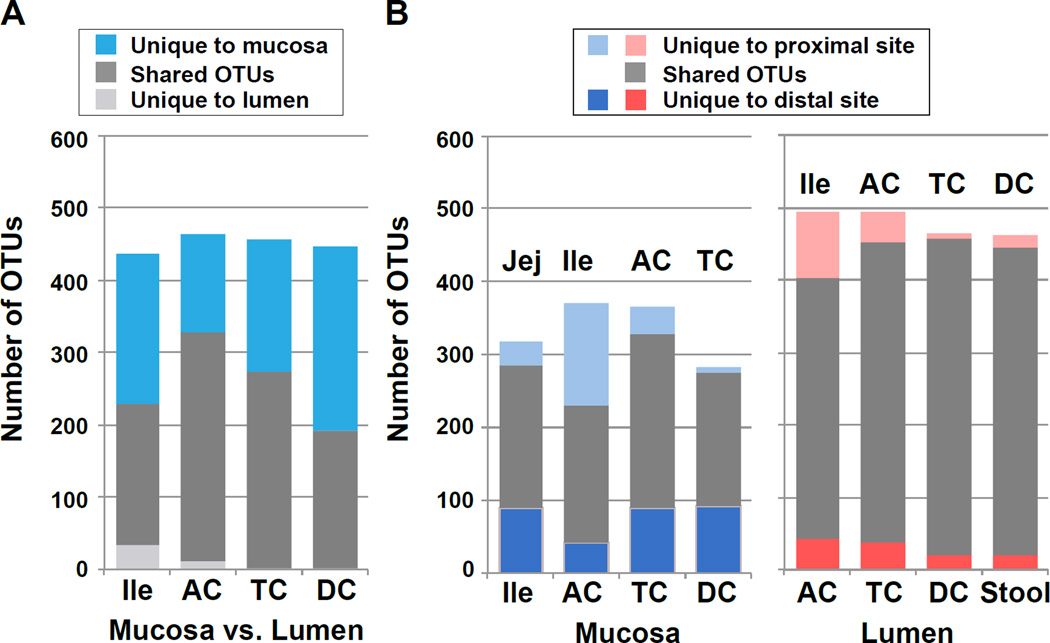
We found that ~40–70% of OTUs are typically shared between adjacent mucosal and lumenal sites (Fig. 3). It is unclear to what extent these overlapping taxa are persistent, metabolically-active residents of the mucosa, rather than lumenal residents incidentally trapped on the mucosal surface (or vice versa). Although lumenal communities were generally more homogenous than those of the mucosa (Fig. 2D), 20–30% of OTUs were unique to each lumenal segment. As each mucosal sample contained a similar distribution of organisms within higher-order taxa, the variation we observed here at the genus or species level may be the result of colonization resistance by the more abundant members within similar functional groups. Whether the gut microbiota undergoes such nonrandom assembly remains unclear.
Figure 3. Microbial overlap between adjacent mucosal and lumenal sites / Differences in mucosal and lumenal community function are driven by Helicobacter.
A) Mean total, shared, and unique OTUs between the mucosa and lumen across all individuals at each paired site. B) Mean total, shared, and unique OTUs between adjacent mucosal (left) and lumenal (right) sites, averaged across individuals. Most lumenal taxa are shared with the adjacent mucosa and lumenal sites, with a gradient of unique mucosal taxa occurring along the intestine. See also Table S2
Using logistic regression to distinguish mucosally and lumenally-enriched taxa and to predict bacterial flow
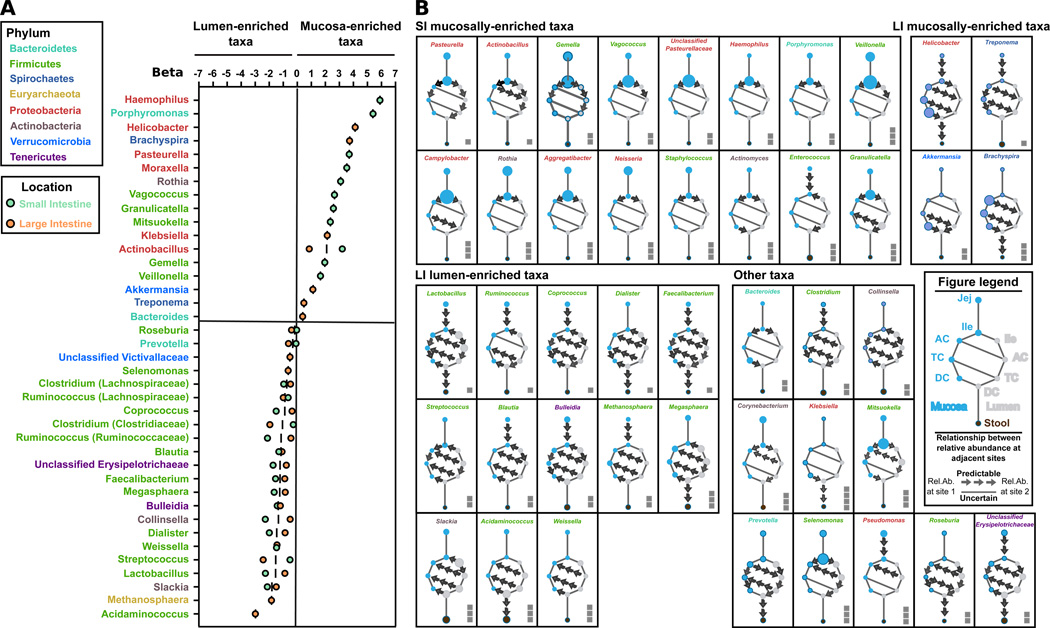
To understand microbial niches and potential bacterial flow within individuals, we used logistic regression to model the extent to which the abundance of a genus in mucosa and lumen (Fig. 4A) and at one site was predictive its abundance at an adjacent site (Fig. 4B, see Experimental Methods). Of 56 genera identified in our cohort, 30% were mucosally-enriched, 40% were lumenally-enriched, and 30% showed no consistent enrichment. The proteobacteria comprised none of the lumenally-enriched taxa but one third of the mucosally-enriched taxa (6/17 genera). Conversely, nearly 70% of the lumenally-enriched taxa were Firmicutes. The mucosally-enriched genera were primarily gram-negative (13/17 genera) and frequently facultatively anaerobic (8/17 genera), while the lumenally-enriched taxa were primarily gram-positive (16/ 22 genera) and obligately anaerobic (19/22 genera). Most obligately anaerobic genera were not abundant in the mucosa; only Treponema, which is well-adapted to oxidative stress, showed a modest mucosal preference (Giacani et al., 2013; Jovanovic et al., 2000). This suggests that oxygen availability is a major, but not sole, determinant of mucosal composition (Albenberg et al., 2014).
Figure 4. A logistic regression model of bacterial taxa site enrichment and flow through the macaque gut.
A) Average within-small-intestine and within-large-intestine β values (regression slopes) for each genus in each biogeographic region. β corresponds to the magnitude of a difference in relative abundance between two sites (mucosa and lumen), and its consistency across 15 animals, and thus the enrichment of a taxa in mucosa or lumen. Only genera with at least one significant (p < 0.05 and q < 0.05) value for β are shown. B) Intra-intestinal microbial predictability between adjacent sites for each bacterial genus. Points on each clock-like diagram represent biogeographic sites, and point size corresponds to mean relative abundance across all animals for each genus. Adjacent sites with significant non-zero β (indicating that relative abundance at one site can predict that of the other) are connected with an arrow; arrows always start at the site with higher relative abundance. Solid lines indicate non-significant β, and arrows with significant β that point opposite of the physiological flow of intestinal contents inside the intestine (proximal to distal) are also replaced by a solid line. For visualization, taxon relative abundances were adjusted by a factor of 101 to 103; the adjustment is indicated by the number of squares by each diagram (e.g. Pasteurella was adjusted by 101 and has one square). See also Fig S3.
Most mucosally-enriched genera identified here were not identified by univariate analysis because, with the exception of Actinobacillus, they were enriched only in either the SI or LI mucosa, but not both (e.g. Klebsiella in the LI, Gemella in the SI)(Fig. S3); univariate analysis only detected mucosal enrichment consistent in both loc ations. The SI lumen was represented only by ileal samples, but most genera strongly enriched in the ileal lumen relative to the ileal mucosa (e.g. Lactobacillus, Slackia) were also strongly enriched at multiple locations along the LI lumen relative to the LI mucosa (Fig. 4; Fig. S3). This lumenal community similarity may be partially explained by pH similarity between ileum (7.0–7.4) and colon (6.6–7.0) (Mercier et al., 2007)(Fig. S4C).
Fig. 4B summarizes the relationships of β-values (regression slopes) between adjacent sites in the macaque gut. We observed four main patterns of microbial enrichment and potential flow: 1) SI mucosally-enriched taxa 2) LI mucosally-enriched taxa 3) SI and LI lumenally-enriched taxa and 4) clades following no consistent pattern. Actinobacillus and Pasteurella exemplify the pattern typically observed in SI mucosally-enriched clades (Fig S3). They are most abundant in the ileal and jejunal mucosa, much less abundant in LI than SI, and more abundant in the LI mucosa than lumen. The differences in abundance between sites are very consistent, so the abundance at one site can be used to predict the abundance at another. Similarly, Brachyspira and Helicobacter are most abundant in the LI mucosa, and stool is highly predictive of their abundance elsewhere in the LI. Lactobacillus, Ruminococcus, and Dialister are enriched throughout the lumen, and predictably present in the mucosa at much lower abundance. Finally, several clades had predictable but atypical abundance patterns. For example, Granulicatella and Enterococcus were highly abundant in the SI mucosa (and nearly-absent in the SI lumen), and present at very low abundance in the LI lumen (but absent in the mucosa). Pseudomonas was only present in the SI mucosa, while Klebsiella was only present in the distal LI. In summary, using logistic regression modeling allowed us to group bacterial taxa that followed similar predictable ecological patterns across the intestine, and in some cases, we were able to predict where taxa may have originated within the intestine when observed in the stool.
Differences in mucosal and lumenal community function correspond to oxygen and nutrient availability
In order to understand the functional differences between communities at distinct biogeographic sites and their relation to community composition, we used PICRUSt (Langille et al., 2013) to infer community metabolic potential (Supplemental Methods), then used LEfSe (Segata et al., 2011) to identify functions that differed significantly between sites (see Methods; Fig. S4D; Table S1). PICRUSt is particularly useful for determining mucosal microbial community function due to the high host DNA content of mucosal metagenomes.
Many functional differences between mucosa and lumen, such as increased capacity for riboflavin biosynthesis, could be explained by difference in Helicobacter. While Helicobacter and gram-positive bacteria both have fused ribAB genes in their riboflavin biosynthesis operons, Helicobacter also have an additional copy of the ribA gene (Fassbinder et al., 2000). Likewise, glutamate / aspartate transport capacity was mucosally increased; H.pylori preferentially consumes amino acids for energy (Stark et al., 1997), and its glutamate and aspartate transport and deamidase activity are essential for mouse colonization(Leduc et al., 2010). Glycolysis capacity was correspondingly increased in the lumen, where Helicobacter was not dominant. The SI enrichment in the mannose-specific phosphotransferase system is likely explained by Pasteurella (Binet and Bouvet, 1998)(Fig. S4D).
Discussion
In this study, we comprehensively examined the composition of the macaque gut microbiome at 10 different biogeographic locations within 15 individuals. The most similar previous study cross-sectionally compared the gut microbiota of healthy and sick (e.g. SIV-infected) macaques, although it also included several biogeographical locations drawn from distinct individuals (McKenna et al., 2008). In contrast, our study included only healthy individuals and comprehensively examined the microbiota of the same individual at many biogeographic sites at the same time. We quantified the degree to which microbial composition at one biogeographical location within the gut predicts that of another, particularly the extent to which stool samples reflect the mucosal microbiome. We found that between stool and colonic mucosa, both the conservation of taxa and their rank correlation were remarkably high (r>0.85), supporting the use of stool samples for translational studies of colonic mucosal inflammation in human subjects.
The human, macaque, and mouse gut microbiomes are fundamentally similar in containing Bacteroidetes, Firmicutes, and Proteobacteria (Human Microbiome Project, 2012; Kostic et al., 2013b; McKenna et al., 2008). In contrast, the macaque gut mucosa was most remarkable for its abundance of ε-proteobacteria, specifically Helicobacter, which reached up to 80% relative abundance in the LI mucosa of some animals. The macaque gut also included a substantial component of the Spirochaetes Treponema and Brachyspira, which comprised ~3% of the mucosal microbiota. A recent study of the gorilla, chimpanzee, and bonobo microbiomes found that they also contained Brachyspira and Treponema, but no Helicobacter was detected in their stool (Moeller et al., 2013). While Spirochaetes carriage is typically associated with intestinal pathology in humans, it can be asymptomatic and is more typically found in stool in residents of non-developed countries (Tsinganou and Gebbers, 2010). Recent studies have detected Brachyspira spp. and Treponema spp OTUs in the stool of Malawians, Amerindians, and children from rural Burkina Faso and Bangladesh slums, but not in comparison cohorts from the USA (De Filippo et al., 2010; Lin et al., 2013; Yatsunenko et al., 2012). The disappearance of these taxa in residents of developed countries may be associated with modern sanitation practices, including use of antibiotics and pesticides.
Integrating these results with our prior knowledge of intestinal ecology and microbial metabolism (Human Microbiome Project, 2012; McKenna et al., 2008; Qin et al., 2010; Stearns et al., 2011; Yatsunenko et al., 2012) refines our insights into the ecological dynamics of the gut microbiome. Relative to the inter-individual differences observed in human populations, inter-individual differences in this study were minimal, yet they were still a significant source of variation despite all animals being uniformly fed and housed for at least two years prior to sampling. Intestinal oxygen content appeared to determine the dominant taxa colonizing the mucosa and lumen, as the mucosa was colonized primarily by facilitative anaerobes, while the lumen was colonized mainly by obligate anaerobes. This in turn dictates the main patterns of community functionality, as lumenal bacteria (Prevotellaceae Lachnospiraceae, Ruminococcaceae) are as a result primarily carbohydrate fermenters (Duncan et al., 2007). There were correspondingly large differences between small and large-bowel mucosal communities, potentially corresponding to difference in pH, bile salt, and/or mucin composition (Croix et al., 2011; Walter and Ley, 2011), although the difference between the lumenal microbiota of the SI and LI was much smaller.
The relationship between the stool and mucosal microbiota is highly relevant to human clinical studies, as disease may localize to specific locations while only stool remains readily accessible (Darfeuille-Michaud et al., 2004). Patient biopsies are the current gold standard for study of human-associated mucosal communities, but invasiveness and expense limit the frequency with which they can be performed. At the same time, an increasing body of data (Gevers et al., 2014; Morgan et al., 2012) underscores the importance of studying the microbiome longitudinally during the development of disease, a near-impossibility with mucosal biopsies. Our results quantify the extent to which the stool microbiota reflects the SI and LI mucosa and is highly correlated with the colonic lumen and mucosa. Thus, although biopsies are optimal for studying the SI mucosa, our results in a primate model representative of the human gut microbiome show that stool is still surprisingly representative of the colonic mucosa.
Experimental Procedures
Sample collection and sequencing
Stool and paired intestinal lumenal and mucosal samples from 10 segments distal to jejunum were collected from 15 clinically-healthy female rhesus macaques, and the V4 region of the16S rRNA gene was sequenced by Illumina MiSeq (Fig. S4A; Fig. S4B). A mean sequence depth of 29,914/sample was obtained; samples with fewer than 3,000 filtered sequences and those OTUs with fewer than 15 reads were excluded from further analysis. All further details of animal husbandry, sample collection, preparation, sequencing, and bioinformatic processing are outlined in the Supplemental Data.
External Data
For all comparisons to previously-published studies, taxonomic or raw sequencing data were obtained from publically-available sources (Handley et al., 2012; Human Microbiome Project, 2012; Yatsunenko et al., 2012) or directly from the investigator (McKenna et al., 2008). Taxonomic tables were summarized to genus-level clades and merged. Further details are outlined in Supplemental Data.
OTU overlap between sites
For each pair of adjacent sites, the number of OTUs observed in both adjacent sites was counted in each individual and subsequently averaged across all 15 animals. To minimize the influence of low prevalence OTUs and differences in sequencing depth, only OTUs with 15 reads per OTU in 15 animals were considered in this analysis.
Identification of microbial taxa enrichment sites and predictability by logistic regression
We built a logistic regression model for each taxa between each adjacent biogeographical site pair, as described in Supplemental Methods. The regression slope β between each pair of locations is calculated as the contrast between the coefficients of indicator variables corresponding to the locations. P-values of all β are adjusted for multiple hypothesis testing using the Benjamini-Hocheberg procedure (false discovery rate, FDR=0.05). Cytoscape was used to visualize Figure 4B.
Sequence accession numbers and availability
Sequences generated in this study are publicly available (NCBI BioProject ID number PRJNA259224).
Supplementary Material
Acknowledgements
We thank L. Waldron, E. Schwager, and J. Moon for assistance with statistical analyses; W. Garrett and the Garrett and Huttenhower research groups for comments on this manuscript; S. Macri, E. Curran, L. Kattenhorn, and B. Assaf for assistance with sample collection; and T. Poon for assistance with sequencing. This work was supported by the Robert F. Naka fellowship (KY), Base Grant (P51 RR000168-48) (NEPRC), NIH R01HG005969 (CH), NSF DBI-1053486 (CH), and ARO W911NF-11-1-0473 and W911NF-11-1-0429 (CH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
CH is a member of the scientific advisory board for SeresHealth™.
Authors’ contributions
KY, KO, SVW, KGM, EJV, GM, JKR, DG, CH, and XCM designed the study. KY, SVW, KGM, DG processed samples and DNA sequenced. KY, KO, BR, TLT, EAF, CH, and XCM analyzed the data. KY, CH and XCM prepared the manuscript. All authors read and approved the final manuscript.
References
- 1.Ahmed, et al. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73:7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albenberg, et al. Correlation Between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota in Human Beings and Mice. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, et al. Obesity in rhesus and cynomolgus macaques: a comparative review of the condition and its implications for research. Comp Med. 2011;61:514–526. [PMC free article] [PubMed] [Google Scholar]
- 4.Binet, et al. Transport of glucose by a phosphoenolpyruvate:mannose phosphotransferase system in Pasteurella multocida. Res Microbiol. 1998;149:83–94. doi: 10.1016/s0923-2508(98)80024-7. [DOI] [PubMed] [Google Scholar]
- 5.Croix, et al. On the relationship between sialomucin and sulfomucin expression and hydrogenotrophic microbes in the human colonic mucosa. PLoS ONE. 2011;6:e24447. doi: 10.1371/journal.pone.0024447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darfeuille-Michaud, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 7.De Filippo, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Applied and environmental microbiology. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fassbinder, et al. Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS microbiology letters. 2000;191:191–197. doi: 10.1111/j.1574-6968.2000.tb09339.x. [DOI] [PubMed] [Google Scholar]
- 11.Fox, et al. Isolation and characterization of a novel helicobacter species, "Helicobacter macacae," from rhesus monkeys with and without chronic idiopathic colitis. J Clin Microbiol. 2007;45:4061–4063. doi: 10.1128/JCM.01100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, et al. Helicobacter canadensis sp. nov. isolated from humans with diarrhea as an example of an emerging pathogen. J Clin Microbiol. 2000;38:2546–2549. doi: 10.1128/jcm.38.7.2546-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gevers, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giacani, et al. Identification of the Treponema pallidum subsp. pallidum TP0092 (RpoE) regulon and its implications for pathogen persistence in the host and syphilis pathogenesis. J Bacteriol. 2013;195:896–907. doi: 10.1128/JB.01973-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handley, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Human Microbiome Project. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huse, et al. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome. 2014;2:5. doi: 10.1186/2049-2618-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovanovic, et al. Neelaredoxin, an iron-binding protein from the syphilis spirochete, Treponema pallidum, is a superoxide reductase. J Biol Chem. 2000;275:28439–28448. doi: 10.1074/jbc.M003314200. [DOI] [PubMed] [Google Scholar]
- 19.Kostic, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013a;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostic, et al. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013b;27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langille, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leduc, et al. Coupled amino acid deamidase-transport systems essential for Helicobacter pylori colonization. Infection and immunity. 2010;78:2782–2792. doi: 10.1128/IAI.00149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lertpiriyapong, et al. Pathogenic properties of enterohepatic Helicobacter spp. isolated from rhesus macaques with intestinal adenocarcinoma. J Med Microbiol. 2014;63:1004–1016. doi: 10.1099/jmm.0.072462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, et al. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One. 2013;8:e53838. doi: 10.1371/journal.pone.0053838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone, et al. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marini, et al. Persistent infection of rhesus monkeys with 'Helicobacter macacae' and its isolation from an animal with intestinal adenocarcinoma. J Med Microbiol. 2010;59:961–969. doi: 10.1099/jmm.0.019117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercier, et al. Oral immunization of rhesus macaques with adenoviral HIV vaccines using enteric-coated capsules. Vaccine. 2007;25:8687–8701. doi: 10.1016/j.vaccine.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moeller, et al. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome research. 2013;23:1715–1720. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 33.Segata, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobhani, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark, et al. Amino acid utilisation and deamination of glutamine and asparagine by Helicobacter pylori. J Med Microbiol. 1997;46:793–800. doi: 10.1099/00222615-46-9-793. [DOI] [PubMed] [Google Scholar]
- 36.Stearns, et al. Bacterial biogeography of the human digestive tract. Scientific reports. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsinganou, et al. Human intestinal spirochetosis--a review. Ger Med Sci. 2010;8:Doc01. doi: 10.3205/000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallender, et al. Nonhuman primate models in the genomic era: a paradigm shift. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2013;54:154–165. doi: 10.1093/ilar/ilt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annual review of microbiology. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 40.Whitlock, et al. Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force. Annals of internal medicine. 2008;149:638–658. doi: 10.7326/0003-4819-149-9-200811040-00245. [DOI] [PubMed] [Google Scholar]
- 41.Yatsunenko, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.